1. Introduction
Rice is the main ration crop in China, and more than 65% of the population in China eat rice as their main food source [
1]. Each year, the global rice production nears 750 million tons, with major producers such as China, India, Indonesia, and Bangladesh playing a critical role in global food security [
2]. However, rice is highly susceptible to various diseases during its growth cycle, with leaf diseases being particularly prominent. These diseases often appear first on the leaves and spread rapidly to the entire plant, significantly affecting both the yield and quality of rice [
3]. It is estimated that global annual yield losses due to rice diseases are approximately 10-15%, with diseases posing a particular threat during critical growth stages, leading to severe reductions in yield [
4]. Therefore, early and accurate detection of rice leaf diseases is essential for ensuring rice yield, disease control, and food security.
Traditional methods of detecting rice leaf diseases rely heavily on manual experience and visual inspection by agronomy experts to identify symptoms. However, this approach is prone to subjective misjudgments and is costly, making it unsuitable for large-scale field diagnostics. With the rapid advancement of image processing and computer technology, computer vision, machine learning, and deep learning have shown significant potential in the field of agricultural disease detection [5-7]. Traditional computer vision methods rely on color, shape, and texture features of disease regions for segmentation and disease type identification of RGB images. However, due to the vast variety of diseases and similar symptoms, these methods struggle to accurately distinguish between different disease types under natural conditions, especially in complex agricultural environments [
8]. In recent years, Convolutional Neural Networks (CNNs) have been widely applied to crop disease detection tasks due to their end-to-end structure, which eliminates the need for traditional image preprocessing and feature extraction steps, significantly improving both efficiency and accuracy [
9,
10].
At present, some studies have tried to use convolutional neural network (CNN) to detect the common diseases of rice leaves, such as white leaf blight, rice blast, stripe leaf spot, etc. Based on the improved ResNet network, Stephen et al. [
11] realized the accurate identification of various rice leaf diseases with an average accuracy of more than 95%. This approach uses a deep learning model to train a large number of labeled disease images so that the model has the ability to distinguish between different disease types. However, limited by image resolution, lighting conditions and other factors, the accuracy of disease recognition in complex environments is still low. Therefore, the network structure based on attention mechanism has gradually emerged in the field of agricultural disease detection in recent years. The attention mechanism enables the network to focus on key features by identifying discriminating areas in images, thereby ignoring irrelevant information and improving the accuracy of disease recognition [
12].
The disease detection network based on attention mechanism has achieved remarkable results in practice. Qian et al. [
13] added the grouping attention module into the ResNet18 model to achieve high-precision segmentation of cucumber leaf diseases in complex environments, with a pixel accuracy of 93.9%. Wang et al. [
14] improved ShuffleNet based on the attention module, and successfully increased the recognition rate of grape diseases in PlantVillage data set to 98.86%. In addition, Yang et al. [
15] improved the GoogleNet model based on the channel attention module to achieve high-precision identification of rice leaf diseases, which significantly improved the model's adaptability in the natural environment.
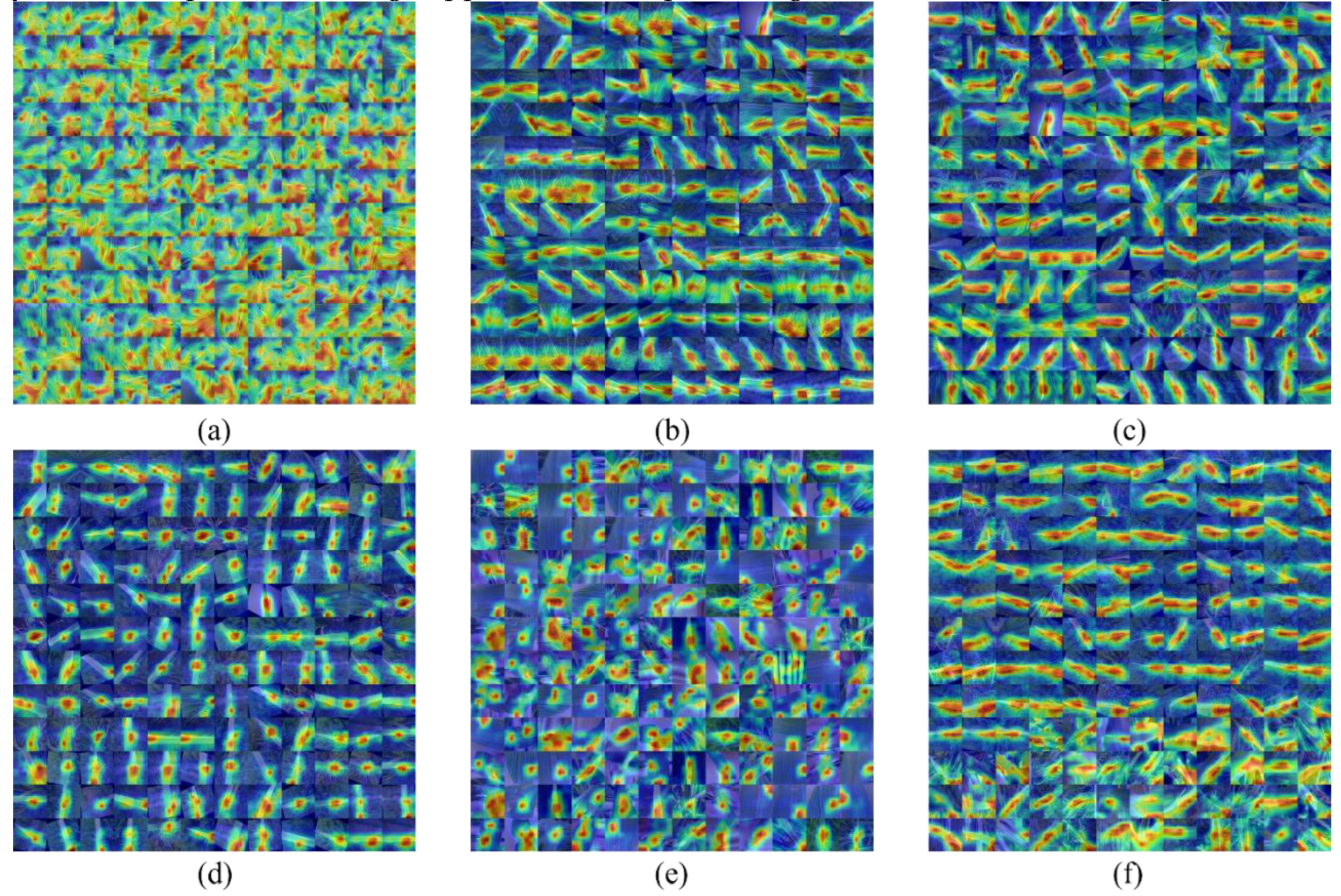
For rice leaf disease detection, the disease area usually only accounts for a small part of the image, and the background information is easy to introduce interference. Therefore, in this paper, an attention mechanism is added to the convolutional neural network (CNN) structure to automatically focus on key disease regions in the leaves, eliminate interference features, and improve the accuracy and robustness of recognition. With the introduction of the attention module, the model can automatically learn the feature differences of different diseases and effectively separate the disease regions according to the complex background. The improved model proposed in this paper is based on the combination of CNN and attention mechanism, which not only improves the accuracy of disease identification, but also has strong robustness, providing an efficient and reliable solution for rice disease detection. In the future, this technology is expected to play an important role in the early diagnosis of diseases, intelligent disease prevention and control, and precision agriculture management.
The main contributions of this paper are as follows:
In order to meet the need of high precision diagnosis of various rice leaf diseases in natural environment, a dataset covering 6 major rice leaf diseases and healthy leaves was constructed. By using data enhancement techniques, this paper extends the data set to enhance the generalization ability of the model in different environments and improve the adaptability in practical applications. Data enhancement includes rotation, translation, brightness adjustment, etc., which makes the disease detection effect of the model more robust under different illumination, viewing Angle and complex background.
• A multi-scale convolutional neural network (CNN) structure integrating deeply separable convolution and attention mechanisms is proposed for rice leaf disease detection. The model is based on the ResNeXt-50 structure, which is optimized and incorporates the Convolutional Block Attention Module (CBAM) to further improve the effect of disease feature extraction. The improved ResNeXt-50 uses deep separable convolution instead of the traditional convolution layer to reduce the computational load and improve the operational efficiency of the model. By integrating CBAM module into the network, the model can adaptively focus on the disease feature region and strengthen the significant expression of the disease region, thus effectively improving the robustness and accuracy of disease detection, especially in complex environments.
• In this study, a three-dimensional dependency relationship (channel C, height H and width W) was constructed for the feature map of rice leaf disease to enhance the ability to express significant features of the disease region. Through multi-scale channel and spatial attention mechanism, CBAM module firstly weights the disease-related features of channel dimension, and then further optimizes the model's attention to key areas through dynamic spatial attention. This multi-scale and dynamic attention mechanism can adjust the focus area adaptively according to different feature layers, thus effectively improving the detection ability of the model in complex background. Combined with ResNeXt's packet convolution feature, the network can efficiently extract and fuse disease features of different scales, making disease detection more accurate.
Through the combination of deep separable convolution and attention mechanisms, the model proposed in this paper achieves a good balance between computational efficiency and feature extraction capabilities, enabling the model not only to accurately diagnose diseases in complex contexts, but also to improve speed and accuracy in practical applications.
The structure of this paper is arranged as follows. The second part introduces the construction and expansion method of rice leaf disease data set, and describes the model improvement process of CBAM module and deep separable convolution fusion into ResNeXt-50 in detail. In the third part, the performance of the proposed model was verified by experiments, and its applicability and popularization potential in other crop disease detection tasks were discussed. In the fourth part, the results of this model are compared with the existing literatures, and the effects and advantages of CBAM module and depth-separable convolution on disease recognition accuracy are analyzed. Finally, the fifth part summarizes the research conclusion and puts forward the prospect of the future research direction.
2. Materials and Methods
2.1 Build the Dataset
The image Data of rice leaf health and disease in this paper came from Kaggle open source database(https://www.Kaggle.com). After the initial collection of rice leaf images, strict manual screening and data cleaning were carried out to ensure data quality and avoid image duplication and classification errors in the data set. The result was a high-quality dataset of nearly 1,500 rice leaf images, each with a fixed size of 224×224 pixels to ensure consistency and standardization when input into the model.
The dataset covers six major rice leaf disease and healthy leaf categories, including yellow dwarf, rice thrips, leaf char, brown spot, rice blast, bacterial leaf blight, and healthy leaves. The leaf disease symptoms of each species of rice are shown in
Figure 1. These images provide clear visual features for a variety of disease symptoms, including spots, leaf discoloration, tissue necrosis, etc., to facilitate accurate classification during subsequent model training.
2.2 Data Enhancement
In deep learning, the diversity of data sets can effectively enhance the generalization ability and robustness of models [
16]. In order to improve the model's recognition performance of rice leaf disease in natural environment, this paper implemented a variety of image enhancement techniques based on Pytorch framework and OpenCV, and expanded and diversified the data set.
1. Rotation: The image is rotated randomly by 90°, 180° and 270° without changing the relative position of the diseased area and the healthy part, so as to simulate the change of different shooting angles under natural conditions and make the model more robust.
2. Zoom: The image is reduced by a certain scale so that the model can identify the disease area on different scales. The scaled image is filled with 0 pixels, expanding the image size to 224×224 pixels, maintaining the consistency of the input image size.
3. Add noise: Add salt and pepper noise and Gaussian noise to the image to simulate shooting conditions with different sharpness, which helps to enhance the adaptability of the model to fuzzy images in the natural environment.
4. Color dithering: Adjust the brightness, saturation and contrast of the image to simulate the difference of visual effects under different light intensity, so that the model can still maintain a high recognition accuracy in the face of light changes.
Through the above data enhancement method, the sample number of each category was expanded, so that the rice leaf disease dataset finally contained 8750 images. The data set is randomly divided into the training set and the verification set according to the ratio of 8:2 to ensure the rationality and effectiveness of the training and verification of the model. Details of the data set are shown in
Table 1.
2.3. Deep learning model
2.3.1. Feature extraction network
Feature extraction is a crucial step in deep learning, and different feature extraction networks differ in parameters, speed and performance. At present, many widely used convolutional neural network models have been proposed, such as AlexNet [
17], VGGNet [
18] and GoogleNet [
19]. However, these CNN models are slow in training and detection due to the large number of parameters and large amount of calculation [
20]. In order to overcome these problems, He et al. [
21] proposed a residual network (ResNet), which effectively solved the problem of gradient disappearance and degradation by introducing residual connections, and won the champion in the ImageNet Large-scale Visual Recognition Challenge in 2015. Compared to AlexNet, VGGNet, and GoogLeNet, ResNet maintains high performance while reducing computational effort.
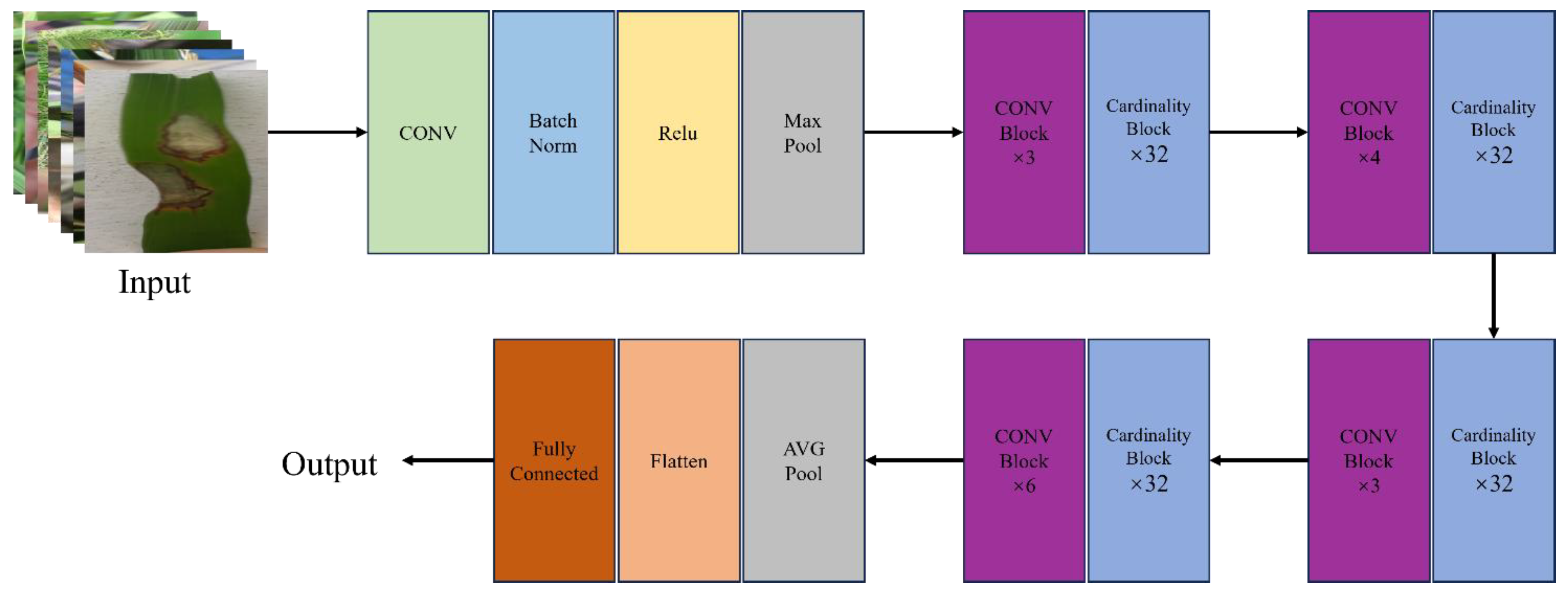
In this paper, we use the improved ResNeXt-50 as the feature extraction network. The ResNeXt-50 is an improvement on the traditional ResNet structure, introducing the idea of packet convolution while optimizing computational efficiency and presentation power. The improved ResNeXt-50 introduces deep separable convolution in each residual module to reduce computational effort and improve computational efficiency. In addition, the Convolutional Block Attention Module (CBAM) is integrated, enabling the model to better focus on the key features of the disease region, thereby improving the accuracy and robustness of disease detection in complex contexts.
By introducing depth-separable convolution and CBAM modules, the improved ResNeXt-50 not only retains the residual structure of ResNet, but also significantly improves the feature representation capability of the network without significantly increasing the computational amount, and can better capture multi-scale features of rice leaf diseases.
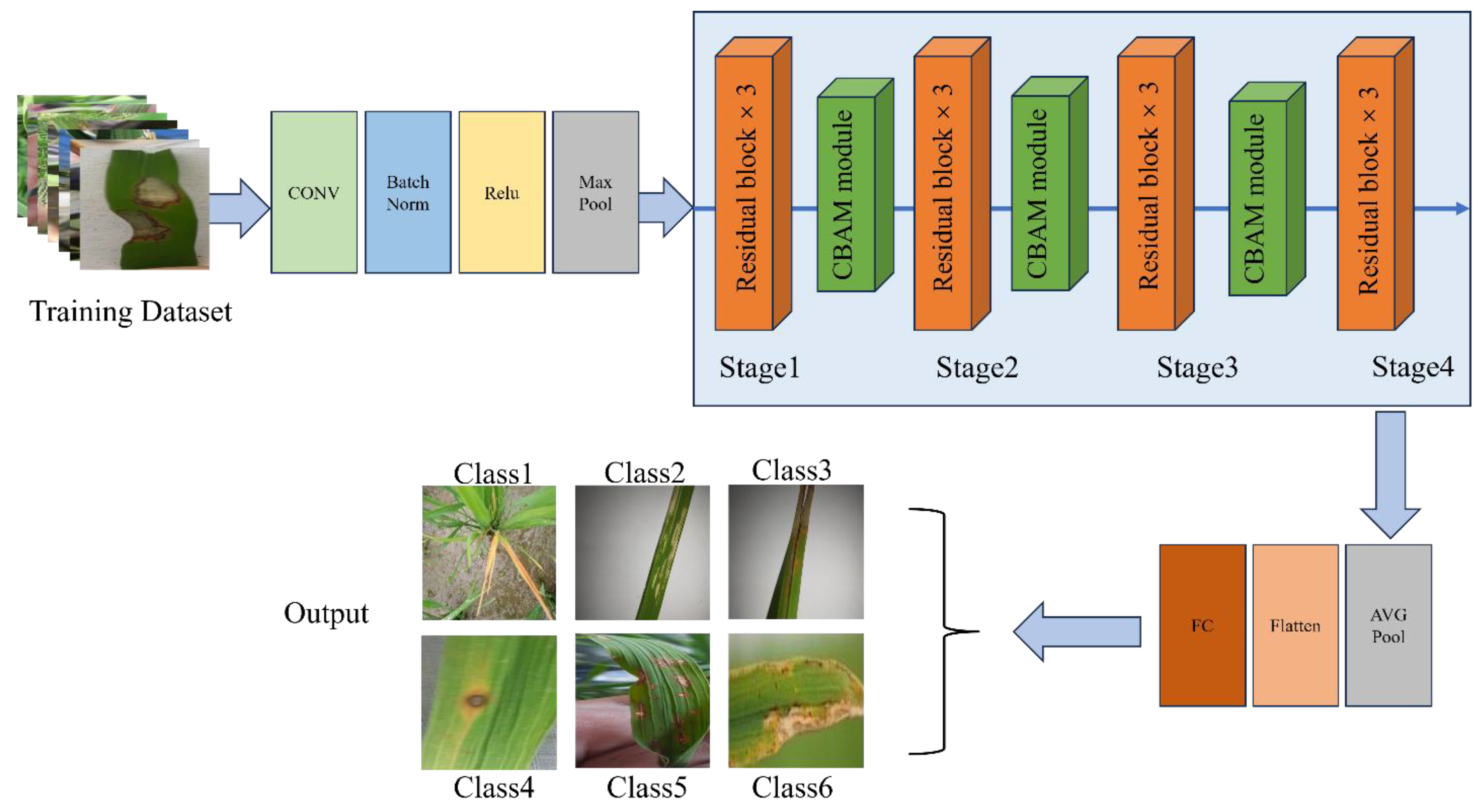
In
Figure 2, the rice leaf disease image was first input into the ResNeXt-50 network structure. After processing the initial convolution layer, batch normalization (BN) layer and activation layer, the feature maps are downsampled by the maximum pooling layer. The ResNeXt-50 model is mainly composed of four stages (Stages 1-4), each of which includes a sampling module and multiple identity mapping modules. At each stage, deep separable convolution replaces traditional convolution operations, effectively reducing the computational effort while enhancing the network's ability to capture important features. After these processing, the output feature map is processed by AVG Pooling, and the multi-dimensional features are reduced to one-dimensional feature vectors through the Flatten layer. Finally, the disease classification result is obtained through the fully connected layer.
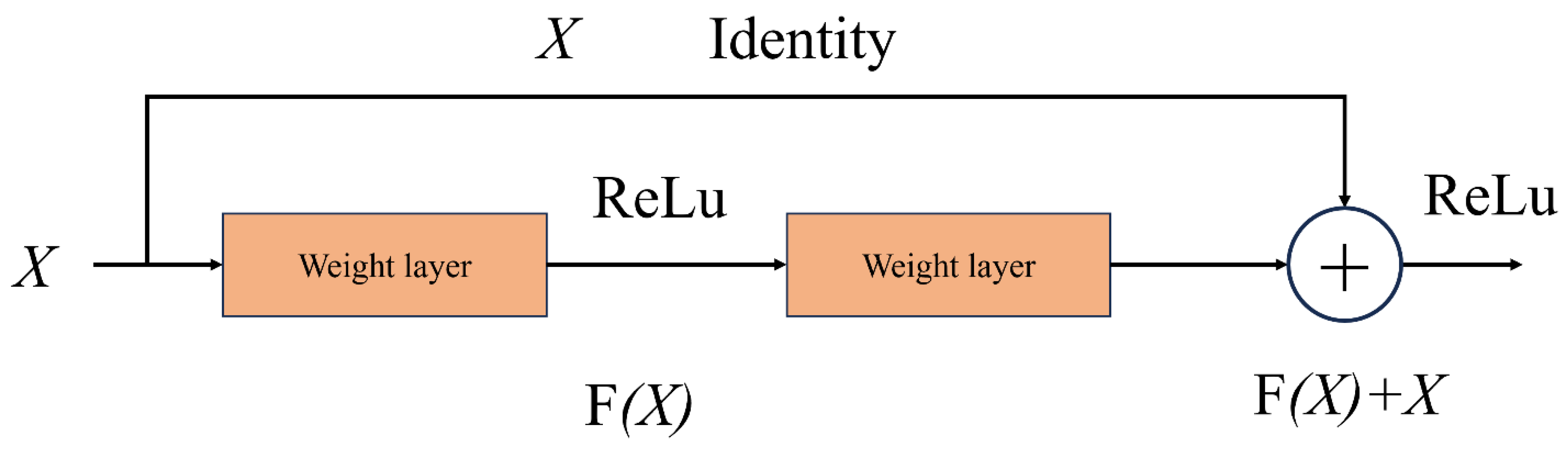
The residuals module of ResNet (
Figure 3) uses standard convolution operations, and the core idea of this architecture is to mitigate the gradient disappearance problem in deep network training by residuals joining. Although this method can guarantee the stable training of the network, with the increase of the number of layers, the amount of computation and the number of parameters will increase, which may lead to the efficiency bottleneck.
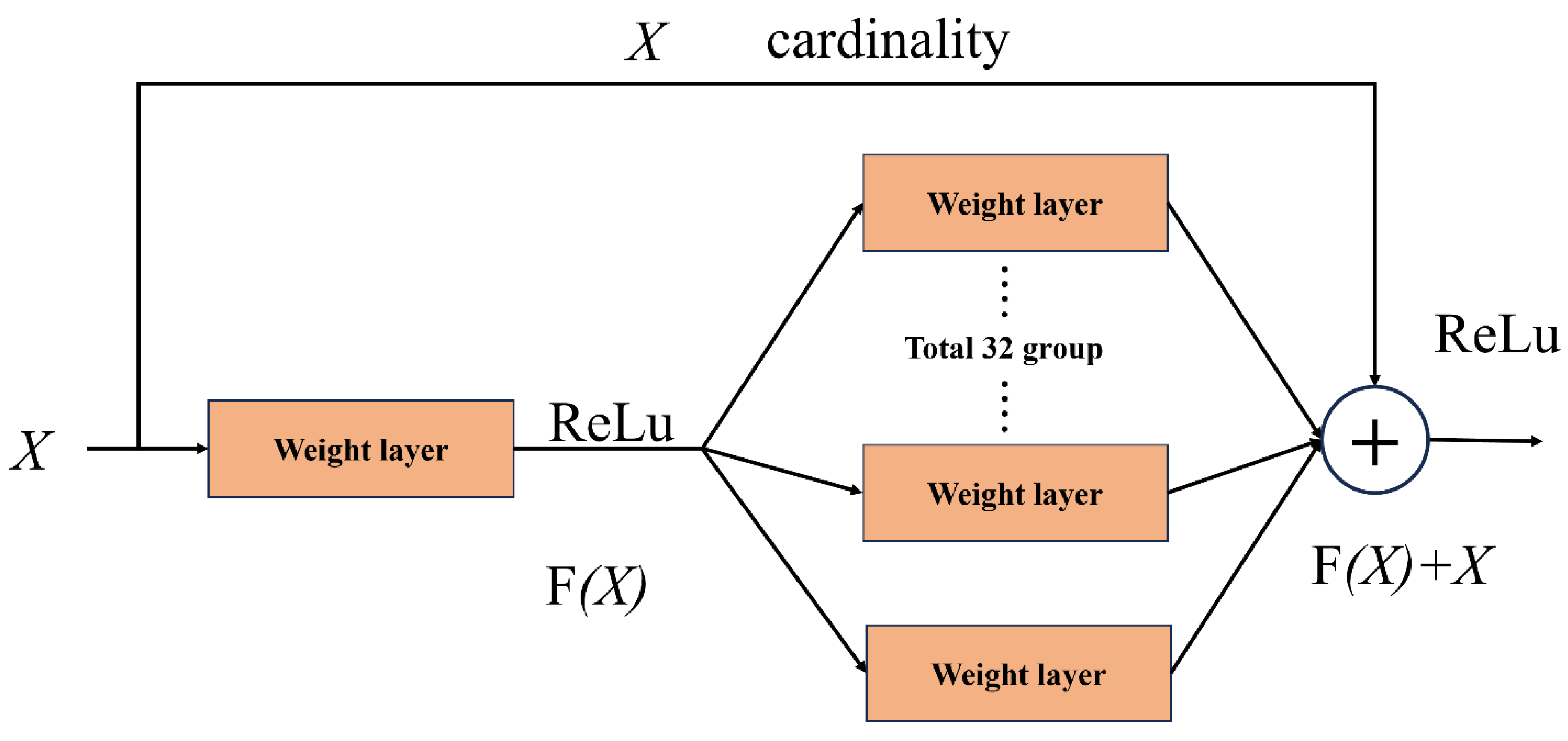
ResNeXt-50 (
Figure 4) optimizes traditional convolution operations by introducing the idea of grouping convolution. The advantage of packet convolution is that it divides the input channels into multiple subgroups for convolution computation, thus reducing the computational complexity and improving the expressiveness of the network without significantly increasing the number of parameters. With this structure, ResNeXt-50 can be more efficient in processing complex disease features and has stronger feature extraction capabilities.
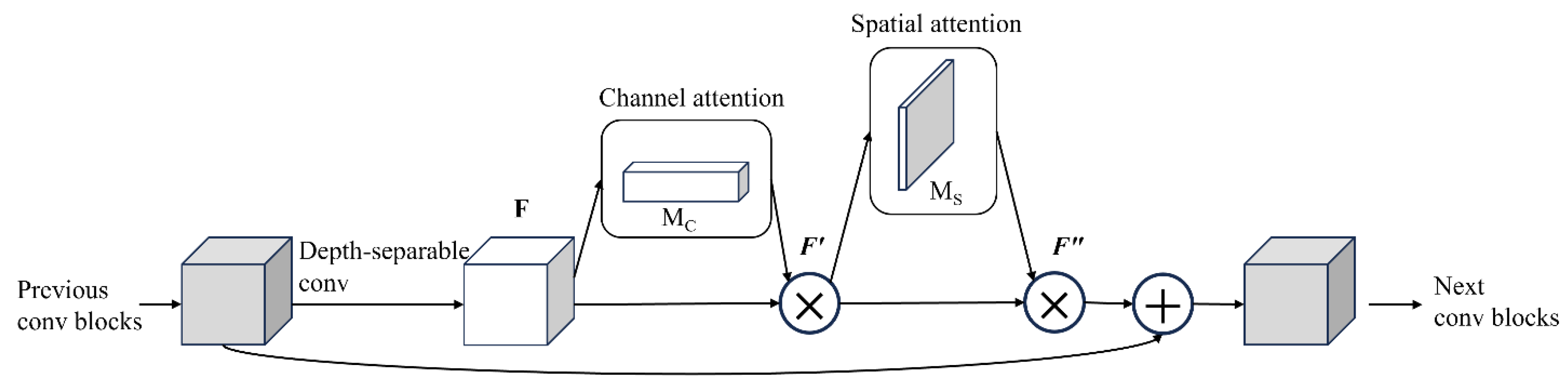
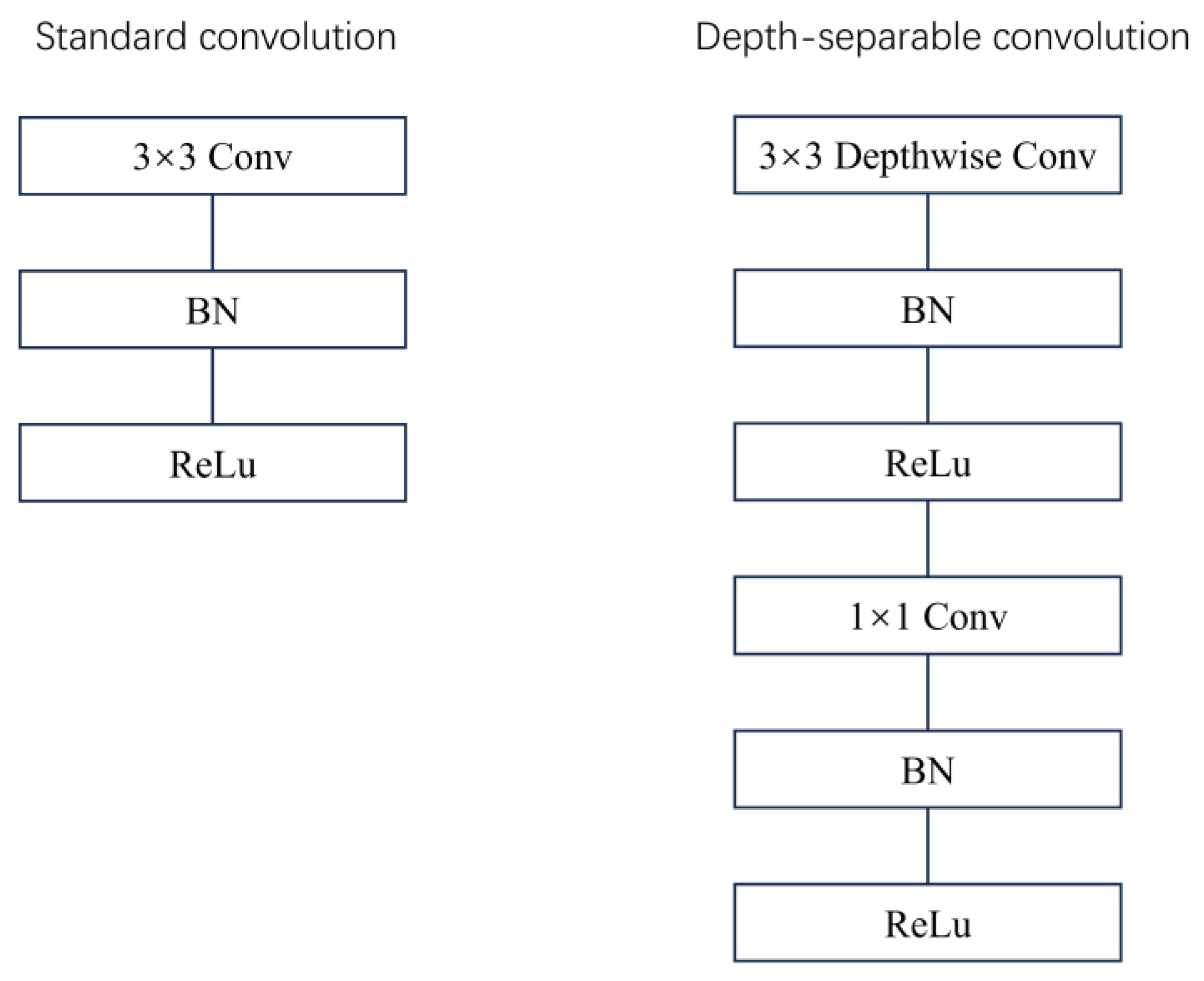
In order to further improve the efficiency and accuracy of the network, the improved ResNeXt-50 is adopted in this paper. In this version, we introduced Depthwise separable convolution (DSC). By decomposing standard convolution into deep convolution and point-by-point convolution (
Figure 5), depth separable convolution consists of depth-by-depth convolution, where depth convolution is used to extract spatial features, and point-by-point convolution, where channel features are extracted. Depth-separable convolution groups convolution in feature dimensions, performs independent depthwise convolution for each channel, and aggregates all channels using a 1x1 pointwise convolution before output.
In
Figure 3,
Figure 4, and
Figure 5, the convolutional structures of ResNet, ResNeXt-50 residuals, and the improved ResNeXt-50 are shown, respectively. The progressive evolution of these structures enables the network to achieve a better balance between performance, computational efficiency and feature extraction capabilities, especially in rice leaf disease detection tasks.
2.3.2 Attention module
CBAM (Convolutional Block Attention Module) [
23] is a lightweight convolutional neural network (CNN) attention mechanism designed to improve the model's performance by enhancing the network's focus on important features. CBAM module combines channel attention mechanism and spatial attention mechanism, which can effectively help the model automatically select important feature channels and spatial locations, thus improving the accuracy and robustness of feature extraction.
Channel attention mechanism: The channels of the input feature map are aggregated through global average pooling and global maximum pooling to generate the weights of each channel. This mechanism can suppress irrelevant channel information and highlight characteristic channels that are helpful for disease detection.
Spatial attention mechanism: the features of each spatial location are weighted to emphasize the information of key regions such as disease regions, so as to improve the model's ability to capture local disease features.
In this study, CBAM modules were integrated into the improved ResNeXt-50 structure to improve the accuracy, robustness and adaptability of rice leaf disease detection by deeply integrating channels and spatial attention mechanisms. Especially in the face of irregular illumination and poor image quality, CBAM module can help the model to extract key disease features more effectively, thus improving the diagnostic accuracy and speed.
2.4 ResNeXt rice leaf disease detection model with depth separable convolution and CBAM modules
In order to improve the efficiency and accuracy of rice leaf disease detection, In this paper, a new ResNeXt network architecture combining Depthwise Separable Convolution and CBAM Convolutional Block Attention Module is proposed. By fusing deep separable convolution with CBAM modules, the model not only reduces the amount of computation, but also improves the accuracy and robustness of disease detection by automatically focusing important features.
Figure 6.
CBAM integrated with a ResBlock in ResNeXt.
Figure 6.
CBAM integrated with a ResBlock in ResNeXt.
Depthwise Convolution is a method to decompose traditional Convolution operations into two steps, namely Depthwise convolution and Pointwise Convolution. This method reduces the computational complexity while maintaining the validity of the model.
1. Deep convolution: Each input channel is convolved only with an independent convolution kernel, the number of channels in the output feature map is the same as the number of input channels, and the calculation of each channel is carried out independently. The formula is as follows:
Where is the input feature map, is the convolution kernel, is the input channel and is the output channel.
2. Point-by-point convolution: 1×1 convolution is used to fuse the feature graphs after deep convolution between channels. The formula is:
Through the application of depth-separable convolution, we can significantly reduce the amount of computation and the number of parameters in the model, while maintaining a high feature extraction capability.
In the channel attention stage, CBAM reduces the spatial dimension of feature graph to by global average pooling. In this process, the feature dimension is compressed to of the input. Then, after being processed by the ReLU activation function, the feature map is entered into the second fully connected layer. At this point, the scale is , and the number of feature channels returns to the input size .
In the compression stage, CBAM module introduces a parameter W into the feature graph
obtained by global average pooling to generate a weight for each feature channel. These weights reflect the importance of the different feature channels and are the core of the entire CBAM module. The weights are applied to the input feature map to achieve feature recalibration, a process called gating mechanism. The excitation process follows formula (3):
Where is the result of the compression process, is the function, and and are the dimension reduction layer with parameter and the dimension increase layer with parameter , respectively. Reweighting is a recalibration process that uses the active output weights to represent the importance of each feature channel after feature selection.
According to the degree of importance, CBAM module weights the channels into the original features by formula (2), while keeping the number of feature channels unchanged and not introducing new feature dimensions. The specific formula is:
Where refers to the channel-level multiplication between the scalar and the feature graph.
By embedding deep separable convolution and CBAM modules into the ResNeXt-50 network, the network architecture uses the feature channel recalibration strategy combined with the residual network, which effectively improves the network performance and significantly reduces the computing cost. Deep separable convolution reduces the computational effort by splitting the convolution process into deep convolution and point-by-point convolution, while CBAM enhances the network's focus on key features through channel attention and spatial attention mechanisms, especially in the learning of complex disease features. This structure can significantly improve the recognition accuracy in rice leaf disease diagnosis tasks, and improve the robustness and adaptability of the model. The whole network structure is shown in
Figure 7.
2.5 Experimental environment configuration
The running environment of this experiment is Windows 10. The central processing unit (CPU) is Intel's 10th generation Core i7, the graphics processing unit (GPU) is NVIDIA GeForce RTX 3070, the memory is 16GB, and the storage is 1TB SSD. The training environment was created by Anaconda3 and configured as Python 3.8.5 and PyTorch 1.7.1 and torchvision 0.8.2 artificial neural network libraries. CUDA 11.0 deep neural network acceleration library is also used.
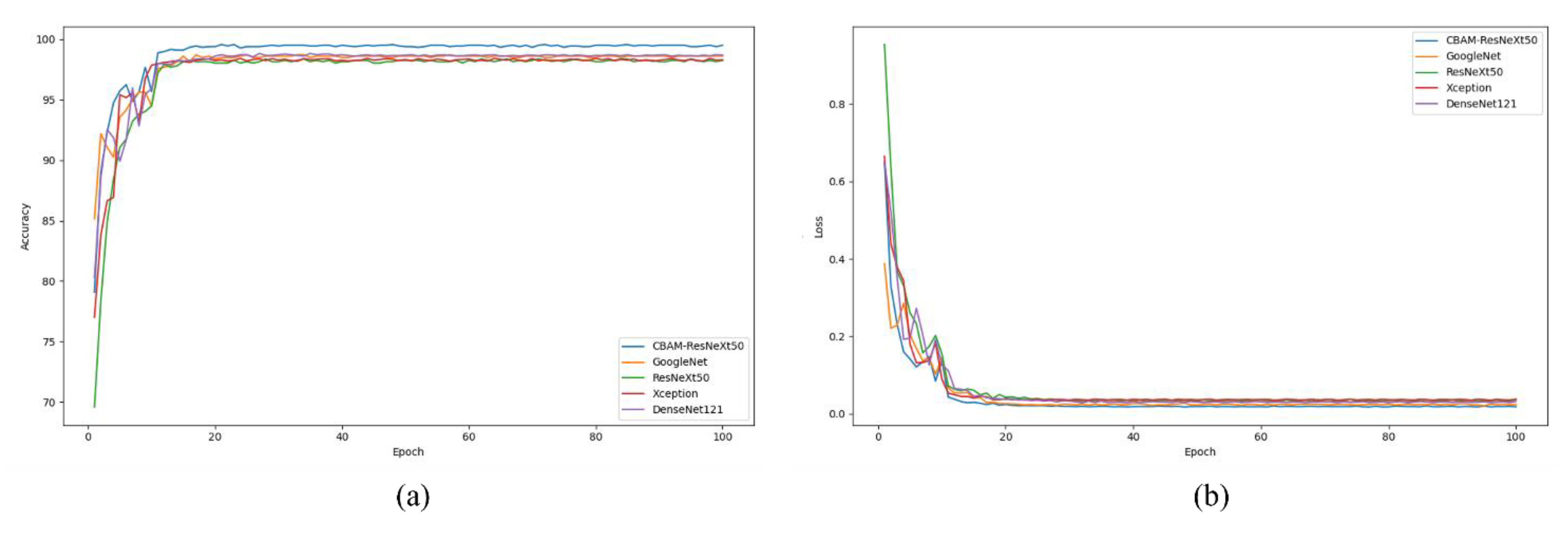
To train the model, the input image is adjusted to 256*256*3, using CrossEntropyLoss and the Adam optimizer. For the training and classification of the model, we set the batch size to 32 and trained 100 epochs with a learning rate of 0.001, using the StepLR scheduler to reduce the learning rate to 0.1 times after every 10 epochs. The dropout layer is defined to reduce overfitting.
The weight values of the feature extraction network use the parameters of the pre-trained ImageNet classification model. This method can greatly reduce the computational cost and training time of the model. After each training, the validation set is tested and the model is saved, and the model with the highest accuracy is selected as the final output.
2.6 Evaluation Indicators
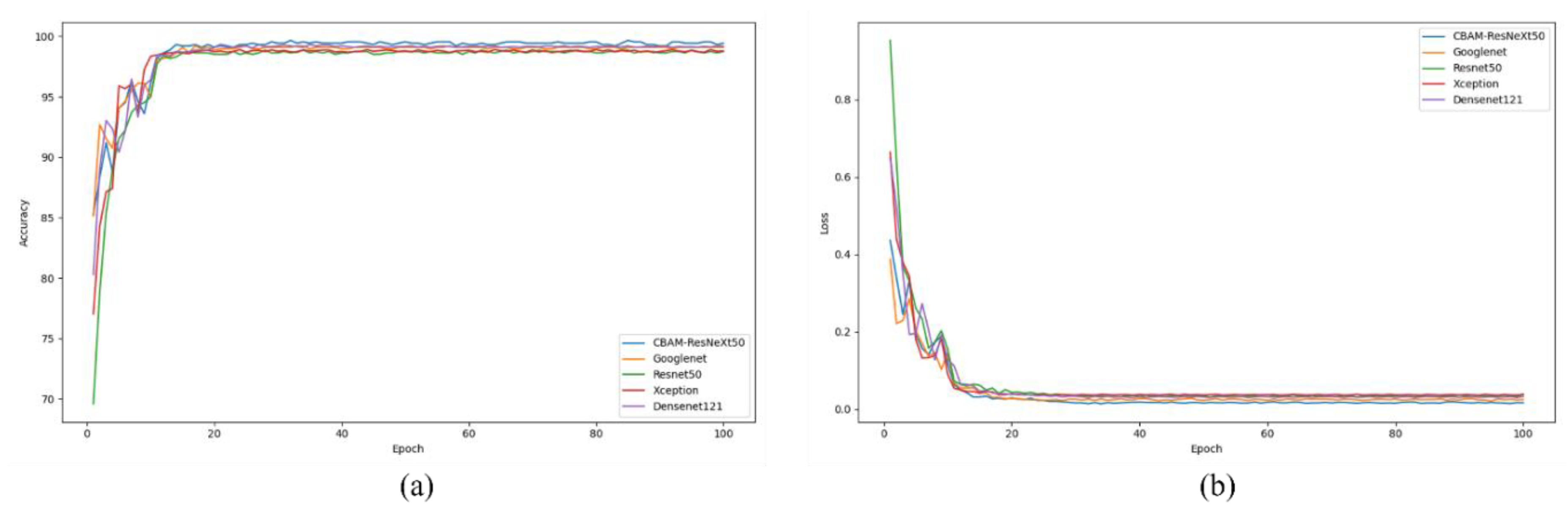
To evaluate the performance of the proposed network, we compared it to several well-known convolutional neural networks (CNNS), including VGG-19, Xception, ResNet-50, and GoogleNet. When evaluating classification results, the average accuracy, which is widely used in the field of image classification, is adopted as the evaluation index. These metrics include Positive Predictive Value (PPV), recall Rate (True Positive Rate (TPR)), F1 Score, and Throughput (TA).
Specific definitions are as follows:
True Positive (TP): The number of samples predicted to be positive that are actually positive.
False positive (FP): The number of samples predicted to be positive that are actually negative.
False negatives (FN): The number of samples predicted to be negative that are actually positive.
From these definitions, the following assessment indicators can be calculated:
Where, is the total detection time of the verification set, and is the total number of samples of the verification set.
Through these evaluation indexes, the classification performance of the proposed network can be comprehensively analyzed and compared with other advanced CNN models to verify its effectiveness.
4. Discussion
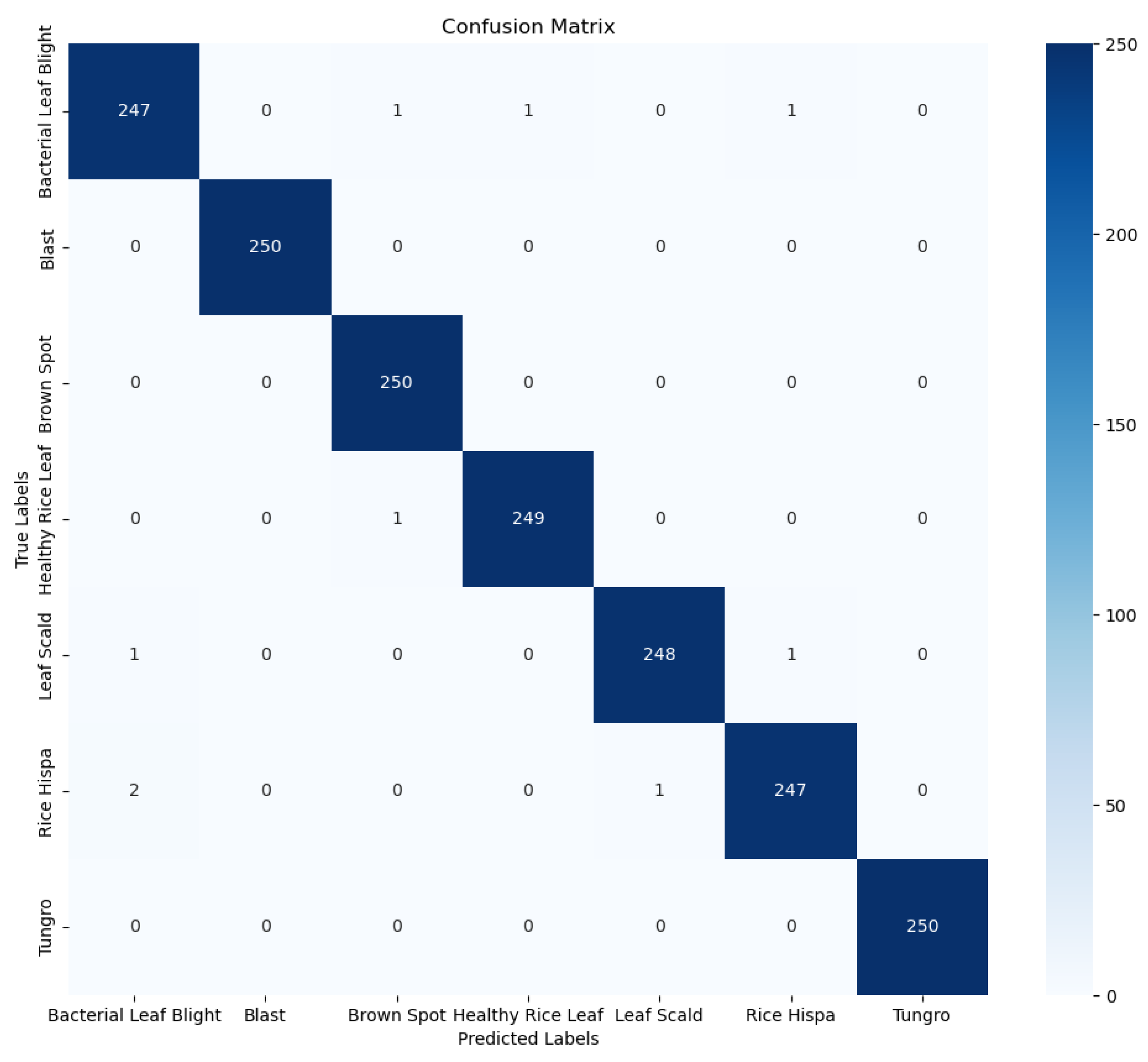
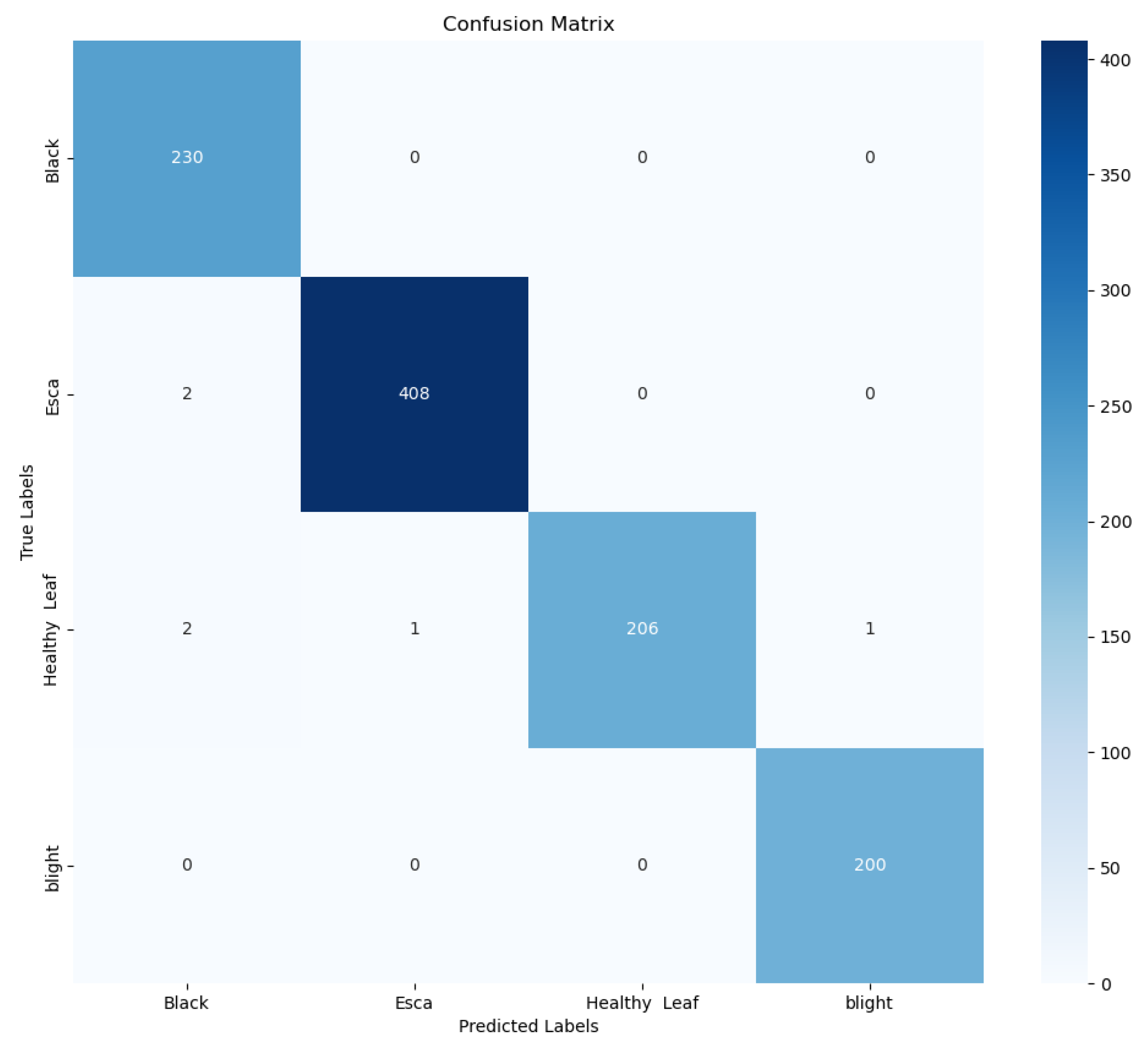
Crop diseases are a major threat to the security of the global vegetable supply and the latest technologies need to be applied to the agricultural sector to control them. Deep learning-based disease detection has been widely studied because of its features such as long-term continuous operation, convenient data collection, good robustness and fast computing speed. In view of the complex characteristics of tomato leaf diseases, a multi-scale diagnostic model was designed to extract disease characteristics. In this study, the dataset was divided into 6 categories (e.g., yellow dwarf, rice thrips, leaf scorch, brown spot, rice blast, and bacterial leaf blight). The CBAM_ResNeXt50 model proposed in this paper achieves an average detection accuracy of 99.66%, which is 3.68% higher than the original ResNet50 network accuracy. The model has a diagnostic accuracy of more than 99% for these four diseases, and the average diagnostic time of a single disease image is only 30.23 milliseconds, which is faster and can meet the needs of real-time operation.
The results of this study were compared with those summarized in
Table 5. As shown in
Table 5, Latif G[
24], Simhadri C G[
27], Yang L[
15] et al. used the same data set, all of which had lower accuracy than the model proposed in this paper. The accuracy of other data sets used by Narmadha R P[
23] and Pandian J A[
25] et al. was also lower than ours, and the accuracy of the model proposed by Upadhyay S K[
24] et al. was higher than ours, because they studied fewer disease classification categories (3 categories). All in all, our model has good general performance and high diagnostic performance for rice leaf diseases.
5. Conclusion
In this study, we successfully developed a multi-scale feature extraction model for rice leaf disease diagnosis. The model deeply integrated the residual block and CBAM attention module, and systematically trained the healthy and different disease rice leaf images. Our results show that using the widely available Kaggle dataset, our model outperforms some recent deep learning studies.
Compared with other models, CBAM_ResNeXt50 showed the best performance in the diagnosis of rice leaf diseases. In addition, when trained with more images from different environments, the performance of the CBAM_ResNeXt50 model typically improves significantly. The trained model can be used effectively in the early automatic diagnosis of rice and other crop diseases. Therefore, this work provides strong support for the early and automated disease diagnosis of rice crops using modern technologies such as smartphones, drone cameras and robotic platforms.
The next research plan is to explore ways to integrate multimodal deep learning, where we will combine image data with other sensor data, such as temperature and humidity, soil nutrients, and meteorological information, and train them through deep learning models. This multi-modal learning will enable us to obtain information from multiple dimensions, enhance the model's adaptability to environmental changes, and improve its performance in complex agricultural scenarios. This innovative approach will provide more reliable data support for precision agriculture, helping farmers identify and address disease problems in a timely manner, thereby improving crop yield and quality. Through these efforts, we expect to achieve higher technical breakthroughs in the field of rice disease diagnosis and promote the development process of agricultural intelligence.