Introduction
The introduction to the deep learning method for rice leaf disease identification using Convolutional Neural Networks (CNNs) shows the important role of rice as a main food and the major risks made by different diseases to its output. With the growing world population, efficient handling of rice fields is important for food security. Traditional methods of disease identification are often labor-intensive and prone to mistakes, requiring the adoption of advanced technologies. CNNs have developed as a useful tool for automating the identification and classification of rice leaf diseases, leveraging their ability to learn complex patterns from pictures. This introduction sets the stage for addressing the methods, results, and effects of using CNNs in rice disease identification.
Rice is a main food source in many Asian countries, making its health important for food security (Akter et al., 2024). Diseases like bacterial blight and blast can greatly lower crop output, necessitating early detection (Banerjee & Sadhukhan, 2023). Advancements in Deep Learning CNNs have changed picture classification tasks, allowing automatic and accurate disease diagnosis (Rashed & kakon., 2025). Various CNN designs, such as ResNet50 and DenseNet, have shown better success in finding rice leaf diseases (Kumar & Bhowmik, 2023). Research Findings Studies show good accuracy rates, with some models getting over 99% accuracy in disease classification (Bhanu et al., 2024) (Akter et al., 2024). The use of big datasets for training improves the stability of CNN models, leading to better generalization across different disease types (Banerjee & Sadhukhan., 2023). While the improvements in CNNs for rice disease detection are positive, challenges remain in terms of dataset variety and model flexibility to different weather conditions. Further study is needed to improve the usefulness of these models in real-world farming settings.
Literature Review
All over the world, experts, agricultural scientists committed to agricultural development have combined varied methods to identify rice leaf diseases, with the aim of improving farming practices. There has been a noticeable rise in interest among researchers lately, especially with regards to studies that show the use of advanced deep learning methods like as Convolutional Neural Networks (CNNs) for the identification of diseases affecting rice leaf.
Addressing the problem of rice disease detection (Ahmed et al., 2023) a biphasic method based on CNN is suggested, which can reduce training data. The collection of 200 pictures consists of three classes - False Smut, Neck Blast, and healthy grain class. This method includes a range of rice diseases and a prediction accuracy of 88.9%. In their work, (Gogoi et al.,2023) suggested a 3-Stage CNN design with a transfer learning method that uses a pre-trained CNN model fine-tuned on a small collection of rice disease pictures. The method was tested on a collection of 8883 disease pictures and 1200 healthy rice leaf images, getting an amazing 94% accuracy in a 10-fold cross-validation process.
Six popular CNN architectures, proposed by (NG et al., 2023) including AlexNet, VGG-19, VGG-16, InceptionV3, MobileNet, and ResNet-50 are trained and tested on a Plant Village dataset for classifying the paddy plant images into one of the four classes namely, Healthy, Brown Spot, Hispa, or Leaf Blast, based on the disease condition. The results noticed, AlexNet with best accuracy of 89.4%, VGG-16, VGG-19 and ResNet-50 showed similar accuracy.
(Ujawe et al., 2023) suggested, CNN Inception V3 and SqueezeNet model. Datasets of 241 pictures. Inception V3 model accuracy is 95% with 12% loss. SqueezeNet model on same dataset precision is 92% with 27% loss.
(Latif et al., 2022) suggested updated system can correctly find and identify six different groups. The changed suggested approach includes a modified VGG-19-based transfer learning method. The best average accuracy using the updated suggested method is 96.08% using the non-normalized expanded dataset.
(Chavan et al., 2022) shows a 6 Layered CNN based model. Achieved total Accuracy 97.23%. The method gets high accuracy for different diseases, with 98.85% for Brown Spot, 98.09% for Rice Blast, 99.17% for Hispa, and 99.25% for healthy leaves.
(Tejaswini et al., 2022) they suggested automated method to identify the fungus disease of rice plants which is a major cause for the loss of rice plants. later processed using several standard deep learning methods like VGG-19, VGG16, Xception, Resnet, along with a handmade 5-layer neural network. 5-layer neural network performs the best at identifying rice leaves out of all of them. 5-layer convolution got 78.2%. (Upadhyay et al., 2022) CNN was taught using 4000 picture samples uses plant leaf images to get defining features based on the size, shape, and color of tumors in the leaf. Result analysis showed that the second model with background removal works better than the first model with 99.7%accuracy.
(Narmadha et al., 2022) suggested a new deep learning-based rice plant disease detection method called DenseNet169-MLP. Bacterial Leaf Blight, Brown Spot, and Leaf Smut. aims to group rice plant illnesses into three types. DenseNet169-MLP model has achieved highest sensitivity of 96.40%, specificity of 98.27%, precision of 96.82%, accuracy of 97.68% and F-score of 96.43%.
(Shaikh et al., 2022) created a Faster-RCNN based model for the early and accurate discovery of rice leaf diseases. Rice Leaf Disease Dataset (RLDD) from an internet library and an individual dataset unique to the project. The accuracy reached for each disease group using the Faster R-CNN method is as follows: Brown Spot - 98.85%, Rice Blast - 98.09%, Hispa - 99.17%, and Healthy Rice Leaf - 99.25%.
(Prottasha et al., 2022) suggested a depthwise separable convolutional neural network for identifying 12 types of rice plant diseases. Total of 500 pictures has been studied including healthy and sick images and taught using the convolutional neural network (CNN) model. The suggested model performs considerably well in contrast to current state- of-the-art CNN designs. approval and testing accuracy of 96.5% and 95.3% respectively while having a greatly smaller model size. So, the finding given in the paper shows an accuracy of 95.38% while identifying the rice plant diseases.
(Ritharson et al., 2024) introduced a custom method for better generalization, having a custom VGG 16 model with 13 convolutional layers, 5 max pooling layers, and 7 fully connected layers. The suggested model showed excellent performance with a better accuracy of 99.94% and maximum precision and memory scores across 9 different disease class labels of rice leaves.
Materials and Methods
Rice, a staple food for much of the world, is subject to several leaf diseases such as Bacterial Leaf Blight, Brown Spot, Hispa, Leaf Smut, and Tungro. This study focuses on creating an accurate and efficient monitoring method using Convolutional Neural Networks (CNNs) to spot these illnesses at an early stage. CNNs, known for their good success in picture recognition tasks, are well-suited for identifying rice leaf diseases. The tests were performed on Google Colab using Keras with TensorFlow as the base, leveraging a T4 GPU for rapid training. The application utilized Python along with tools like NumPy, OpenCV, and Scikit-learn.
Data Collection
I have collected about 711 picture data out of which 591 are taken by myself in the field and rest from websites Kaggle. And labeled with five class bacterialleafblight, brownspot, hispa, leafsmut and tungro.
Figure 1.
Five types of rice leaf disease Images.
Figure 1.
Five types of rice leaf disease Images.
Statistical Analysis
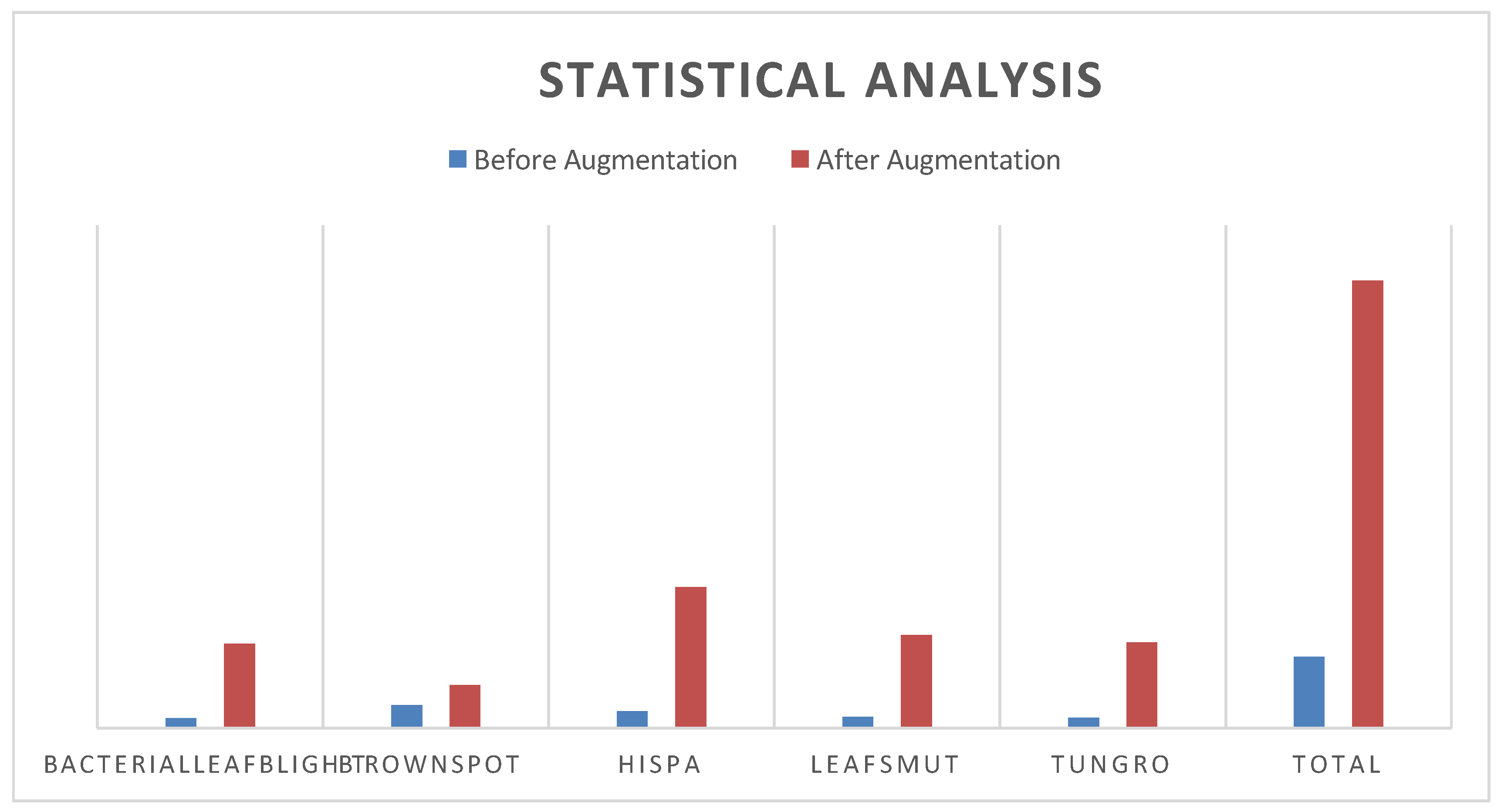
Through examining data from a total of 711 pictures on five different disease types: bacterialleafblight, brownspot, hispa, leafsmut, and tungro. With the addition method, I turned data into 4449 overall pictures.
Table 1.
Rice Leaf Disease Distribution Dataset.
Table 1.
Rice Leaf Disease Distribution Dataset.
| Class Name |
Before Augmentation |
After Augmentation |
| bacterialleafblight |
100 |
840 |
| brownspot |
230 |
429 |
| hispa |
168 |
1404 |
| leafsmut |
111 |
924 |
| tungro |
102 |
852 |
| Total |
711 |
4449 |
Figure 2.
Statistical Analysis of dataset.
Figure 2.
Statistical Analysis of dataset.
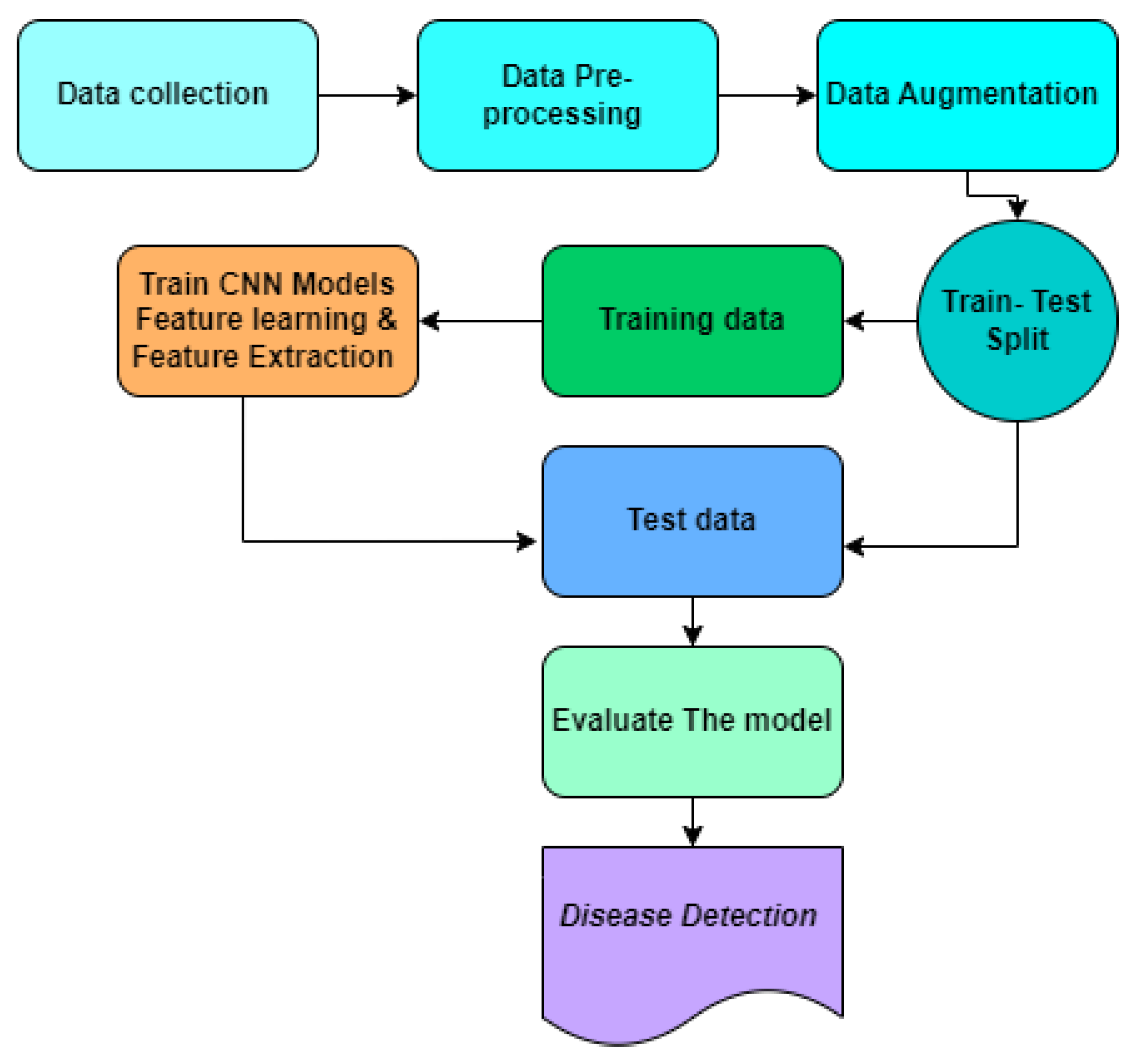
Figure 3.
Proposed Methodology.
Figure 3.
Proposed Methodology.
Using Convolutional Neural Networks (CNNs) is a successful way in the complicated area of rice leaf disease identification. The difference is not just in how it is applied, but also in the way the CNN design was built. Considering the previously taught algorithm EfficientNetB7 that I used, VGG-19, ResNet-101, and InceptionResNetV2 allowed me to accomplish my main goal of disease identification.
Our pick of CNN designs is based on models, each of which adds unique features to the larger architecture of disease identification.
EfficientNetB7
EfficientNetB7, created by Google AI, is a powerful CNN system intended for picture recognition and classification jobs. It stands out for its efficient scaling method that combines depth, width, and resolution to improve both accuracy and speed. The model uses leftover links, dynamic channel scaling, and Swish activation to improve feature representation. With 813 layers—including 16 convolutional layers, batch normalization, activation, and fully linked layers—EfficientNetB7 provides high accuracy while keeping processing speed.
VGG-19
VGG-19 is a deep CNN with 19 layers, including 16 convolutional and 3 fully linked layers. It uses stacked 3×3 convolutional filters and max-pooling layers in a rotating pattern to catch both small and complex features. Max-pooling helps in downsampling and keeping important spatial information. The final fully connected layers perform classification based on the retrieved features.
ResNet-101
ResNet-101, created by Microsoft Research, is a deep CNN with 101 levels that shines in computer vision tasks. It uses residual learning with skip links to solve vanishing gradient problems, allowing successful training of very deep networks. Its design includes convolutional layers, batch normalization, ReLU activations, and residual units, allowing it to record both fine and complex features—making it ideal for picture classification and object recognition.
InceptionResNetV2
Inception-ResNet-v2 is a convolutional neural network that has received training using a large dataset containing over a million pictures taken from the ImageNet database. The best parts of both Inception and ResNet are merged in this smart mixed design. Similar to Inception V3, InceptionResNetV2 is equipped with the Softmax function in its final layer. The InceptionResNetV2 design is marked by its large depth, made of a total of 42 levels. The input layer of the network is designed to handle pictures with sizes of 299 × 299 pixels. This system uses advanced design concepts to improve its success in different computer vision jobs.
Table 2.
Comparison Of The CNN Network Designs Applied In This Project.
Table 2.
Comparison Of The CNN Network Designs Applied In This Project.
| Architecture Name |
Publ.
Year |
Contribution |
Parameters
(M) |
Layers |
| EfficientNetB7 |
2019 |
Scaling network depth, width, and resolution uniformly for better accuracy and efficiency |
66 |
607 |
| VGG-19 |
2014 |
Deeper network with small 3x3 filters for improved accuracy |
143.7 |
19 |
| ResNet101 |
2015 |
Residual blocks to address vanishing 55.9gradients and enable deeper networks |
44.7 |
101 |
| InceptionResNetV2 |
2016 |
Combines Inception blocks with residual connections for further accuracy gains |
55.9 |
449 |
Data Preprocessing
The success of dataset change is usually tied to the details of data preparation. Effectively preparing the data not only allows better research but also improves the accuracy of the results. In the area of this study, getting ideal preparation results is especially important for a full understanding of disease trends in rice fields. In early November, I directly collect Rice leaf from the rice field and collect some data from online and split into five classes and set into files. We removed a number of unclear, needless, and unrelated pictures from our data. So, it’s ready to be used and gives the result.
Data Augmentation
Image enhancement is widely used in deep learning to grow datasets by adding changes to current photos while retaining their names. This increases dataset unpredictability, improves model adaptation, and boosts precision. In this study, we applied different enhancement methods such as brightness, contrast, saturation changes, scaling, cutting, flipping, rotation (±15° to 90°), distortion, shear, skew, and intensity transformations. Each original picture was used to create ten enhanced copies. Images were trimmed to 265×265 and center-cropped (rows and columns 64–192). We used the “imgaug” library to perform augmentations sequentially and in random order, including horizontal flips (50% chance), Gaussian blur (σ: 0–0.5), contrast adjustment (0.75–1.5), Gaussian noise, and brightness changes (20% chance per channel).
Figure 4.
Before and after augmentation.
Figure 4.
Before and after augmentation.
Results and Discussion
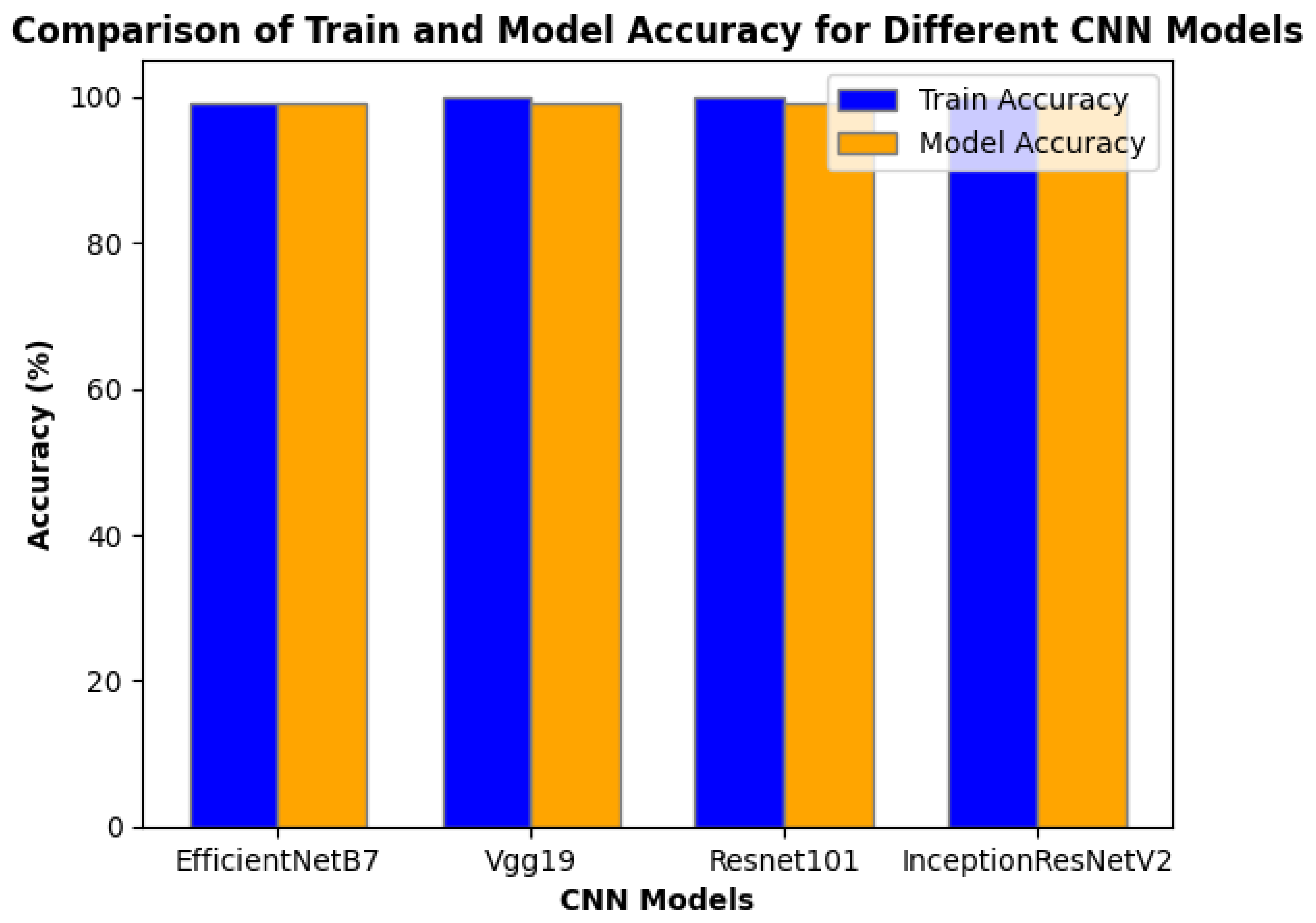
This part shows the results of our four CNN networks: EfficientNetB7, VGG- 19, ResNet-101, and InceptionResNetV2. The models’ sorting ability is first presented. The classification ability of the models is first given. Interestingly all our four models provide 99% -100% train accuracy because its input was pre-processed using GaussianBlur, linearContrast, removal noise from pixels and change the color and increase the brightness of the pictures. It should be mentioned that all four of our models achieve 99% model accuracy given their input was pre-processed using GaussianBlur, GaussianNoise, noise removal from pixels, and picture brightness and color changes.
Table 3.
Train And Model Accuracy Of CNN Models.
Table 3.
Train And Model Accuracy Of CNN Models.
| CNN Models |
Train Accuracy |
Model Accuracy |
| EfficientNetB7 |
99% |
99% |
| VGG-19 |
100% |
99% |
| ResNet-101 |
100% |
99% |
| InceptionResNetV2 |
100% |
99% |
Figure 5.
Comparison of Train and Model Accuracy for Different CNN Models.
Figure 5.
Comparison of Train and Model Accuracy for Different CNN Models.
Except for EfficientNetB7, all models achieve 100% accuracy on the training data, showing great learning of the training patterns. This might, however, indicate possible overfitting, in which a model memorizes the training data too closely and experiences with generalization. However, all models also have a 99% Model Accuracy, which likely still refers to the test accuracy. This is a high result for all models on unknown data.
Table 4.
Precision, Recall, F1-Score, And Model Support for Four Models.
Table 4.
Precision, Recall, F1-Score, And Model Support for Four Models.
| EfficientNetB7 |
| |
bacterialleafblight |
brownspot |
hispa |
leafsmut |
tungro |
| Precision |
100% |
94% |
100% |
98% |
99% |
| Recall |
99% |
94% |
100% |
100% |
98% |
| F1- Score |
99% |
94% |
100% |
99% |
99% |
| Support(N) |
165 |
90 |
279 |
173 |
165 |
| VGG-19 |
| |
bacterialleafblight |
brownspot |
hispa |
leafsmut |
tungro |
| Precision |
98% |
98% |
100% |
99% |
99% |
| Recall |
99% |
92% |
100% |
99% |
100% |
| F1- Score |
99% |
95% |
100% |
99% |
99% |
| Support(N) |
165 |
90 |
279 |
173 |
165 |
| ResNet-101 |
| |
bacterialleafblight |
brownspot |
hispa |
leafsmut |
tungro |
| Precision |
99% |
99% |
99% |
99% |
99% |
| Recall |
100% |
93% |
100% |
100% |
99% |
| F1- Score |
100% |
96% |
100% |
100% |
99% |
| Support(N) |
165 |
90 |
279 |
173 |
165 |
| InceptionResNetV2 |
| |
bacterialleafblight |
brownspot |
hispa |
leafsmut |
tungro |
| Precision |
99% |
99% |
99% |
98% |
99% |
| Recall |
100% |
92% |
100% |
99% |
99% |
| F1- Score |
100% |
95% |
100% |
99% |
99% |
| Support(N) |
165 |
90 |
279 |
173 |
165 |
Here Table shows the Precision, Recall, F1-score, and model support. The performance of the EfficientNetB7, VGG-19, ResNet-101, and InceptionResNetV2 models for spotting rice leaf disease was examined.
Precision, which measures the accuracy of the model’s good results, varies among groups. EfficientNetB7 regularly gets the highest accuracy for the bacterialleafblight (100%), hispa (100%), and tungro (99%) classes among the models. In comparison, VGG- 19 has the lowest accuracy for brown spot (98%) while InceptionResNetV2 has the best precision for all classes except leafsmut (98%).
Recall, showing the model’s ability to correctly spot positive instances, displays swings. EfficientNetB7 excels in recall for bacterialleafblight (99%), hispa (100%), tungro (98%) and leafsmut (100%) classes. However, it has a relatively lower memory for brownspot (94%). VGG-19 gets the best recall for hispa (100%), while its lowest recall is found for brownspot (92%).
F1-Score, a mix between precision and memory, shows trends similar to precision and recall. EfficientNetB7 regularly beats other models, getting the best F1- score for bacterialleafblight (99%), hispa (100%), and tungro (99%). VGG-19 shows the lowest F1-score for brown spot (95%).
Support, showing the number of cases for each class, stays pretty stable across models. However, it’s worth mentioning that the support numbers for brownspot (90%) are the lowest among the classes for all models.
So, we can say that EfficientNetB7 constantly outperforms on numerous measures, with the best accuracy, recall, and F1-score for multiple classes. VGG-19 and ResNet-101 also show close performance, but InceptionResNetV2 falls somewhat behind on several measures.
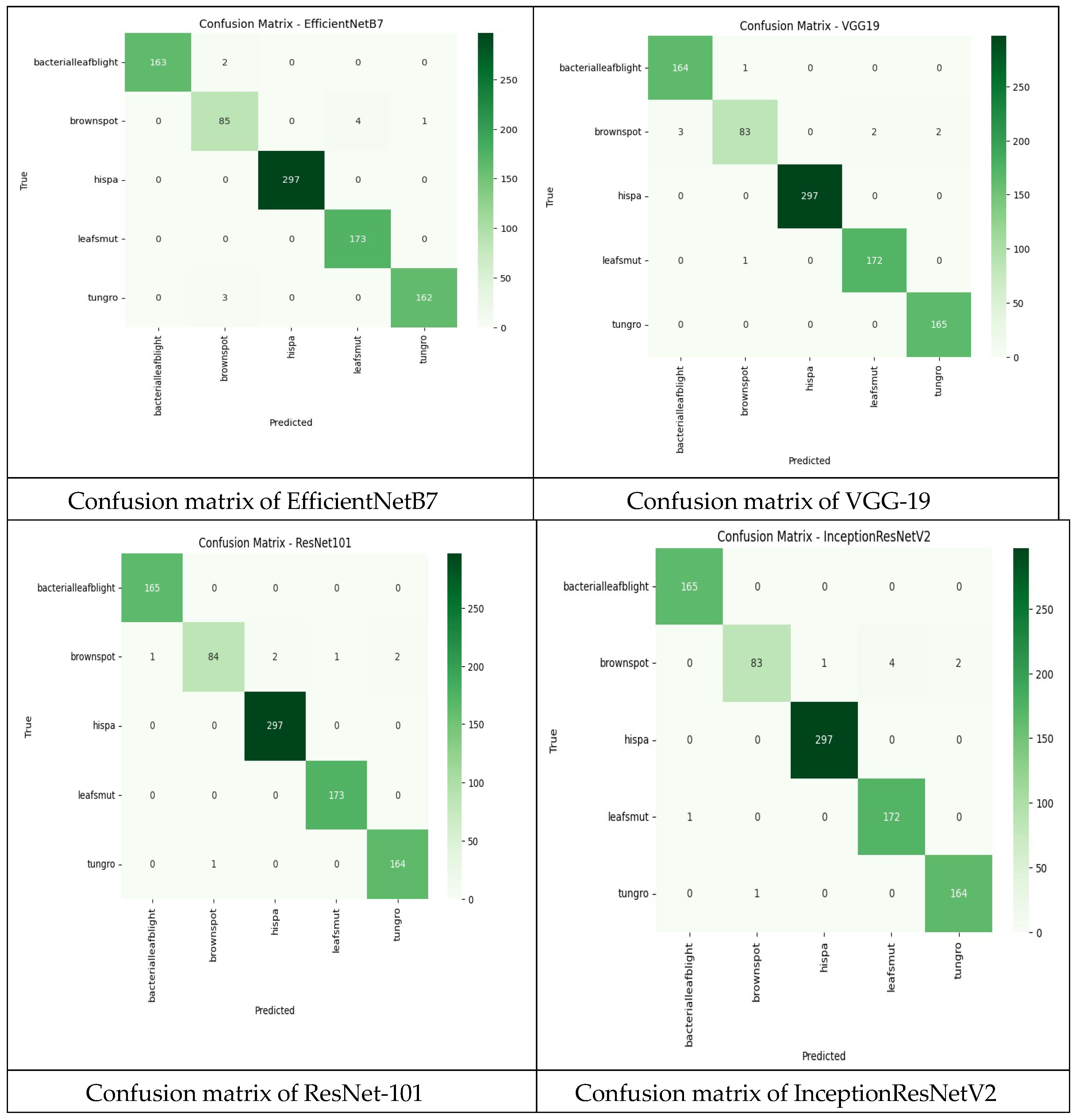
Confusion matrix is a useful tool in judging the classification performance of a model, giving a more detailed view beyond simple precision. Because they allow for direct comparisons between factors such as True Positive, False Positive, True Negative, and False Negative. It takes a test sample and compares the model’s expected labels to the real labels. True Positive (TP): The model correctly identifies a damaged leaf as sick. False Positive (FP): The model wrongly predicted positive cases. True Negative (TN): The model correctly predicted negative cases. False Negative (FN): The model wrongly predicted negative cases.
Figure 6.
Confusion matrix of four CNN.
Figure 6.
Confusion matrix of four CNN.
Table 5.
Efficientnetb7, Vgg-19, Resnet-101& Inceptionresnetv2 Different Parameters.
Table 5.
Efficientnetb7, Vgg-19, Resnet-101& Inceptionresnetv2 Different Parameters.
| Parameter |
EfficientNetB7 |
VGG-19 |
ResNet-101 |
InceptionResNetV2 |
| Number of Image’s |
2848 |
2848 |
2848 |
2848 |
| Image size |
224x224 |
224x224 |
224x224 |
224x224 |
| Epoch |
46/50 |
26/50 |
26/50 |
34/50 |
| Train Accuracy |
99.54% |
100% |
100% |
100% |
| Validation Accuracy |
99.44% |
98.45% |
99.58% |
99.02% |
| Validation loss |
0.0151 |
0.0740 |
0.0279 |
0.0446 |
Here are some thoughts based on the given information:
best Validation Accuracy: VGG-19 got 98.45% validation accuracy, which is the second best among the models. If getting the highest accuracy is a top concern, ResNet-101 would be the best worker in this area with 99.58% confirmation accuracy.
Training Accuracy: All models got 100% training accuracy, showing that they learned the training data well. However, perfect training accuracy doesn’t necessarily promise the best results on new, unknown data.
Validation Loss: Lower validation loss values are usually desired as they suggest better adaptation to unknown data. In this situation, EfficientNetB7 has the lowest validation loss (0.0151), followed closely by ResNet-101 (0.0279).
Epochs Required: The number of epochs each model was trained for changes. A model that gets good results with fewer epochs may be called more efficient. EfficientNetB7 achieved high accuracy in 46 epochs, whereas VGG-19 and ResNet-101 got their claimed accuracy in 26 epochs.
Considering these factors, if we value a mix between high accuracy, low validation loss, and training speed, EfficientNetB7 might be a good candidate.
Model Evolution
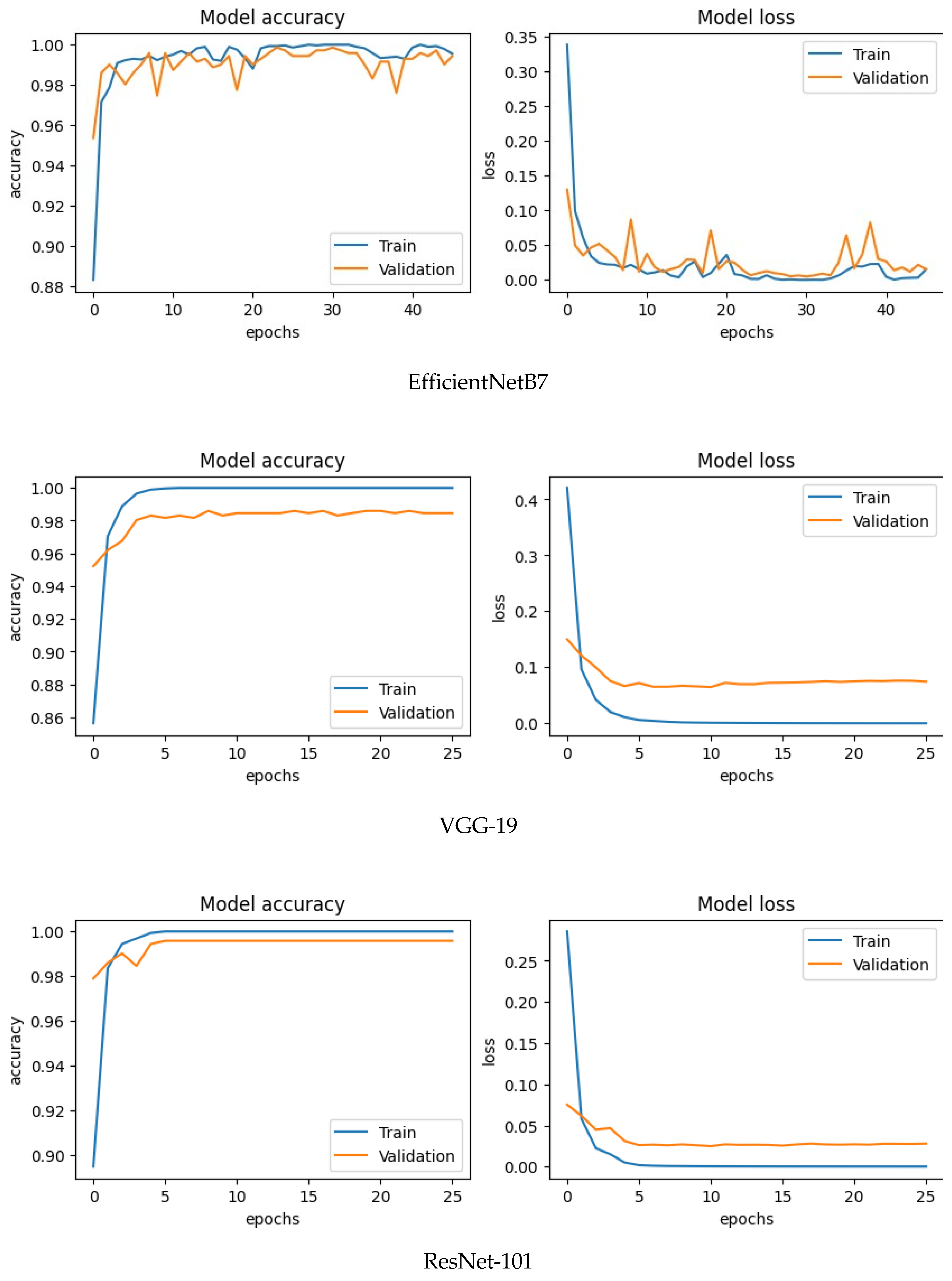
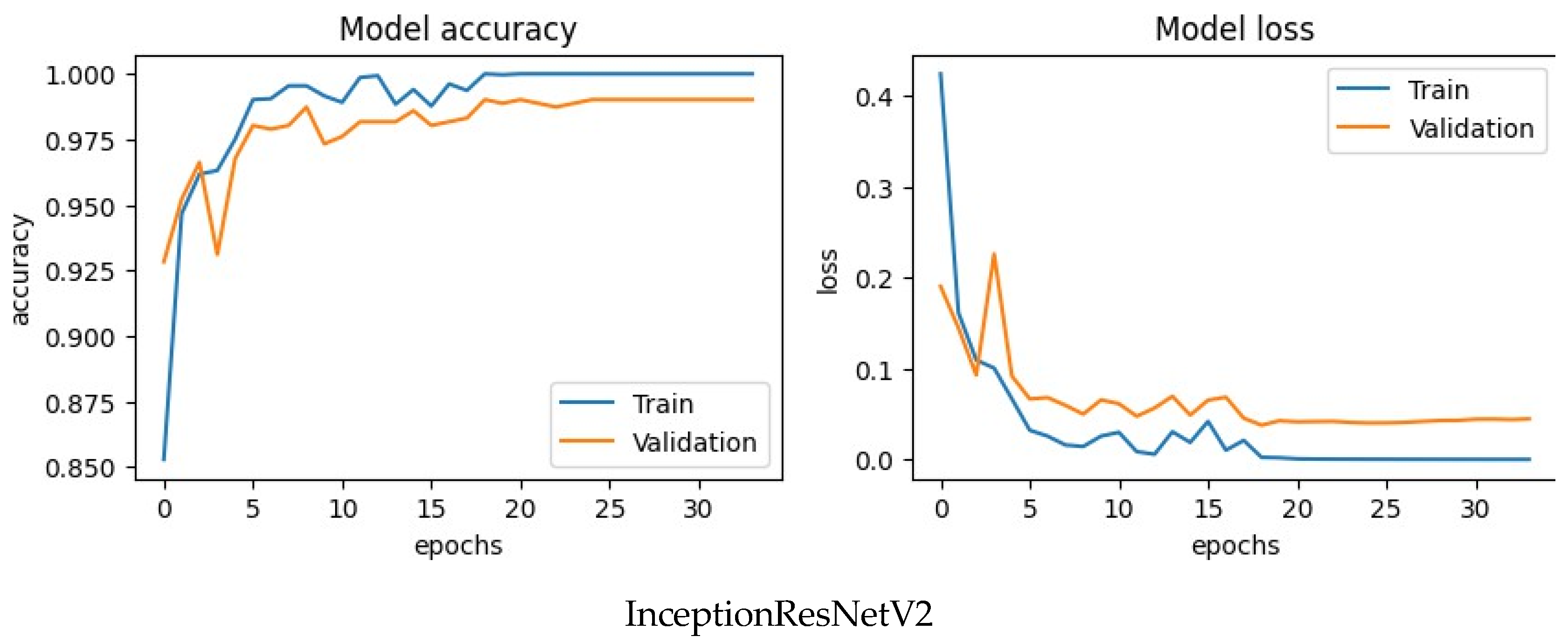
This above picture shows the accuracy and loss graph of train and validation of four CNN model. These graph shows the accuracy of the model on the training data (blue line) and the validation data (orange line) as the number of epochs (training iterations) increases for all CNN model and shows the loss of the model on the training data (blue line) and the validation data (orange line) as the number of epochs increases. Here x-axis denotes the number of epochs and y-axis represents accuracy and loss. Whenever the number of epochs rises the train accuracy quickly climbs up to 1,000 for most of the models and shows that the training data can be used to make accurate predictions. Validation accuracy originally starts at 0.975 and grows slowly to 0.99, showing a strong ability to apply to new data for most of the model. The gap between the train and confirmation accuracy is small, and it does not suggest major overfitting. For model loss, Train loss shows successful learning on the training data, as it drops quickly and constantly. Validation loss also drops gradually although a little more slowly, in line with the validation accuracy trend.
Figure 7.
Accuracy and loss curve of CNN.
Figure 7.
Accuracy and loss curve of CNN.
Discussion
In our project we fully examined how effectively CNN worked in finding diseases of rice leaf with 5 classes: bacterialleafblight, brownspot, hispa, leafsmut, and tungro. We apply CNN models for identifying Rice leaf disease. Applied augmentation on our datasets that make an additional total 4449 pictures. We split our data sets into 80% for training and 20% for test reasons and then build a functional model to train our dataset with pre-trained CNN models EfficientNetB7, VGG-19, ResNet-101, and InceptionResNetV2. By individual classification report we found that EfficientNetB7 model got best results among those models.
Limitations
While this study shows good accuracy in spotting rice leaf diseases using deep learning models, several limitations remain. First, the dataset, although expanded to 4,449 pictures, came from only 711 original samples, which may limit the variety and extension of the models in real-world situations. Additionally, the collection includes only five disease groups and may not catch the full range of rice illnesses or differences across different geographical and weather conditions. The study was performed in a controlled setting using cloud-based training (Google Colab with a T4 GPU), which may not fully reflect success in real-time or resource-constrained field operations. Moreover, lighting conditions, picture quality, and background noise were not clearly changed, which could impact model stability in real applications.
Future Work
Future study should focus on increasing the collection with more diverse and real-world examples from different areas and situations. Incorporating extra disease groups and healthy leaf pictures would improve model stability and flexibility. The merging of mobile or edge-based rollout methods can help bring these models closer to actual, on-field use by farmers. Further improvements could include group learning methods, mixed models, or attention processes to improve performance. Exploring explainable AI (XAI) methods can also make the models clearer and more trustworthy for end-users. Lastly, combining this recognition system into a user-friendly program for real-time disease tracking and early warning systems could greatly help farming production.
Conclusions
In order to effectively control illnesses and keep crop health, rice leaf diseases must be identified as soon as possible. The success of many CNN models, such as EfficientNetB7, VGG-19, ResNet-101, and InceptionResNetV2, for both the identification and classification of rice leaf diseases has been the major focus of this work. We found that EfficientNetB7 beats other models in accuracy, recall, and F1-score for different sickness classes. Specifically, EfficientNetB7 has the highest F1-score for bacterial leaf blight (99%), hispa (100%), and tungro (99%). EfficientNetB7 has the lowest confirmation loss (0.0151), followed closely by ResNet101 (0.0279). This shows the efficiency of EfficientNetB7 & ResNet-101 in successfully recognizing and sorting different forms of rice leaf diseases. In terms of total model accuracy, these two models come out with an amazing accuracy rate of 99%. This shows the model’s power to make correct predictions and lessen confusion, showing its reliability for useful use in real-world situations.
References
- Akter, S.; Sumon, R.I.; Ali, H.; Kim, H.C. Utilizing Convolutional Neural Networks for the Effective Classification of Rice Leaf Diseases Through a Deep Learning Approach. Electronics 2024, 13, 4095. [Google Scholar] [CrossRef]
- Banerjee, S.; Sadhukhan, B. Identification of Rice Leaf Diseases Using CNN and Transfer Learning Models. 2023 International Conference on Electrical, Electronics, Communication and Computers (ELEXCOM) 2023; IEEE; pp. 1–6.
- Kumar, A.; Bhowmik, B. Automated rice leaf disease diagnosis using CNNs. 2023 IEEE Region 10 Symposium (TENSYMP) 2023; IEEE; pp. 1–6.
- Jeejo, A.; Joseph, J.; Byju, A.; KB, D. Rice Leaf Disease Detection using Deep Learning. Grenze International Journal of Engineering Technology (GIJET) 2024, 10. [Google Scholar]
- Rashed, M.; kakon, M.I. A Comparison of CNN Performance in Skin Cancer Detection. Preprints 2025. [Google Scholar] [CrossRef]
- Ahmed, T.; Rahman, C.R.; Abid, M.F.M. Rice disease detection based on dual-phase convolution neural network. Geographical Research Bulletin 2023, 2, 128–143. [Google Scholar]
- Gogoi, M.; Kumar, V.; Begum, S.A.; Sharma, N.; Kant, S. Classification and Detection of Rice Diseases Using a 3-Stage CNN Architecture with Transfer Learning Approach. Agriculture 2023, 13, 1505. [Google Scholar] [CrossRef]
- NG, P.; Prathima, S.; S Nath, S. Generic Paddy Plant Disease Detector (GP2D2): An Application of the Deep-CNN Model. International journal of electrical and computer engineering systems 2023, 14, 647–656. [Google Scholar]
- Ujawe, P.; Gupta, P.; Waghmare, S. Identification of Rice Plant Disease Using Convolution Neural Network Inception V3 and Squeeze Net Models. International Journal of Intelligent Systems and Applications in Engineering 2023, 11, 526–535. [Google Scholar]
- Latif, G.; Abdelhamid, S.E.; Mallouhy, R.E.; Alghazo, J.; Kazimi, Z.A. Deep learning utilization in agriculture: Detection of rice plant diseases using an improved CNN model. Plants 2022, 11, 2230. [Google Scholar] [CrossRef] [PubMed]
- Chavan, T.; Lokhande, D.B.; Patil, D.P. Rice Leaf Disease Detection using Machine Learning. Network 2022, 37, 43. [Google Scholar]
- Tejaswini, P.; Singh, P.; Ramchandani, M.; Rathore, Y.K.; Janghel, R.R. Rice leaf disease classification using CNN. IOP Conference Series: Earth and Environmental Science 2022, 1032, 012017. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Kumar, A. A novel approach for rice plant diseases classification with deep convolutional neural network. International Journal of Information Technology 2022, 1–15. [Google Scholar] [CrossRef]
- Narmadha, R.P.; Sengottaiyan, N.; Kavitha, R.J. Deep Transfer Learning Based Rice Plant Disease Detection Model. Intelligent Automation Soft Computing 2022, 31. [Google Scholar] [CrossRef]
- Shaikh, J.A.; Paradeshi, K.P. Faster R-CNN Based Rice Leaf Disease Detection method. Telematique 2022, 5890–5901. [Google Scholar]
- Prottasha, S.I.; Reza, S.M.S. A classification model based on depthwise separable convolutional neural network to identify rice plant diseases. International Journal of Electrical Computer Engineering 2022, 12, 2088–8708. [Google Scholar] [CrossRef]
- Ritharson, P.I.; Raimond, K.; Mary, X.A.; Robert, J.E.; Andrew, J. DeepRice: A deep learning and deep feature based classification of Rice leaf disease subtypes. Artificial Intelligence in Agriculture 2024, 11, 34–49. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).