Submitted:
18 February 2025
Posted:
19 February 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
Sample Collection and Morphological Study
DNA Extraction, Amplification, and Sequencing
Alignment and Phylogenetic Analysis
Molecular Dating and Diversification Analysis
Reconstruction of Ancestral Morphologies
3. Results
3.1. Phylogenetic Analyses
3.2. Estimation of Divergence Times and Diversification Using the SSU rRNA Gene
3.3. Reconstruction of Ancestral Character State
- Vestibular kineties (VK): Group III retains the ancestral state of three rows of VK, whereas Group I and II show increases to four and five rows, respectively. Notably, F. canadensis shows transitions from three to five and back to three rows, consistent with the ancestral state of the genus Frontonia. Group IV undergoes significant changes, expanding to four, five, and even six rows (Figure 3A).
- Postoral kineties (PK): Group I shows various increases in PK number (six to eight rows), Group II retains the ancestral state PK number except for F. magna, Group III decrease PK number except for F, anatolica, whereas Group IV shows both decreasing and increasing tendencies, with some species reaching up to six rows (Figure 3B).
- Peroral membrane (PM): Groups II and III show an increases in PM ciliary rows, although species such as F. sinica, F. salmastra, and F. lynni revert from two rows to one (Figure 3C).
- Peniculi 1 and 2 (P1 and P2): Group I shows a marked increase from four to five rows, while other groups, with a few exceptions, retain the ancestral state (Figure 3D-E).
- Peniculi 3 evolution: Groups I and II show an increases in ciliary rows from three to four or five, whereas F. mengi and F. sinica uniquely decrease to two rows. Group IV members retain the ancestral state with several species increasing to four or five rows, while Group III decreases from the ancestral three ciliary rows to two. Most Frontonia species retain the ancestral linear structure of peniculi 3, with shortening of peniculi 3 considered an ancestral character (Figure 3F-H).
- Habitat adaptation: The genus Frontonia originates from brackish habitats before adapting to freshwater environments. Group III remains in the brackish environment and adapts to marine habitats for several species, while Group I and IV retain a freshwater adaptation, except for F. acuminata, which adapts to a marine environment. Groups II evolves from freshwater to marine and to brackish habitats (Figure 4).
- Contractile vacuole (CV): The ancestral analysis suggests that the genus originally possessed a single CV, a character often retained. Group III, excluding some species, increases from one to two CVs, while unique species in Group IV show up to 10 CVs per individual (Figure 4).
- Contractile vacuolar pores (CVP) and collecting canals (CC): The evolution of CVPs and CCs is not closely correlated with the evolution of CVs (Figure 4).
4. Discussion
4.1. Evolutionary Patterns of Morphological Characters in the Genus Frontonia
4.2. Evolutionary History of Genus Frontonia
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1
| Species | Accession No. | Origin | Molecular information (Source by publication based on SSU rRNA gene) | Morphological information |
|---|---|---|---|---|
| SSU | ||||
| Frontonia anatolica | MG456578 | Turkey | Kizildang & Yildiz, 2019 | Kizildang & Yildiz, 2019, Yildiz & Senler, 2013 |
| Frontonia anatolica | MG456581 | Turkey | ||
| Frontonia acuminata | MG456579 | Turkey | Kizildang & Yildiz, 2019 | Kizildang & Yildiz, 2019, Foissner, et al., 1994, Kahl, 1931, Pernard, 1922 |
| Frontonia angusta | MG456580 | Turkey | Kizildang & Yildiz, 2019 | Kizildang & Yildiz, 2019, Foissner, et al., 2002, Kahl, 1931, Foissner, et al., 1994 |
| Frontonia canadensis | KJ475313 | Shenzen, China | Zhao et al., 2016 | Pan, X., et al., 2013, Roque. and de Puytorac, 1972. |
| Frontonia canadensis | KJ475311 | Huizhou, China | ||
| Frontonia canadensis | KJ475309 | Huizhou, China | ||
| Frontonia canadensis | KJ475310 | Nansha, China | ||
| Frontonia canadensis | KJ475312 | Hongkang, China | ||
| Frontonia canadensis | FJ868196 | Nansha, China | Fan et al., 2013 | |
| Frontonia cf. acuminata | PV016905 | Gwangju, Korea | present study | present study |
| Frontonia cf. acuminata | PV016906 | Ulsan, Korea | ||
| Frontonia cf. acuminata | PV016907 | Gunsan,Korea | ||
| Frontonia cf. acuminata | PV016908 | Ulsan, Korea | ||
| Frontonia cf. atra | PV016909 | Daejon, Korea | ||
| Frontonia cf. atra | PV016910 | Gunsan, Korea | ||
| Frontonia cf. atra | PV016911 | Masan, Korea | ||
| Frontonia cf. atra | PV016912 | Chungcheongbukdo, Korea | ||
| Frontonia cf. atra | PV016913 | Gyeongsangnamdo, Korea | ||
| Frontonia cf. atra | PV016914 | Ulsan, Korea | ||
| Frontonia cf. atra | PV016917 | Gyeongju, korea | ||
|
Frontonia atra |
MT040844 | Serchio river, Italy | Serra et al., 2021 | Serra et al., 2021, Dragesco & Dragesco-Kerneis, 1986, Foissner, et.al., 1994 |
| Gangneung-si | Omar and Jung, 2021 | |||
| Frontonia didieri | KJ475297 | Qingdao, China | Zhao et al., 2016 | Long et al., 2008 |
| Frontonia didieri | KJ475299 | Qingdao, China | ||
| Frontonia didieri | KJ475298 | Shenzen, China | ||
| Frontonia didieri | DQ885986 | Qingdao, China | Long et al., 2008 | |
|
Frontonia ocularis |
FJ868198 | Guangzhou, China | Pan et al., 2013 | Pan et al., 2013 |
| Gangneung-si | Jung, 2021 | |||
| Frontonia elegans | FJ868200 | Guangzhou, China | Fan et al., 2013 | Fan et al., 2013 |
| Frontonia elegans | KJ475301 | Huizhou, China | Zhao et al., 2016 | |
| Frontonia elegans | KJ475302 | Huizhou, China | ||
| Frontonia elegans | KJ475303 | Huizhou, China | ||
| Frontonia cf. leucas | PV016904 | Gwangju, Korea | present study | present study |
| Frontonia leucas | MG437395 | Turkey | Yildiz & Kizildang , 2017 | Yildiz & Kizildang, 2019 |
| Frontonia leucas | MG437396 | |||
| Frontonia leucas | AM072622 | Italy | Fokin et al., 2006 | Serra et al., 2021, Dragesco & Dragesco-Kerneis, 1986, Foissner, et.al., 1994, Kahl, 1931 |
| Frontonia leucas | KY855558 | Andhra Pradesh, India | Serra et al., 2021 | |
| Frontonia lynni | DQ190463 | Qingdao, China | Long et al., 2008 | Long et al., 2005 |
| Frontonia magna | FJ868199 | Zhuhai, China | Zhao et al., 2016 | Fan et al., 2011 |
| Frontonia magna | FJ876953 | Shenzen, China | Fan et al., 2011 | |
| Frontonia mengi | FJ875141 | Qingdao, China | Fan et al., 2011 | Fan et al., 2011 |
| Frontonia paramagna | KJ475304 | Guangzhou, China | Zhao et al., 2016 | |
| Frontonia paramagna | KJ475306 | Sichuan, China | Zhao et al., 2016 | |
| Frontonia paramagna | JQ868786 | Harbin, China | Chen et al., 2014 | Chen et al., 2014 |
| Frontonia paramagna | KJ475305 | Qingdao, China | Zhao et al., 2016 | |
| Frontonia paramagna | MF279207 | Harbin, China | Cai et al., 2018 | Cai et al., 2018 |
| Frontonia paramagna | KY855554 | Andhra Pradesh, India | Serra et al., 2021 | Serra et al., 2021 |
| Frontonia paramagna | MW031789 | Heilojiang, China | Sun et al.,2021 | Sun et al.,2021 |
| Frontonia shii | MF279208 | Harbin, China | Cai et al., 2018 | Cai et al., 2018 |
| Frontonia sinica | KJ475308 | Qingdao, China | Zhao et al., 2016 | Fan et al., 2013 |
| Frontonia sinica | FJ868197 | Shenzen, China | Fan et al., 2013 | |
| Frontonia subtropica | FJ868202 | Shenzen, China | Fan et al., 2013 | Pan et al., 2013 |
| Frontonia tchibisovae | KJ475318 | Qingdao, China | Zhao et al., 2016 | Long et al., 2008 |
| Frontonia tchibisovae | KJ475316 | Qingdao, China | ||
| Frontonia tchibisovae | KJ475319 | Qingdao, China | ||
| Frontonia tchibisovae | DQ883820 | Yantai, China | Long et al., 2008 | |
| Frontonia terricola | MF926593 | Guangzhou, China | Xu et al., 2018 | Xu et al., 2018, Foissner, 1987, Foissner, et al., 2002 |
| Frontonia pusilla | FJ868201 | Guangzhou, China | Fan et al., 2013 | Fan et al., 2013 |
| Frontonia vernalis | U97110 | UK | Hirt et al., 1997 | |
| Frontonia vernalis | MT040840 | Tusacany, Italy | Serra et al., 2021 | Serra et al., 2021 |
| Frontonia paravernalis | MT040839 | Tusacany, Italy | Serra et al., 2021 | Serra et al., 2021 |
| Frontonia salmastra | MH319376 | Tusacany, Italy | Fokin et al., 2019 | Fokin et al., 2019, Dragesco & Dragesco-Kerneis, 1986 |
| Frontonia vesiculosa | MT040850 | Andhra Pradesh, India | Serra et al., 2021 | Serra et al., 2021, Dragesco & Dragesco-Kerneis, 1986, Kahl, 1932 |
| Frontonia minuta | MT040846 | Serchio river, Italy | Serra et al., 2021 | Serra et al., 2021, Dragesco & Dragesco-Kerneis, 1986 |
| Frontonia fusca | MT040845 | Serchio river, Italy | Serra et al., 2021 | Serra et al., 2021, Fokin, 2008, Kahl, 1932 |
| Frontonia sp. | MT040843 | India | Serra et al., 2021 | Serra et al., 2021 |
| Frontonia sp. | MT040841 | Serchio river, Italy | Serra et al., 2021 | Serra et al., 2021 |
Appendix A.2
| DivBAyes 1.1 | SubT 1.1 | ||
|---|---|---|---|
| Estimated taxaa | Diversification date | Data of taxab | Subtitution taxac |
| 69 species | 450 Mya | 33 species | 36 species |
References
- Long, H.; Song, W.; Gong, J.; Hu, X.; Ma, H.; Zhu, M.; Wang, M. Frontonia Lynni n. Sp., a New Marine Ciliate (Protozoa, Ciliophora, Hymenostomatida) from Qingdao, China. Zootaxa 2005, 57–64. [Google Scholar] [CrossRef]
- Long, H.; Song, W.; Al-Rasheid, K.A.S.; Wang, Y.; Yi, Z.; Al-Quraishy, S.A.; Lin, X.; Al-Farraj, S.A. Taxonomic Studies on Three Marine Species of Frontonia from Northern China: F. Didieri n. Sp., F. Multinucleata n. Sp. and F. Tchibisovae Burkovsky, 1970 (Ciliophora: Peniculida). Zootaxa 2008, 50, 35–50. [Google Scholar] [CrossRef]
- Fan, X.; Chen, X.; Song, W.; Al-Rasheid, K.A.S.; Warren, A. Two Novel Marine Frontonia Species, Frontonia Mengi Spec. Nov. and Frontonia Magna Spec. Nov. (Protozoa; Ciliophora), with Notes on Their Phylogeny Based on Small-Subunit RRNA Gene Sequence Data. Int. J. Syst. Evol. Microbiol. 2011, 61, 1476–1486. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Lin, X.; Liu, W.; Xu, Y.; Al-Farraj, S.A.; Al-Rasheid, K.A.S.; Warren, A. Morphology of Three New Marine Frontonia Species (Ciliophora; Peniculida) with Note on the Phylogeny of This Genus. Eur. J. Protistol. 2013, 49, 312–323. [Google Scholar] [CrossRef]
- Pan, X.; Liu, W.; Yi, Z.; Fan, X.; Al-Rasheid, K.A.S.; Lin, X. Studies on Three Diverse Frontonia Species (Ciliophora, Peniculida), with Brief Notes on 14 Marine or Brackish Congeners. Acta Protozool. 2013, 52, 35–49. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, Y.; Pan, X.; Ding, W.; Al-Rasheid, K.A.S.; Qiu, Z. Morphology and Phylogeny of a New Frontonia Ciliate, F. Paramagna Spec. Nov. (Ciliophora, Peniculida) from Harbin, Ortheast China. Zootaxa 2014, 3827, 375–386. [Google Scholar] [CrossRef]
- Cai, X.; Wang, C.; Pan, X.; El-Serehy, H.A.; Mu, W.; Gao, F.; Qiu, Z. Morphology and Systematics of Two Freshwater Frontonia Species (Ciliophora, Peniculida) from Northeastern China, with Comparisons among the Freshwater Frontonia Spp. Eur. J. Protistol. 2018, 63, 105–116. [Google Scholar] [CrossRef]
- Zhao, Y.; Yi, Z.; Gentekaki, E.; Zhan, A.; Al-Farraj, S.A.; Song, W. Utility of Combining Morphological Characters, Nuclear and Mitochondrial Genes: An Attempt to Resolve the Conflicts of Species Identification for Ciliated Protists Q. Mol. Phylogenet. Evol. 2016, 94, 718–729. [Google Scholar] [CrossRef]
- Kizildag, S.; Yildiz, I. Morphology and Molecular Phylogeny of Four Frontonia Species from Turkey (Protista, Ciliophora). Zootaxa 2019, 4609, 548–564. [Google Scholar] [CrossRef]
- Li, T.; Pan, X.; Lu, B.; Miao, M.; Liu, M. Taxonomy and Molecular Phylogeny of a New Freshwater Ciliate Frontonia Apoacuminata sp. nov. (Protista, Ciliophora, Oligohymenophorea) from Qingdao, PR China. Int. J. Syst. Evol. Microbiol. 2021, 71. [Google Scholar] [CrossRef]
- Serra, V.; D’Alessandro, A.; Nitla, V.; Gammuto, L.; Modeo, L.; Petroni, G.; Fokin, S.I. The Neotypification of Frontonia Vernalis (Ehrenberg, 1833) Ehrenberg, 1838 and the Description of Frontonia Paravernalis sp. nov. Trigger a Critical Revision of Frontoniid Systematics. BMC Zool. 2021, 6, 1–33. [Google Scholar] [CrossRef]
- Sun, M.; Li, Y.; Cai, X.; Liu, Y.; Chen, Y.; Pan, X. Further Insights into the Phylogeny of Peniculid Ciliates (Ciliophora, Oligohymenophorea) Based on Multigene Data. Mol. Phylogenet. Evol. 2021, 154, 107003. [Google Scholar] [CrossRef] [PubMed]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The Characterization of Enzymatically Amplified Eukaryotic 16S-like RRNA-Coding Regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Wang, C.; Pan, X.; El-Serehy, H.A.; Mu, W.; Gao, F.; Qiu, Z. Morphology and Systematics of Two Freshwater Frontonia Species (Ciliophora, Peniculida) from Northeastern China, with Comparisons among the Freshwater Frontonia Spp. Eur. J. Protistol. 2018, 63, 105–116. [Google Scholar] [CrossRef]
- Kim, S.Y.; Choi, J.K.; Dolan, J.R.; Shin, H.C.; Lee, S.; Yang, E.J. Morphological and Ribosomal DNA-Based Characterization of Six Antarctic Ciliate Morphospecies from the Amundsen Sea with Phylogenetic Analyses. J. Eukaryot. Microbiol. 2013, 60, 497–513. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Posada, D. JModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More Models, New Heuristics and High-Performance Computing Europe PMC Funders Group. Nat. Methods 2015, 9, 772. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian Evolutionary Analysis by Sampling Trees. BMC Evol. Biol. 2007, 7, 1–8. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Schönborn, W.; Dörfelt, H.; Foissner, W.; Krienitz, L.; Schäfer, U. A Fossilized Microcenosis in Triassic Amber. J. Eukaryot. Microbiol. 1999, 46, 571–584. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Ryberg, M.; Nilsson, R.H.; Brandon Matheny, P. Divbayes and SubT: Exploring Species Diversification Using Bayesian Statistics. Bioinformatics 2011, 27, 2439–2440. [Google Scholar] [CrossRef] [PubMed]
- Maddison, W.P. and D.R.M. Mesquite: A Modular System for Evolutionary Analysis. Version 3.61. http://www.mesquiteproject.org 2019.
- Miller, M.A., Pfeiffer, W. and Schwartz, T. No TitleThe CIPRES Science Gateway: A Community Resource for Phylogenetic Analyses. Proc. 2011 TeraGrid Conf. Extrem. Digit. Discov. (pp. 1-8).
- Ho, S.Y.W.; Duchêne, S. Molecular-Clock Methods for Estimating Evolutionary Rates and Timescales. Mol. Ecol. 2014, 23, 5947–5965. [Google Scholar] [CrossRef]
- Rajter, Ľ.; Vďačný, P. Rapid Radiation, Gradual Extinction and Parallel Evolution Challenge Generic Classification of Spathidiid Ciliates (Protista, Ciliophora). Zool. Scr. 2016, 45, 200–223. [Google Scholar] [CrossRef]
- Vďačný, P. Estimation of Divergence Times in Litostomatean Ciliates (Ciliophora: Intramacronucleata), Using Bayesian Relaxed Clock and 18S RRNA Gene. Eur. J. Protistol. 2015, 51, 321–334. [Google Scholar] [CrossRef]
- Vďačný, P.; Breiner, H.-W.; Yashchenko, V.; Dunthorn, M.; Stoeck, T.; Foissner, W. The Chaos Prevails: Molecular Phylogeny of the Haptoria (Ciliophora, Litostomatea). Protist 2014, 165, 93–111. [Google Scholar] [CrossRef]
- Cai, X.; Wang, C.; Pan, X.; El-Serehy, H.A.; Mu, W.; Gao, F.; Qiu, Z. Morphology and Systematics of Two Freshwater Frontonia Species (Ciliophora, Peniculida) from Northeastern China, with Comparisons among the Freshwater Frontonia Spp. Eur. J. Protistol. 2018, 63, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Gao, F.; Liu, W.; Fan, X.; Warren, A.; Song, W. Morphology and SSU RRNA Gene Sequences of Three Frontonia Species, Including a Description of F. Subtropica Spec. Nov. (Ciliophora, Peniculida). Eur. J. Protistol. 2013, 49, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Clamp, J.; Xu, D.; Huang, B.; Shin, M.K. An Integrative Approach to Phylogeny Reveals Patterns of Environmental Distribution and Novel Evolutionary Relationships in a Major Group of Ciliates. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Syberg-Olsen, M.J.; Irwin, N.A.T.; Vannini, C.; Erra, F.; Di Giuseppe, G.; Boscaro, V.; Keeling, P.J. Biogeography and Character Evolution of the Ciliate Genus Euplotes (Spirotrichea, Euplotia), with Description of Euplotes Curdsi Sp. Nov. PLoS One 2016, 11, 1–18. [Google Scholar] [CrossRef]
- Zhao, Y.; Yi, Z.; Warren, A.; Song, W.B. Species Delimitation for the Molecular Taxonomy and Ecology of the Widely Distributed Microbial Eukaryote Genus Euplotes (Alveolata, Ciliophora). Proc. R. Soc. B Biol. Sci. 2018, 285. [Google Scholar] [CrossRef]
- Lahr, D.J.G.; Laughinghouse, H.D.; Oliverio, A.M.; Gao, F.; Katz, L.A. How Discordant Morphological and Molecular Evolution among Microorganisms Can Revise Our Notions of Biodiversity on Earth. BioEssays 2014, 36, 950–959. [Google Scholar] [CrossRef] [PubMed]
- Babcock, L.E.; Peng, S.C.; Brett, C.E.; Zhu, M.Y.; Ahlberg, P.; Bevis, M.; Robison, R.A. Global Climate, Sea Level Cycles, and Biotic Events in the Cambrian Period. Palaeoworld 2015, 24, 5–15. [Google Scholar] [CrossRef]
- Sepkoski, J.J. Environmental Trends in Extinction During the Paleozoic. American Association for the Advancement of Science Stable 2010, 235, 64–66. [Google Scholar]
- Stanley, E.L.; Bauer, A.M.; Jackman, T.R.; Branch, W.R.; Mouton, P.L.F.N. Between a Rock and a Hard Polytomy: Rapid Radiation in the Rupicolous Girdled Lizards (Squamata: Cordylidae). Mol. Phylogenet. Evol. 2011, 58, 53–70. [Google Scholar] [CrossRef]
- Jenkyns, Hugh, C. Evidence for Rapid Climate Change in the Mesozoic-Palaeogene Greenhouse World. Philosophical Transactions of the Royal Society of London. Series A: Mathematical, Physical and Engineering Sciences 2003, 361, 1885–1916. [CrossRef]
- Price, G.D.; Twitchett, R.J.; Wheeley, J.R.; Buono, G. Isotopic Evidence for Long Term Warmth in the Mesozoic. Sci. Rep. 2013, 3, 1–5. [Google Scholar] [CrossRef] [PubMed]
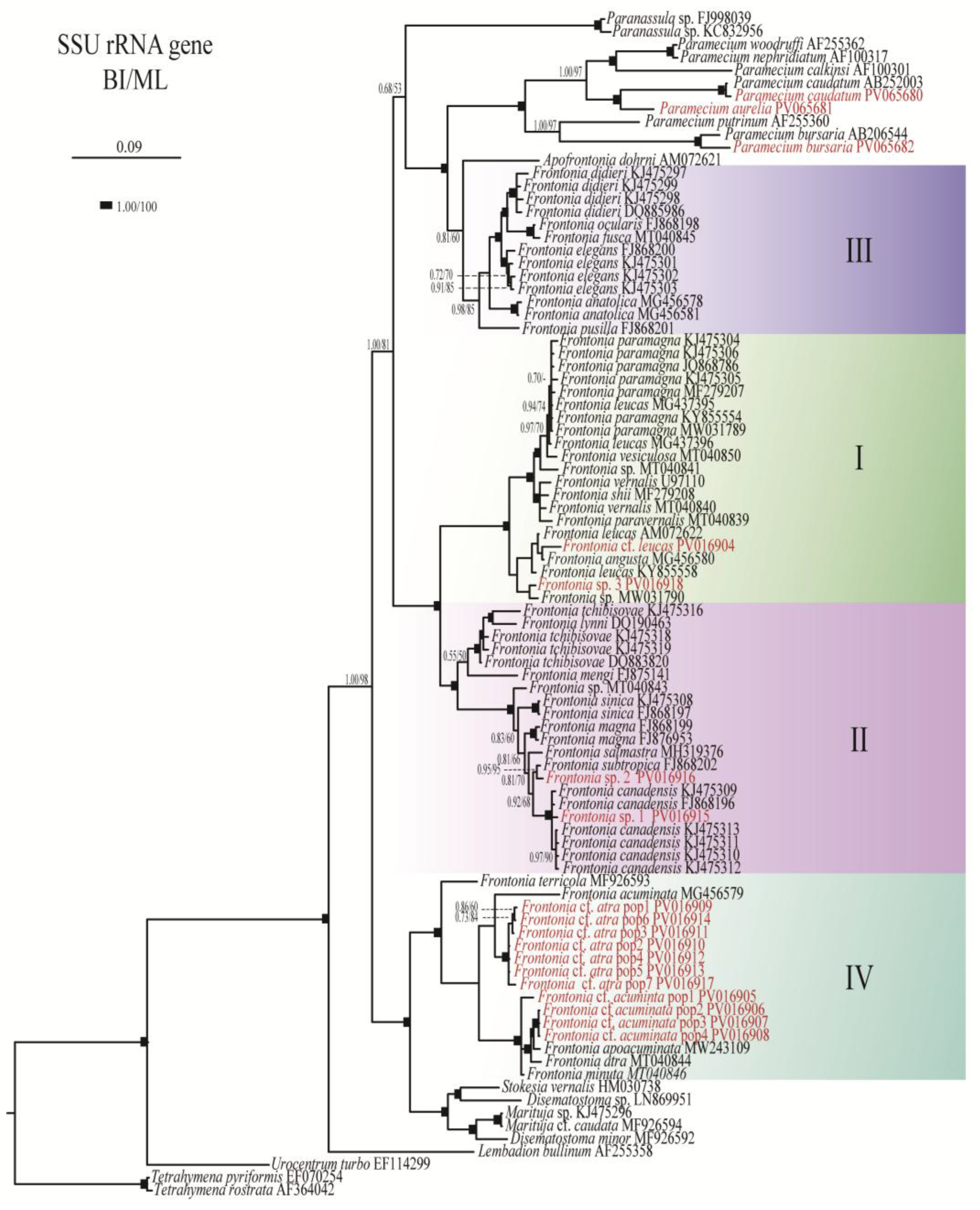
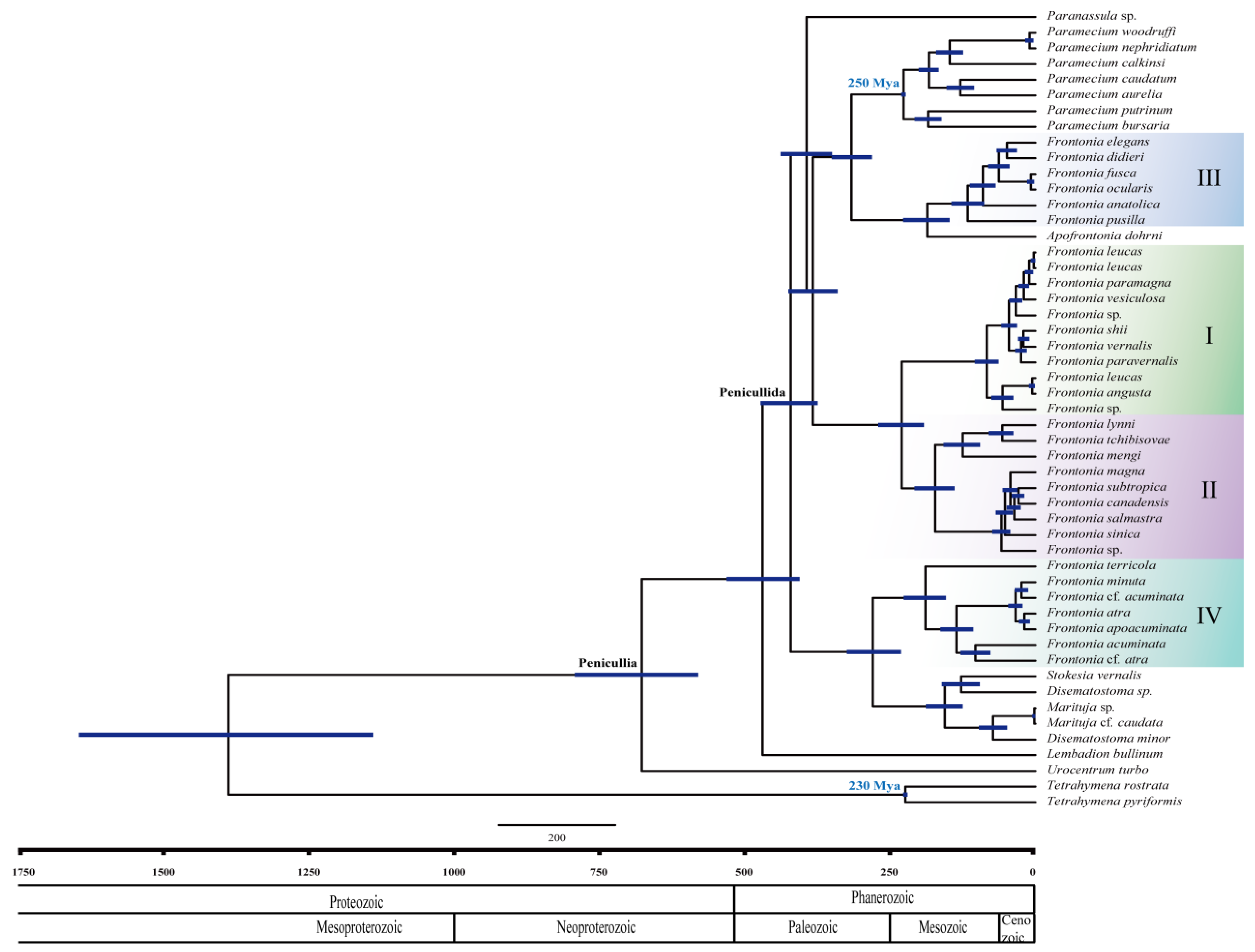
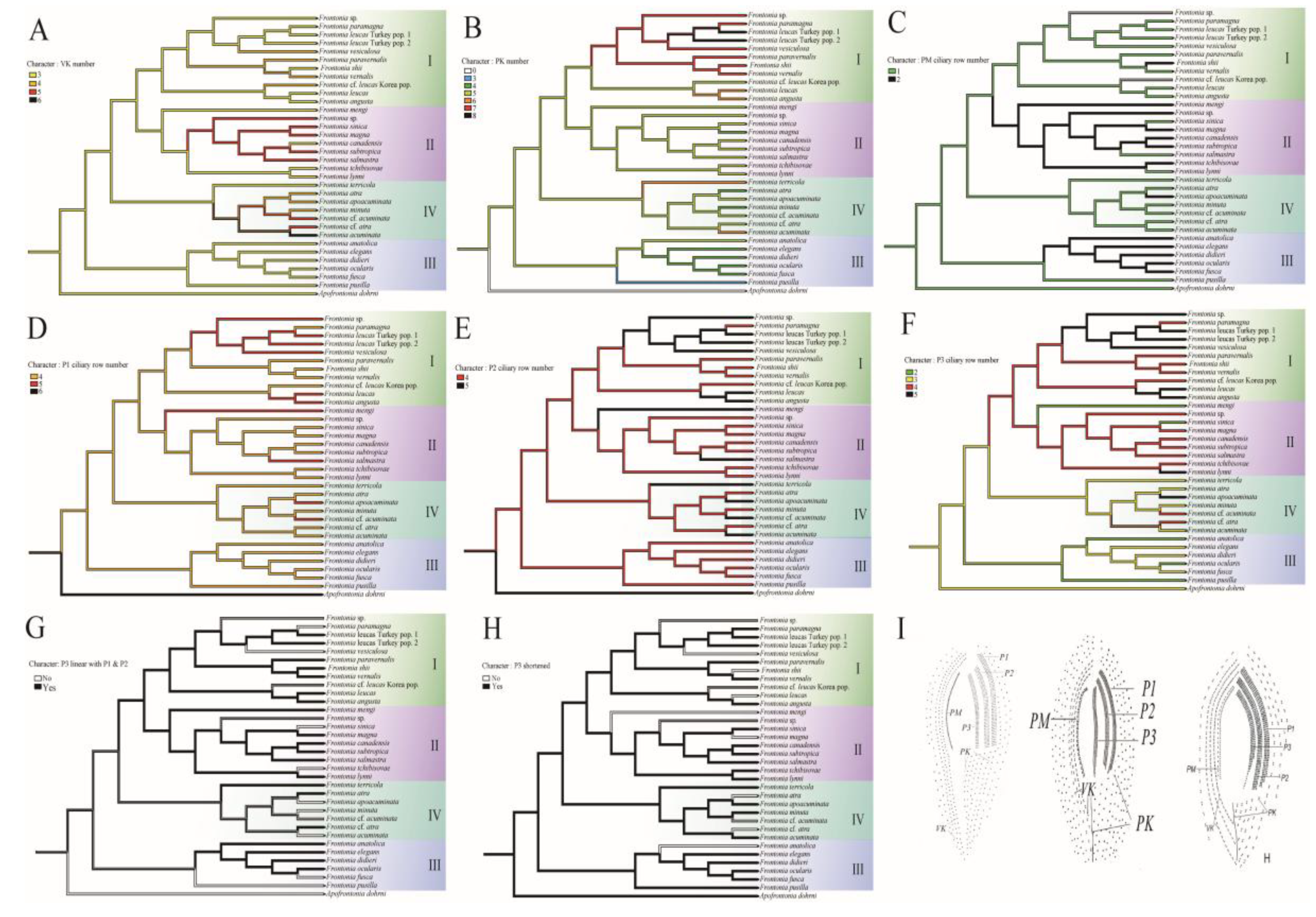
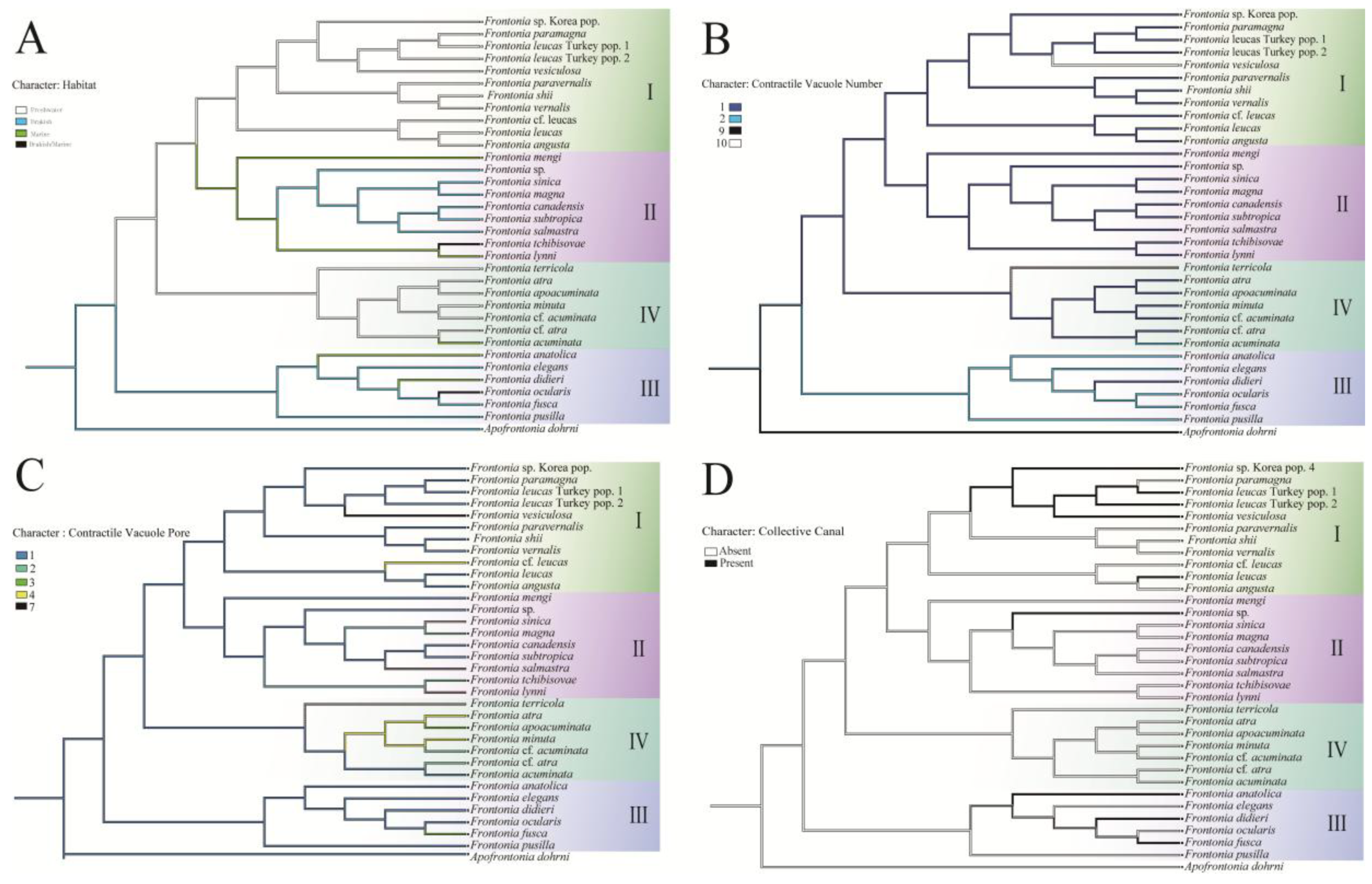
| Taxon | Collection site | accession no. |
|---|---|---|
| Frontonia cf. acuminata pop1 | Gwangju, Korea | PV016905 |
| Frontonia cf. acuminata pop2 | Ulsan, Korea | PV016906 |
| Frontonia cf. acuminata pop3 | Gunsan, Korea | PV016907 |
| Frontonia cf. acuminata pop4 | Ulsan, Korea | PV016908 |
| Frontonia cf. atra pop1 | Daejon, Korea | PV016909 |
| Frontonia cf. atra pop2 | Gunsan, Korea | PV016910 |
| Frontonia cf. atra pop3 | Masan, Korea | PV016911 |
| Frontonia cf. atra pop4 | Chungcheongbukdo, Korea | PV016912 |
| Frontonia cf. atra pop5 | Gyeongsangnamdo, Korea | PV016913 |
| Frontonia cf. atra pop6 | Ulsan, Korea | PV016914 |
| Frontonia cf. atra pop7 | Gyeongju, Korea | PV016917 |
| Frontonia cf. leucas | Gwangju, Korea | PV016904 |
| Frontonia sp. 1 | Ulsan, Korea | PV016915 |
| Frontonia sp. 2 | Ulsan, Korea | PV016916 |
| Frontonia sp. 3 | Gyeongju, Korea | PV016918 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





