1. Introduction
Nile tilapia
Oreochromis niloticus (Linnaeus, 1758) global production has increased over the last thirty years due to the increasing demand for animal protein [
1,
2]. In Kenya, this species accounts for 80% of total aquaculture production and gained popularity in fish farming, including regions like Central Kenya, where fish consumption is not traditionally common [
3,
4,
5]. Due to the increase in inland aquaculture practices, fish disease outbreaks, mortality, and higher parasite burdens are highly likely to occur [
6], prompting attention from researchers and fisheries stakeholders. In Kenya, fish parasitological work is steadily growing; approximately 119 species of fish parasites have been reported, with only 83 identified at the species level [
7].
Clinostomidae Lühe, 1901, comprises digenetic trematodes with a heteroxenous life cycle that involves multiple hosts. The adult stages are commonly found in fish-eating birds' buccal cavities and oesophagus as definitive hosts [
8,
9]. Their life cycle begins when birds release eggs into aquatic environments, which subsequently hatch into free-swimming miracidia. Miracidia infect freshwater snails as first intermediate hosts and various fish species as second intermediate hosts harbouring the metacercarial stages [
10]. Although rarely infected, humans and mammals have occasionally been reported as accidental hosts of representatives of Clinostomidae. [
11,
12]. Due to the increased number of reports of
Clinostomum Leidy, 1856, in aquaculture conditions, it has recently been the subject of many studies. Advances in genetic research have expanded the
Clinostomum species catalogue, addressing the challenges posed by relying solely on morphological characteristics, which often show high similarity and minimal variation between species. These genetic tools, therefore, allow the identification of previously unrecognized species and enable linking larval stages to their corresponding adult forms [
9,
13,
14,
15,
16,
17].
Despite more than 50 species being reported worldwide as members of
Clinostomum, only 15 species are considered valid to date [
9,
13,
18,
19]. The diversity of
Clinostomum species in the Afrotropical region, on the other hand, is insufficiently explored, with only four species currently recognized:
C. cutaneum Paperna, 1964,
C. phalacrocoracis Dubois,1931,
C. tilapiae Ukoli, 1966 and
C. ukolii Caffara, Locke, Echi, Halajian, Luus-Powell, Benini, Tedesco & Fioravanti, 2020 [
15]. Similarly, the limited understanding of the effects of these parasites on Nile tilapia and the conditions facilitating their emergence makes it difficult for aquaculture stakeholders to access valuable information for informed decision-making to support aquaculture sustainability [
20]. It is interesting to look at the parasitic fauna infecting fish in the Central region of Kenya, as most inland aquaculture production occurs there [
18]. For this reason, we conducted a survey in the Upper Tana River region with the purpose of assessing parasite diversity infecting Nile tilapia reared in fish farms. Regarding clinostomid infections, three species of
Clinostomum:
C. cutaneum,
C. phalacrocoracis and
C. tilapiae, have been reported previously in Nile tilapia in Kenya [
13,
21]. The present study identified the metacercariae using light microscopy, scanning electron microscopy (SEM) and molecular methods. For scanning electron microscopy, surface observations have revealed new ultrastructural features important for the taxonomy and systematics of a wide range of organisms [
22]. The findings in this study contribute to resolving taxonomic ambiguities within
Clinostomum. In particular, we present additional features in
C. cutaneum that were not seen in earlier studies, further enhancing the accuracy of species differentiation within this genus.
2. Materials and Methods
2.1. Study Area and Sample Collection
The Upper Tana River region covers around 15,000 km
2 and is characterized by the highest precipitation rates in Kenya, with a humid or semi-humid climate year-round [
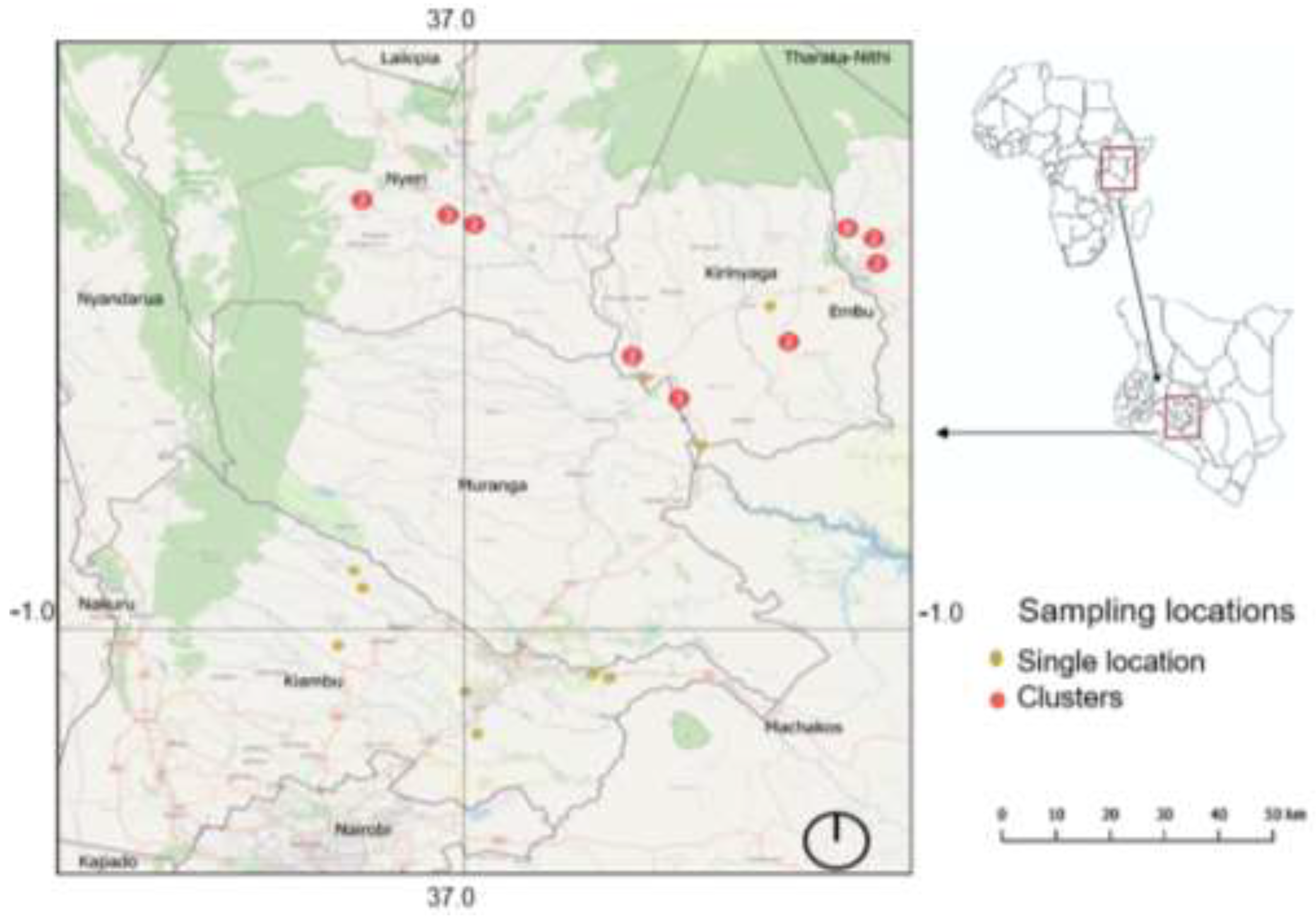
23]. Between mid-January and mid-February 2024, we collected 157 Nile tilapia specimens from fish farms in this area after getting a research permit from the National Commission for Science, Technology and Innovation (NACOSTI), permit: NACOSTI/P/23/31261. The fish hosts were then sacrificed by cervical dislocation, and a fin clip of each specimen preserved in absolute ethanol was deposited at the Royal Belgian Institute of Natural Sciences, Belgium (XXX-XXX). The map showing the sampling localities (
Figure 1) was created using QGIS v3.38.3 (QGIS Development Team 2022, QGIS Information System, Open Source Geospatial Foundation Project.
http://qgis.osgeo.org, accessed on 10 June 2024).
2.2. Parasitological Examination
The external surfaces and internal body organs of fish were carefully inspected using the naked eye and a stereomicroscope to detect encysted metacercariae. The encysted metacercariae were carefully excised using a fine needle, relaxed with boiling water, and preserved in 70% ethanol for further morphological processing. Additional specimens were preserved in absolute ethanol for subsequent molecular analysis. The infection parameters, i.e., prevalence (P) and mean intensity (M.I) were calculated according to Bush et al. [
24].
2.3. Morphological Identification
For morphological identification, we only used a subset of the individuals isolated as some were too small to excise and use or too big to mount on slides. For this, ten specimens were stained with Borax carmine, dehydrated in a graded ethanol series for 30 minutes: 30%, 50%, 70% (three times), 80%, 95%, 100% (three times), cleared with Amman’s lactophenol and mounted on permanent slides using Euparal. The metacercariae were viewed using a Leica DM2500 optical microscope fitted with a Leica DMC4500 camera. The voucher specimens were deposited in the collection of the Research Group Zoology: Biodiversity and Toxicology at Hasselt University (HU) (Diepenbeek, Belgium) (HU XXIII.2.02-2.22).
2.4. Scanning Electron Microscopy
Ten specimens were prepared for scanning electron microscopy. The specimens were post-fixed in 4% osmium tetroxide (OsO4), thoroughly washed in distilled water to remove excess osmium, and dehydrated in graded ethanol series for 30 minutes each: 30%, 50%, 70% (three times), 80%, 95% and 100% (three times). They were then dried using hexamethyldisilazane under a fume hood overnight. Afterwards, they were mounted on aluminium SEM stubs with double adhesive tape and gold coated at 30mA using JEOL JFC-1300 sputter coater. Imaging was done with a Phenom XL G2 Desktop Scanning Electron Microscope (ThermoFisher Scientific) at an accelerating voltage of 5 kV.
2.5. DNA Extraction, Amplification and Sequencing
Sequences of the nuclear gene portions from internal transcribed spacer 1 (ITS1), 5.8S, ITS2, 28S rDNA and mitochondrial cytochrome
c oxidase subunit 1 (COI mtDNA) were obtained using polymerase chain reactions (PCR). These nuclear ribosomal markers evolve at different rates, which makes them suitable for assessing genetic divergence at the interspecific level [
25,
26]. The COI mtDNA, on the other hand, is a promising resource for assessing intraspecific genetic differentiation because it is a fast-evolving marker compared to nuclear rDNA [
27].
The posterior end of the metacercariae was cut, and DNA extraction was done using the protocol adapted by Kmentová et al. [
28]. Samples stored in 99% ethanol were spun down, ethanol removed and left to dry for 30 minutes. 195 µl of TNES buffer (400 mM NaCl, 20 mM EDTA, 50 mM Tris pH 8, 0.5% SDS) and 5 µL of Thermo ScientificTM proteinase
K (20 mg/mL) were added to the samples. After incubation at 55°C overnight, 2 µL of Invitrogen
TM yeast tRNA (10 mg/mL) was added as a carrier and briefly spun down before adding 65 µL of 5 M NaCl and 290 µL of 96% ethanol. The samples were cooled for 60 min at -20°C, then spun down for 15 mins at 18000 rcf to a small white pellet. The supernatant was removed and replaced with 1 mL of chilled 70% ethanol. The samples were centrifuged for 8 minutes at 18000 rcf (this ethanol rinsing step, removing the supernatant, adding ethanol, and centrifuging were repeated once). The supernatant was removed, and the DNA was eluted in 30 µL of 0.1 x TE buffer (0.02% Thermo Scientific
TM Tween-20 washing buffer). The DNA extract was placed overnight at 4°C for resuspension and stored at -20°C.
Partial ITS1, 5.8S, ITS2 and 28S regions were amplified using forward primer NC5 (5’- GTA GGT GAA CCT GCG GAA GGA TCA TT -3’) [
26] and reverse primer NC2 (5’-TTA GTT TCT TTT CCT CCG CT -3’) [
29]. The PCR reaction was performed using MangoMix™. For each 2 μL of DNA extract, 12.50 μL of MangoMix™, 0.50 μL of MgCl
2 (1 mM), 1.25 μL of the forward and reverse primers (0.5 μM), respectively, and 7.50 μL of ddH2O was added, adding up to a total of 25 μL per reaction. The PCR amplification occurred under the following conditions: initial denaturation of 2 min at 94°C, 39 cycles of 1 min at 94°C, 1 min at 52°C, and 1:30 min at 72°C, final elongation of 7 min at 72°C and cooling to 4°C.
Part of the mitochondrial COI gene was amplified using forward ASmit1 (5′- TTT TTT GGG CAT CCT GAG GTT TAT -3′) and reverse Asmit2 (5′-TAA AGA AAG AAC ATA ATG AAA ATG -3′) primers both widely used for digeneans and other flatworms [
30]. For each two μL of DNA extract, 12.50 μL of MangoMix™, 0.50 μL of MgCl
2 (1 mM), 1.25 μL of the forward and reverse primers (0.5 μM), respectively, and 7.50 μL of ddH2O was added, adding up to a total of 25 μL per reaction. The PCR conditions were set as follows: 2 min initial denaturation at 94°C, 37 cycles of 30 sec at 94°C, 40 sec at 48°C, and 50 sec at 72°C, final elongation of 5 min at 72°C and cooling to 4°C. Gel electrophoresis was used to check for successful amplification. The PCR products were excised and purified using GeneJet Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s guidelines. The corresponding primer pairs used for amplification were used for subsequent Sanger sequencing in Macrogen (Netherlands).
2.6. Sequence Analysis
We verified our morphological identification of the sequences generated in this study by BLASTing [
31] the obtained sequences on the NCBI website, which allowed us to identify the closest congeners. Multiple sequence alignments were constructed using MUSCLE v5 [
32] under default parameters, and a maximum-likelihood-based model selection was performed in Molecular Evolutionary Genetics Analysis (MEGA) v11.0.13 [
33]. After choosing a model in MEGA based on the Bayesian Information Criterion, we selected the highest-ranked model available in this software to calculate pairwise distances. As a result, we used the Kimura 2-parameter model [
34] for the ITS sequences and the Tamura-Nei model [
35] for the COI sequences. A haplotype genealogy graph was constructed using Fitchi [
36], using all available sequences of the closest congeneric species. All sequences have been submitted to GenBank, and the corresponding accession numbers will be provided upon acceptance.
3. Results
3.1. Infection Parameters and Metacercariae Isolated
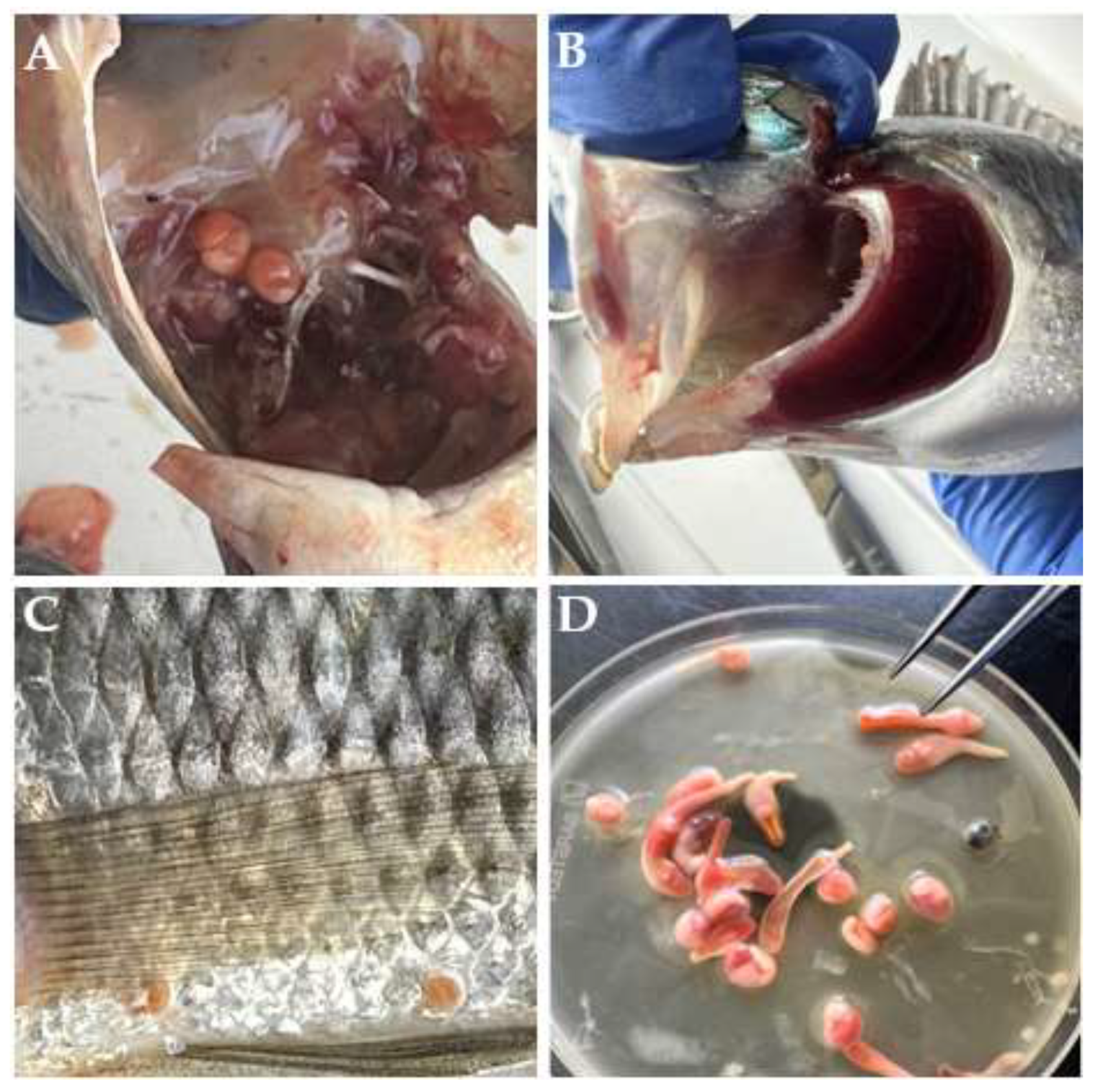
Among the 157 examined hosts, 27 individuals were found to be infected with metacercariae (
Figure 2D), using a subset of the isolated individuals, we identified them to belong to a total of four species: three species of
Clinostomum (
C. cutaneum, C. phalacrocoracis and
C. tilapiae) and one species of
Euclinostomum (
E. heterostomum). The prevalence of infection was 17.2% (27/157), while the mean intensity was 7.3. The minimum number of metacercariae collected from each host was one, while the highest was 38. Mixed infections involving
C. cutaneum and
C. phalacrocoracis,
C. phalacrocoracis and
C. tilapiae, and
C. cutaneum and
E. heterostomum were observed. Most of the clinostomid metacercariae were recovered from the skin (infection frequency = 0.54) of the infected hosts (
Figure 2C), followed by the buccal cavity (infection frequency = 0.45) (
Figure 2A) and occasionally from the gills (infection frequency = 0.01) (
Figure 2B), as shown in
Figure 2.
3.2. Morphological Observations in C. Cutaneum
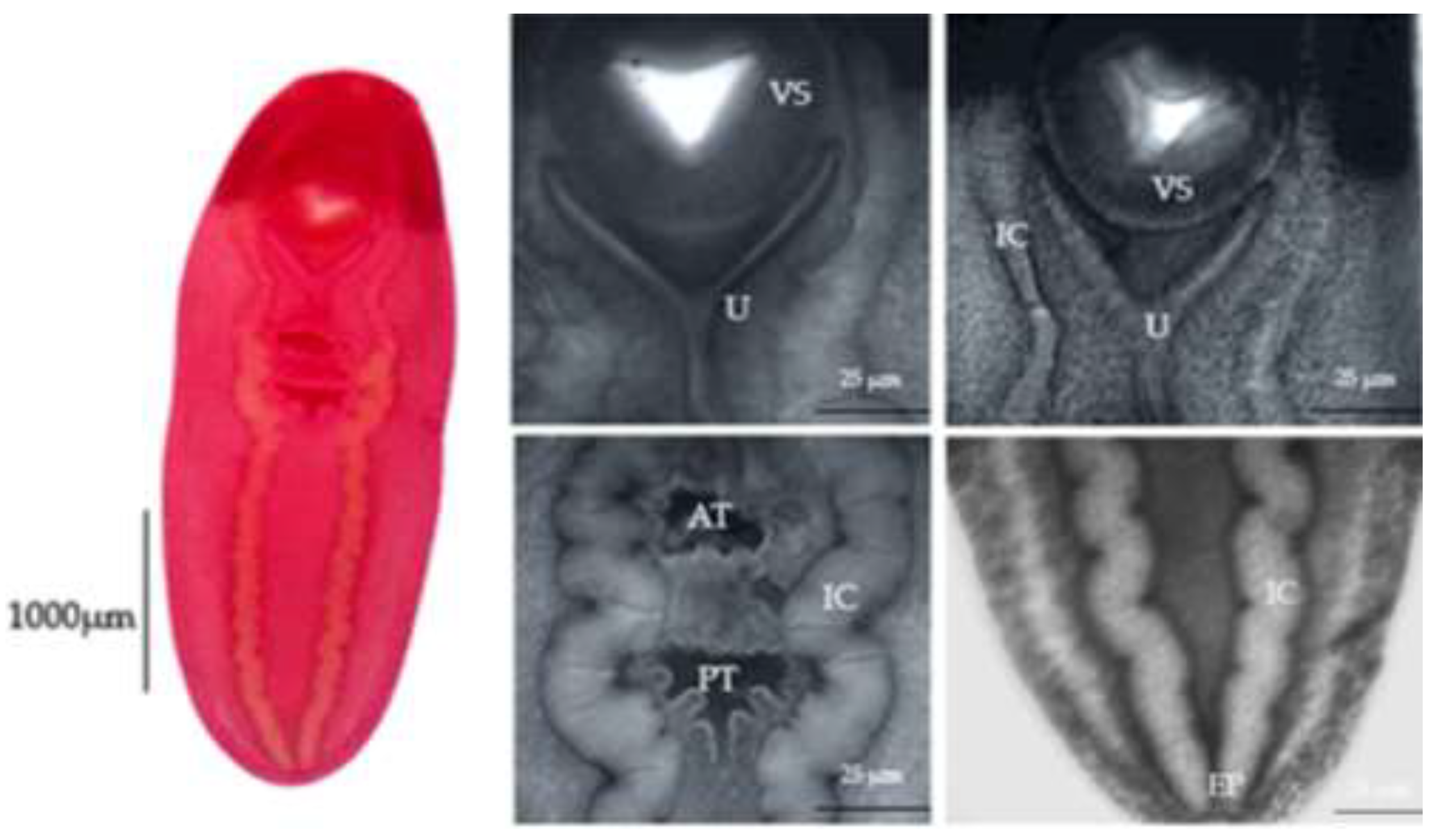
The morphological features of eight of our specimens are consistent with the characterization of
C. cutaneum previously reported by Gustinelli et al. [
9]. The key observations include a distinct Y-shaped uterus, intestinal caeca extending laterally from the anterior to the posterior body ends, and a smaller anterior testis than the posterior testis (
Figure 3).
3.3. Scanning Electron Microscopy Results
Scanning electron microscopy of
C. cutaneum in this study revealed several novel features. The excretory pore (
Figure 4E, 4F), previously undetected in the work of Gustinelli et al. [
9] due to a possible cuticular fold, was clearly visible and surrounded by minute, spiny papillae (white arrow in
Figure 4E). Additionally, an everted cirrus (
Figure 4C) was observed. In the adult stages of
C. cutaneum, Gustinelli et al. [
9] observed basal papillae in the cirrus. However, the cirrus observed in our study lacked basal papillae (encircled in red), highlighting morphological differences between the adult and metacercarial stages. The tegumental area around the genital pore (
Figure 4D) was also surrounded by dome-shaped papillae (white asterisks) never reported before, further enriching the morphological characterization of this species.
3.4. Molecular Analyses
Thirteen out of 15 partial ITS1-5.8S-ITS2 sequences were obtained in this study, ranging in length from 1043 to 1102 bp. These sequences included 554–570 bp corresponding to ITS1, 157 bp to 5.8S and 290–328 bp to ITS2. Eight newly generated sequences (GenBank accession numbers XXX-XXX) showed an average similarity of 99.80% (range: 99.88–-100%) to four published sequences of
C. cutaneum (
Table 1) published by [
9,
14]. Three sequences (GenBank accession numbers XXX-XXX) averaged 99.9% (range: 99.94-100%) to six sequences of
C. phalacrocoracis published by [
9,
14,
37] (KP110567, KP110567, FJ609422-23, KJ786975-76). One sequence (GenBank accession number XXX) averaged 99.7% (range: 99.59-99.7%) to eight sequences of
C. tilapiae published by [
15] (KY649349-55), while one sequence (GenBank accession number XXX) averaged 99.9% (range: 99.16-100%) to 11 sequences of
E. heterostomum published by [
38] (KP721422-25, KP721427, KP721430-31, KP721435, KP721437-39).
A total of 43 sequences of congeners, all identified to species level, were obtained from GenBank (
Table 1) for further molecular analyses.
We provide the intraspecific and interspecific differences in ITS sequences in
Table 2.
The percentage identity of the eight COI mtDNA sequences (579 bp) obtained in this study was not comparable to other sequences in GenBank using BLAST. This is because the primers used in this study [
30] amplified a region different from the published sequences for clinostomids (positions 1–688bp in COI mtDNA gene). The sequences obtained in this study overlapped the region between 699–1287 bp in the COI mtDNA gene from the mitogenome of
Clinostomum complanatum (OP681143) [
39]. The pairwise distances of COI mtDNA generated in this study are shown in
Table 3.
Based on the values provided in
Table 3, the isolates from the same species showed very low genetic distances ( ≤0.009), indicating high similarity within the species. In contrast, comparisons between different species showed much greater distances, with values ranging from 0.081 to 0.143. The smallest interspecific distance (0.081) observed between
C. tilapiae and
C. phalacrocoracis suggests a closer genetic relationship compared to their relationship with
C. cutaneum.
For the haplotype networks of the 807 bp region covering the ITS1, 5.8S and ITS2 regions, we used the minimum spanning tree model [
40] to visualize the genetic structure in the different populations and species (
Figure 5). The haplotype of
C. cutaneum (yellow) is shared with those previously described in Kenya. Similarly,
C. phalacrocoracis shared the same haplotype with Kenyan (blue) and Israeli (light blue) populations, suggesting gene flow or dispersal between these regions. The
C. tilapiae haplotype (pink) is shared with the Nigerian haplotype. For
E. heterostomum (green), one haplotype is shared with published sequences from Israeli populations. However, three additional haplotypes are observed, suggesting intraspecific genetic diversity within this species despite all isolates being collected from the same region in Israel.
4. Discussion
To date, 15 species of
Clinostomum have been identified using a combination of molecular and morphological methods [
18], which provide a reliable basis for diagnosis within this genus [
9,
13,
15,
41]. As part of a comprehensive study aimed at surveying the diversity of parasites infecting Nile tilapia in the Upper Tana River region in Kenya, we identified the isolated metacercariae as
C. cutaneum, C. phalacrocoracis, C. tilapiae and
E. heterostomum. The first three species have been previously reported infecting Nile tilapia and grey herons in Kenya. The report of
C. tilapiae in this region lacked supporting morphological or molecular data. This study provides so far lacking molecular data for
C. tilapiae and
E. heterostomum in Kenya, but since only one specimen was available for each species, no morphological descriptions were possible. For this reason, more work still needs to be done to properly characterize the morphology of
E. heterostomum and
C. tilapiae in
O. niloticus (a cichlid host), with the latter parasite species having its previous report with genetic data from the mochokid catfish
Synodontis batensoda Rüppell, 1832 in Nigeria.
Regarding infection parameters, a high prevalence of
Clinostomum spp. was observed in cichlids in Lake Kinneret, Israel (23.4%) [
37], in cultured
O. niloticus in Sahary fish hatchery, Egypt (25%), in wild
Sarotherodon galilaeus (Linnaeus, 1758) in Lake Nasser, Egypt (33%) [
42] and our study (17.2%). Similarly, research by Mahdy et al. [
43] found that farmed fish had a higher infection rate (32%) than wild tilapia (24%), suggesting that
Clinostomum infections may be widespread in aquaculture systems. Higher parasite burdens in farmed fish can lead to reduced market value and increased mortality, particularly in cases where heavy infestations cause damage to fish tissues, gills and skin [
16,
42,
44]. The ability of metacercariae to impair host health through secondary infections from bacteria or fungi, ultimately resulting in death, has been documented [
42,
45,
46], reinforcing the need for proactive parasite management strategies in tilapia farming.
While SEM helps visualize detailed structural characteristics of the tegument and surface morphology that are otherwise missed with light microscopy, it has been only applied to some species within
Clinostomum. Compared to the surface ultrastructure of
C. ukolii and
C. tilapiae, whose teguments are completely covered with minute spines, the tegument of
C. cutaneum lacks spines. Similarly, dome-like structures were observed only on the genital pore of
C. cutaneum, whereas in
C. ukolii, such structures are present across the tegumental surface and sometimes between suckers [
17]. In the present study on
C. cutaneum, we observed additional diagnostic features that were previously missed by Gustinelli et al. [
9].
5. Conclusions
This study provides new diagnostic features for the metacercariae of C. cutaneum, refining its morphological characterization. It also provides the first molecular data for C. tilapiae, reports the first species occurrence for E. heterostomum in Kenya, and confirms its presence on Nile tilapia in the country. Additionally, we offer a molecular barcoding resource using COI mtDNA sequences, with our selected markers amplifying a previously unsequenced region of the mitochondrial genome. The high prevalence of Clinostomum spp. shows their potential impact on both fish and human health, emphasizing the need for continued monitoring. Finally, we demonstrate the value of SEM as a complementary tool for more precise parasite species identification.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org, Figure S1: Phylogeny based on maximum likelihood method using ITS1-5.8S-ITS2 rDNA generated using Geneious Prime 2025.0 (
https://www.geneious.com). Node support values include SH-aLRT and UFBoot (1000 Ultrafast bootstraps). Black dots indicate strong support (SH-aLRT > 80, UBoost > 95) across analyses. The scale bar represents the number of expected substitutions per site.
Author Contributions
Conceptualization, M.I.S.; methodology, M.I.S., M.T.; software, M.I.S and K.T.; validation, M.I.S, M.P.M.V, N.K. and K.T.; formal analysis, M.I.S., K.T., and N.K.; investigation, M.I.S., D.M., M.T.; resources, M.I.S., D.M.; data curation, M.I.S., K.T.; writing—original draft preparation, M.I.S; writing—review and editing, M.I.S., N.K., and M.P.M.V.; visualization, M.I.S., and K.T.; supervision, M.P.M.V.; project administration, M.I.S., M.P.M.V.; funding acquisition, M.I.S., M.P.M.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Special Research Fund (BOF) of Hasselt University, grant number BOF22DOCLI04 and Hasselt University International and Intersectoral mobility funding - 2024 to M.I.S. This work was supported by the AfroWetMaP project of the Belgian Federal Science Policy Office (4255-FED-tWIN-G3 program, Prf-2022-049).
Institutional Review Board Statement
The Ethical Committee for Animal Experimentation of Hasselt University, Belgium, does not require ethical clearance when animals are euthanized abroad. Fishes were dissected in the field in Kenya. Under Kenyan legislation, no ethical approval for this type of research was required.
Data Availability Statement
Voucher materials from this study have been deposited in the Zoology: Biodiversity and Toxicology collection at Hasselt University (Diepenbeek, Belgium) (catalogue number HU XXIII2.02-XXIII2.22). Additionally, the genetic sequences generated in this study have been made publicly available on GenBank under accession numbers XXX-XXX.
Acknowledgements
The authors sincerely thank Ralph Appy (Cabrillo Marine Aquarium, California, USA) for the great help in scanning electron microscopy and Edward Njagi, Gilbert Kosgei, Gathua Joseph and Justus Ochong’ (Ichthyology section, National Museums of Kenya) for field equipment supply and storage of tissue samples.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Béné, C.; Barange, M.; Subasinghe, R.; Pinstrup-Andersen, P.; Merino, G.; Hemre, G.I.; Williams, M. Feeding 9 Billion by 2050 – Putting Fish Back on the Menu. Food Secur. 2015, 7, 261–274. [Google Scholar] [CrossRef]
- Béné, C.; Arthur, R.; Norbury, H.; Allison, E.H.; Beveridge, M.; Bush, S.; Campling, L.; Leschen, W.; Little, D.; Squires, D.; et al. Contribution of Fisheries and Aquaculture to Food Security and Poverty Reduction: Assessing the Current Evidence. World Dev. 2016, 79, 177–196. [Google Scholar] [CrossRef]
- Munguti, J.; Obiero, K.; Orina, P.; Mirera, D.; Kyule, D.; Mwaluma, J.; Opiyo, M.; Musa, S.; Ochiewo, J.; Njiru, J.; et al. State of Aquaculture Report 2021: Towards Nutrition Sensitive Fish Food Production Systems.
- KMFRI Kenya’s Aquaculture Brief 2017: Status, Trends, Challenges and Future Outlook; Mombasa, Kenya.
- Opiyo, M.A.; Marijani, E.; Muendo, P.; Odede, R.; Leschen, W.; Charo-Karisa, H. A Review of Aquaculture Production and Health Management Practices of Farmed Fish in Kenya. Int. J. Vet. Sci. Med. 2018, 6, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Pillay, T.V.R.; Kutty, M.N. Aquaculture Principles and Practices; Second Edi.; Blackwell Publishing, 2005; ISBN 9781780648972.
- Kibet, C.J.; Donde, O.O.; Okwiri, B.; Otachi, E.O. Taxonomic Status of Fish Parasites in Kenyan Inland Water Systems and Their Significance on the Freshwater Fisheries and Aquaculture Productivity within the Region. Lakes Reserv. Res. Manag. 2019, 24, 402–410. [Google Scholar] [CrossRef]
- Shamsi, S.; Halajian, A.; Tavakol, S.; Mortazavi, P.; Boulton, J. Pathogenicity of Clinostomum complanatum (Digenea: Clinostomidae) in Piscivorous Birds. Res. Vet. Sci. 2013, 95, 537–539. [Google Scholar] [CrossRef]
- Gustinelli, A.; Caffara, M.; Florio, D.; Otachi, E.O.; Wathuta, E.M.; Fioravanti, M.L. First Description of the Adult Stage of Clinostomum cutaneum Paperna, 1964 (Digenea: Clinostomidae) from Grey Herons Ardea cinerea L. and a Redescription of the Metacercaria from the Nile Tilapia Oreochromis niloticus niloticus (L.) in Kenya. Syst. Parasitol. 2010, 76, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.L.; Chen, H.Y.; Shih, H.H. Occurrence and Distribution of Yellow Grub Trematodes (Clinostomum complanatum) Infection in Taiwan. Parasitol. Res. 2017, 116, 1761–1771. [Google Scholar] [CrossRef] [PubMed]
- Park, C.W.; Kim, J.S.; Joo, H.S.; Kim, J. A Human Case of Clinostomum complanatum Infection in Korea. Korean J. Parasitol. 2009, 47, 401–404. [Google Scholar] [CrossRef]
- Ermakova, L.; Kozlov, S.; Nagorny, S.; Golovchenko, N.; Telicheva, V.; Kiosova, J.; Zotova, M.; Pshenichnaya, N. The First Case of Human Invasion by Clinostomum complanatum in the European Part of Russia. IJID Reg. 2024, 11, 100346. [Google Scholar] [CrossRef]
- Caffara, M.; Locke, S.A.; Gustinelli, A.; Marcogliese, D.J.; Fioravanti, M.L. Morphological and Molecular Differentiation of Clinostomum complanatum and Clinostomum marginatum (Digenea: Clinostomidae) Metacercariae and Adults. J. Parasitol. 2011, 97, 884–891. [Google Scholar] [CrossRef]
- Locke, S.A.; Caffara, M.; Marcogliese, D.J.; Fioravanti, M.L. A Large-Scale Molecular Survey of Clinostomum (Digenea, Clinostomidae). Zool. Scr. 2015, 44, 203–217. [Google Scholar] [CrossRef]
- Caffara, M.; Locke, S.A.; Echi, P.C.; Halajian, A.; Benini, D.; Luus-Powell, W.J.; Tavakol, S.; Fioravanti, M.L. A Morphological and Molecular Study of Clinostomid Metacercariae from African Fish with a Redescription of Clinostomum tilapiae. Parasitology 2017, 144, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Li, B.F.; Liu, X.H.; Ge, H.L.; Xie, C.Y.; Cai, R.Y.; Hu, Z.C.; Zhang, Y.G.; Wang, Z.J. The Discovery of Clinostomum complanatum Metacercariae in Farmed Chinese Sucker, Myxocyprinus asiaticus. Aquaculture 2018, 495, 273–280. [Google Scholar] [CrossRef]
- Caffara, M.; Locke, S.A.; Echi, P.C.; Halajian, A.; Luus-Powell, W.J.; Benini, D.; Tedesco, P.; Fioravanti, M.L. A New Species of Clinostomum Leidy, 1856 Based on Molecular and Morphological Analysis of Metacercariae from African Siluriform Fishes. Parasitol. Res. 2020, 119, 885–892. [Google Scholar] [CrossRef]
- Sereno-Uribe, A.L.; García-Varela, M.; Pinacho-Pinacho, C.D.; Pérez-Ponce de León, G. Three New Species of Clinostomum Leidy, 1856 (Trematoda) from Middle American Fish-Eating Birds. Parasitol. Res. 2018, 117, 2171–2185. [Google Scholar] [CrossRef]
- Sereno-Uribe, A.L.; Pinacho-Pinacho, C.D.; García-Varela, M.; De León, G.P.P. Using Mitochondrial and Ribosomal DNA Sequences to Test the Taxonomic Validity of Clinostomum complanatum Rudolphi, 1814 in Fish-Eating Birds and Freshwater Fishes in Mexico, with the Description of a New Species. Parasitol. Res. 2013, 112, 2855–2870. [Google Scholar] [CrossRef]
- Shigoley, M.I.; Antoine-Moussiaux, N.; Jauniaux, T.; Vanhove, M.P.M. Parasitology of One of the World’s Foremost Fisheries Target Species Lacks a One Health Approach. Hydrobiologia 2024. [Google Scholar] [CrossRef]
- Ochieng, V.O.; Matolla, G.K.; Khyria, S.K. A Study of Clinostomum Affecting Oreochromis niloticus in Small Water Bodies in Eldoret-Kenya. Int. J. Sci. Eng. Res. 2012, 3, 1–6. [Google Scholar]
- Bello, M.A.; Ruiz-León, Y.; Sandoval-Sierra, J.V.; Rezinciuc, S.; Diéguez-Uribeondo, J. Scanning Electron Microscopy (SEM) Protocols for Problematic Plant, Oomycete, and Fungal Samples. J. Vis. Exp. 2017, 2017, 1–10. [Google Scholar] [CrossRef]
- Lange, K.; Mogoi, S.; Weert, F. Van The Economics of Ecosystem Services of the Tana River Basin. Wetl. Int. Kenya, Tech. Rep. 2015.
- Bush, A.O.; Lafferty, K.D.; Lotz, J.M.; Shostak, A.W. Parasitology Meets Ecology on Its Own Terms: Margolis et Al. Revisited*. J. Parasitol. 1997, 83, 575–583. [Google Scholar] [CrossRef]
- Hillis, D.M.; Dixon, M.T. Ribosomal DNA : Molecular Evolution and Phylogenetic Inference. Q. Rev. Biol. 1991, 66, 411–453. [Google Scholar] [CrossRef] [PubMed]
- Vanhove, M.P.M.; Tessens, B.; Schoelinck, C.; Jondelius, U.; Littlewood, D.T.J.; Artois, T.; Huyse, T. Problematic Barcoding in Flatworms: A Case-Study on Monogeneans and Rhabdocoels (Platyhelminthes). Zookeys 2013, 365, 355–379. [Google Scholar] [CrossRef] [PubMed]
- Kmentová, N.; Koblmüller, S.; Van Steenberge, M.; Raeymaekers, J.A.M.; Artois, T.; De Keyzer, E.L.R.; Milec, L.; Muterezi Bukinga, F.; Mulimbwa N’sibula, T.; Masilya Mulungula, P.; et al. Weak Population Structure and Recent Demographic Expansion of the Monogenean Parasite Kapentagyrus spp. Infecting Clupeid Fishes of Lake Tanganyika, East Africa. Int. J. Parasitol. 2020, 50, 471–486. [Google Scholar] [CrossRef]
- Kmentová, N.; Hahn, C.; Koblmüller, S.; Zimmermann, H.; Vorel, J.; Artois, T.; Gelnar, M.; Vanhove, M.P.M. Contrasting Host-Parasite Population Structure: Morphology and Mitogenomics of a Parasitic Flatworm on Pelagic Deepwater Cichlid Fishes from Lake Tanganyika. Biology (Basel). 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Newton, L.A.; Chilton, N.B.; Beveridge, I.; Hoste, H.; Nansen, P.; Gasser, R.B. Genetic Markers for Strongylid Nematodes of Livestock Defined by PCR-Based Restriction Analysis of Spacer RDNA. Acta Trop. 1998, 69, 1–15. [Google Scholar] [CrossRef]
- Littlewood, D.T.J.; Rohde, K.; Clough, K.A. Parasite Speciation within or between Host Species? - Phylogenetic Evidence from Site-Specific Polystome Monogeneans. Int. J. Parasitol. 1997, 27, 1289–1297. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the Number of Nucleotide Substitutions in the Control Region of Mitochondrial DNA in Humans and Chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Matschiner, M. Fitchi: Haplotype Genealogy Graphs Based on the Fitch Algorithm. Bioinformatics 2016, 32, 1250–1252. [Google Scholar] [CrossRef]
- Caffara, M.; Davidovich, N.; Falk, R.; Smirnov, M.; Ofek, T.; Cummings, D.; Gustinelli, A.; Fioravanti, M.L. Redescription of Clinostomum phalacrocoracis Metacercariae (Digenea: Clinostomidae) in Cichlids from Lake Kinneret, Israel. Parasite 2014, 21. [Google Scholar] [CrossRef]
- Caffara, M.; Locke, S.A.; Cristanini, C.; Davidovich, N.; Markovich, M.P.; Fioravanti, M.L. A Combined Morphometric and Molecular Approach to Identifying Metacercariae of Euclinostomum heterostomum (Digenea: Clinostomidae). J. Parasitol. 2016, 102, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Monnens, M.; Halajian, A.; Littlewood, D.T.J.; Briscoe, A.G.; Artois, T.; Vanhove, M.P.M. Can Avian Flyways Reflect Dispersal Barriers of Clinostomid Parasites? First Evidence from the Mitogenome of Clinostomum complanatum. Gene 2023, 851. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E. Analysis of Haplotype Networks: The Randomized Minimum Spanning Tree Method. Methods Ecol. Evol. 2018, 9, 1308–1317. [Google Scholar] [CrossRef]
- Nguyen, J.A.; Woodyard, E.T.; McAllister, C.T.; Marcquenski, S. V.; Rose, D.; Slifka, C.M.; Robison, L.R.S.; Griffin, M.J.; Rosser, T.G. Morphological and Molecular Data Establishes Clinostomum dolichorchum n. sp. (Digenea: Clinostomidae) in the Great Blue Heron Ardea herodias L. and American Bullfrog Rana catesbeiana Shaw. Syst. Parasitol. 2024, 101, 1–26. [Google Scholar] [CrossRef]
- Hamouda, A.H.; Younis, A.E. Characterization of Clinostomum cutaneum and Clinostomum phalacrocoracis in Tilapia Species of Aswan Governorate, Egypt: A Morphological, Molecular and Histopathological Study. Aquac. Res. 2021, 52, 6726–6740. [Google Scholar] [CrossRef]
- Mahdy, O.A.; Attia, M.M.; Shaheed, I.B.; Abdelsalam, M.; Elgendy, M.Y.; Salem, M.A. Evaluation of Praziquantel Effectiveness in Treating Nile Tilapia Clinostomid Infections and Its Relationships to Fish Health and Water Quality : By. BMC Vet. Res. 2024, 20, 449. [Google Scholar] [CrossRef]
- Echi, P.C.; Eyo, J.E.; Okafor, F.C.; Onyishi, G.C.; Ivoke, N. First Record of Co-Infection of Three Clinostomatid Parasites in Cichlids (Osteichthyes: Cichlidae) in a Tropical Freshwater Lake. Iran. J. Public Health 2012, 41, 86–90. [Google Scholar]
- Lo, C.F.; Chen, S.C.; Wang, C.H. The Study of Clinostomum Complanatum (RUD., 1814) V. The Influences of Metacercaria of Clinostomum complanatum on Fish. Fish Pathol. 1985, 20, 305–312. [Google Scholar] [CrossRef]
- Eiras, C.; Luiza, M.; Goulart, G.; Pavanelli, G.C.; Machado, M.H. Histological Studies on the Effects of Clinostomum marginatum (Digenea, Clinostomidae) in Its Second Intermediate Host Loricariichthys platymetopon (Osteichthyes, Loricariidae) of the Upper Paraná River, Brazil. Acta Sci. Biol. Sci. 1999, 21, 237–241. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).