Announcement
Dairy products are susceptible to contamination by
Bacillus during processing and production since
Bacillus can form endospores and biofilms [
1], possesses high heat resistance and dispersal abilities, and can grow and multiplies in milk pipelines [
2]. A significantly elevated incidence of
Bacillus cereus contamination was reported in unprocessed milk samples [
3], and
Bacillus altitudinis contamination was detected in pre-pasteurized milk [
4].
We isolated heat-tolerant bacteria from the washed water of the milk pipeline after boiling the sample for 10 min and growing the bacteria on Nutrient Agar with manganese. Most colonies were identified as
Bacillus altitudinis based on nearly complete 16S rRNA gene sequences. We selected strain HH-03 for further genome analysis. Genomic DNA from the pure culture of strain HH-03 was extracted using the MagAttract® HMW DNA Kit (Qiagen, USA). The sequencing library was constructed using the Transposase Enzyme Linked Long-read Sequencing (TELL-Seq™) WGS Library Prep Kit (Universal Sequencing Technology, USA) and the sequencing primers provided by the TELL-Seq™ Illumina Sequencing Primer Kit (Universal Sequencing Technology, USA), following the manufacturer’s instructions [
5]. The libraries were sequenced on an Illumina NovaSeq 6000 150 bp paired-end platform to obtain raw sequencing data. Quality control and splicing of the raw sequencing data of strain HH-03 were conducted using the TELL-Read and TELL-Link processes provided by Universal Sequencing Technology [
5].
Following the removal of contigs exhibiting inadequate sequencing depth and contamination, the strain HH-03 yielded 12 contigs with more than 100 sequencing depth, with the largest one being 3.82 Mbp in length.
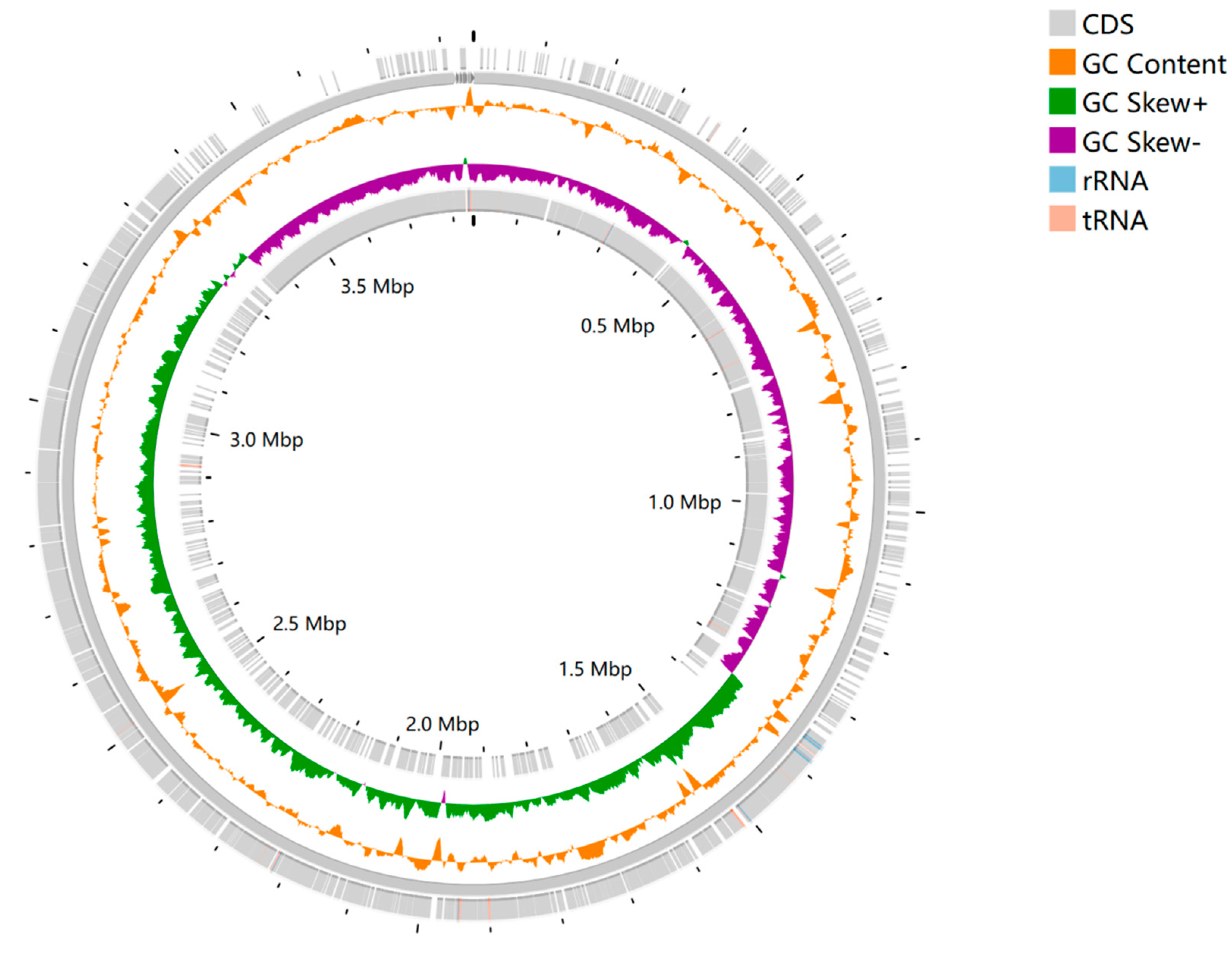
To gain insight into the genome structure, the coding sequences of the positive and negative strands of the genome, the location of non-coding RNAs, and the spliced genome were plotted in a genome feature circle diagram. The spliced FASTA files were uploaded to CGViewer [
6], an online genome feature map website, for online interactive operations, and the genomes were annotated using Prokka [
7]. The genomic circle diagram of strain HH-03 is shown in
Figure 1. The assembled genome size of the sample was 3,846,532 bp with a GC content of 41.2%. Additionally, the sample contained 4,010 protein-coding genes, 17 rRNA genes, and 77 tRNA genes.
Strain HH-03 was subjected to average nucleotide identity (ANI) analysis. The highest ANI value for strain HH-03 genome was 97.85% with Bacillus altitudinis, indicating that strain HH-03 should be identified as Bacillus altitudinis.
The genomic information of Bacillus altitudinis HH-03 will be helpful for the development of clean-in-place processes.
Bacillus altitudinis HH-03 was deposited in Guangdong Microbial Culture Collection Center (GDMCC) under accession number GDMCC 1.5556. The genome sequence of strain HH-03 from the current study have been deposited in eLMSG (an eLibrary of Microbial Systematics and Genomics,
https://www.biosino.org/elmsg/index) under accession number LMSG_G000053875.1 and in NCBI under accession number JBLADY000000000.
Acknowledgments
This work was supported by Key Laboratory of Milk and Dairy Products Detection and Monitoring Technology for State Administration for Market Regulation (MDPDMT-2022-04).
References
- Abee T, Kovács AT, and Kuipers OP, et al. Biofilm formation and dispersal in Gram-positive bacteria. Curr Opin Biotechnol 2011, 22, 172–179. [CrossRef] [PubMed]
- Marchand S, De Block J, De Jonghe V, et al. Biofilm formation in milk production and processing environments; influence on milk quality and safety. Compr Rev Food Sci Food Saf 2012, 11, 133–147. [CrossRef]
- Owusu-Kwarteng J, Akabanda F, et al. Prevalence, virulence factor genes and antibiotic resistance of Bacillus cereus sensu lato isolated from dairy farms and traditional dairy products. BMC Microbiol 2017, 17, 65. [CrossRef] [PubMed]
- Didouh N, Khadidja M, Campos C, et al. Assessment of biofilm, enzyme production and antibiotic susceptibility of bacteria from milk pre- and post-pasteurization pipelines in Algeria. Int J Food Microbiol 2023, 16, 110389.
- Chen Z, Pham L, Wu TC, et al. Ultralow-input single-tube linked-read library method enables short-read second-generation sequencing systems to routinely generate highly accurate and economical long-range sequencing information. Genome Res 2020, 30, 898–909. [CrossRef] [PubMed]
- Stothard P, Grant JR, Van Domselaar G. Visualizing and comparing circular genomes using the CGView family of tools. Briefings Bioinf 2019, 20, 1576–1582. [CrossRef] [PubMed]
- Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).