Submitted:
17 November 2024
Posted:
18 November 2024
You are already at the latest version
Abstract
A Gram-positive, aerobic, rod-shaped and spore forming bacterium strain designation B190/17, was isolated from an air monitoring sample of a Brazilian immunobiological production facility in 2017. The strain was not identifiable by biochemical methodology VITEK® 2 or by MALDI-TOF MS with VITEK® MS RUO and MALDI Biotyper®. The 16S rRNA gene sequencing showed 98.51% similarity with Bacillus wudalianchiensis FJAT 27215T, 98.28% with ‘Bacillus aerolatus’ CX 253T, 97.96% with Bacillus badius MTCC 1458T, 97.63% with Bacillus xiapuensis FJAT 46582T and 97.21% with Bacillus thermotolerans SGZ8T. Biochemical data showed the strain was alanine arylamidase, Ala-Phe-Pro arylamidase, ELLMAN (cysteine residues), leucine arylamidase, phenyalanine arylamidase and tyrosine arylamidase positive. The genomic DNA G+C% content of B190/17 was 41.6 mol%. The phylogenetic, genomic taxonomy and biochemical tests suggested that B190/17 represents a novel species and should be classified as the type strain of a novel Bacillus species. The name Bacillus lumedeiriae sp. nov. is proposed. After characterization, B190/17 was added to the MALDI Biotyper® database as Bacillus lumedeiriae sp. nov.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacterial strains and culture conditions
2.2. Phenotypic tests
2.3. Genotypic identification by 16S rRNA gene sequencing
2.4. Genome sequencing, assembly, and annotation
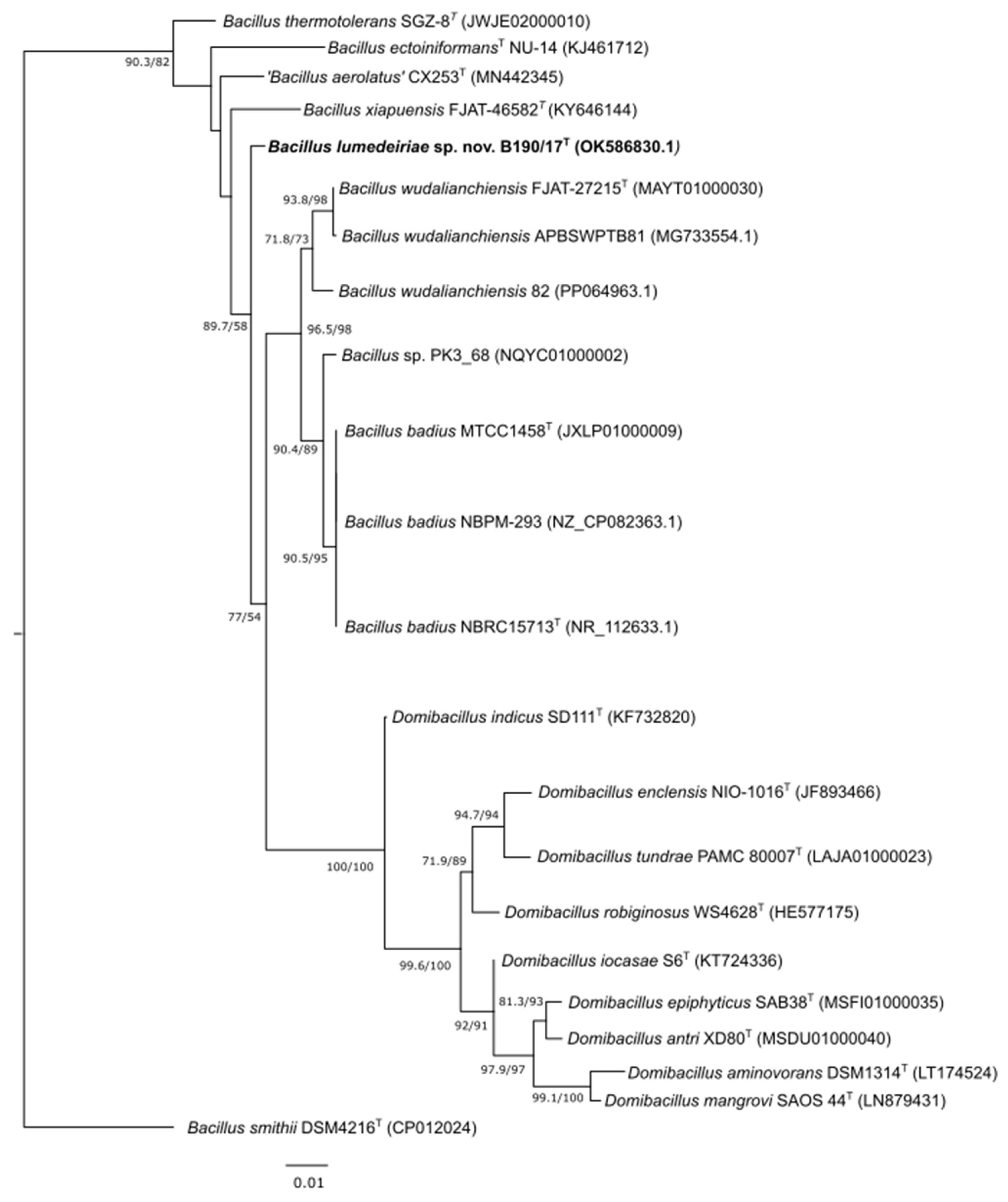
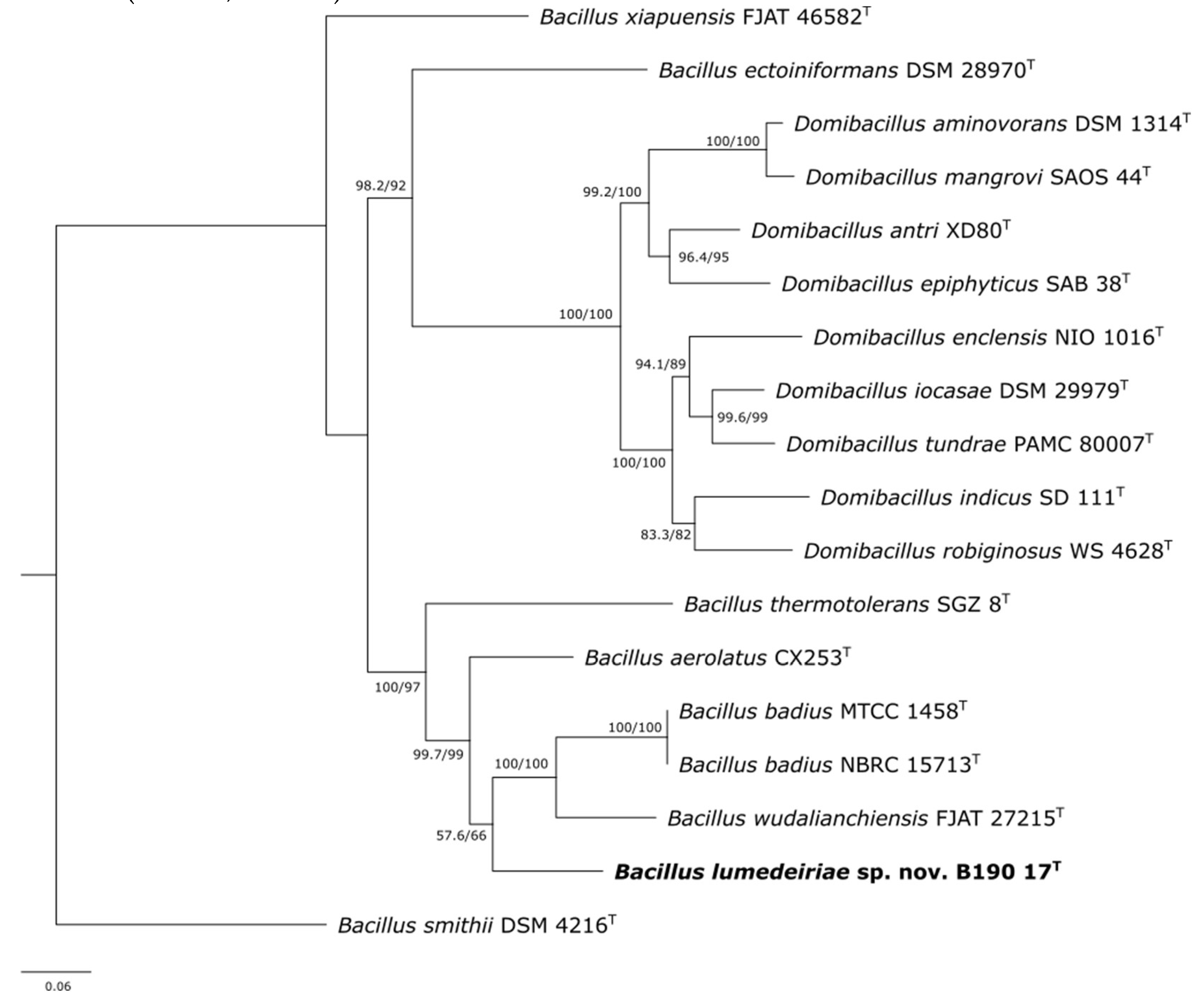
2.5. Phylogenetic analysis of 16S rRNA, rpoB and gyrB genes
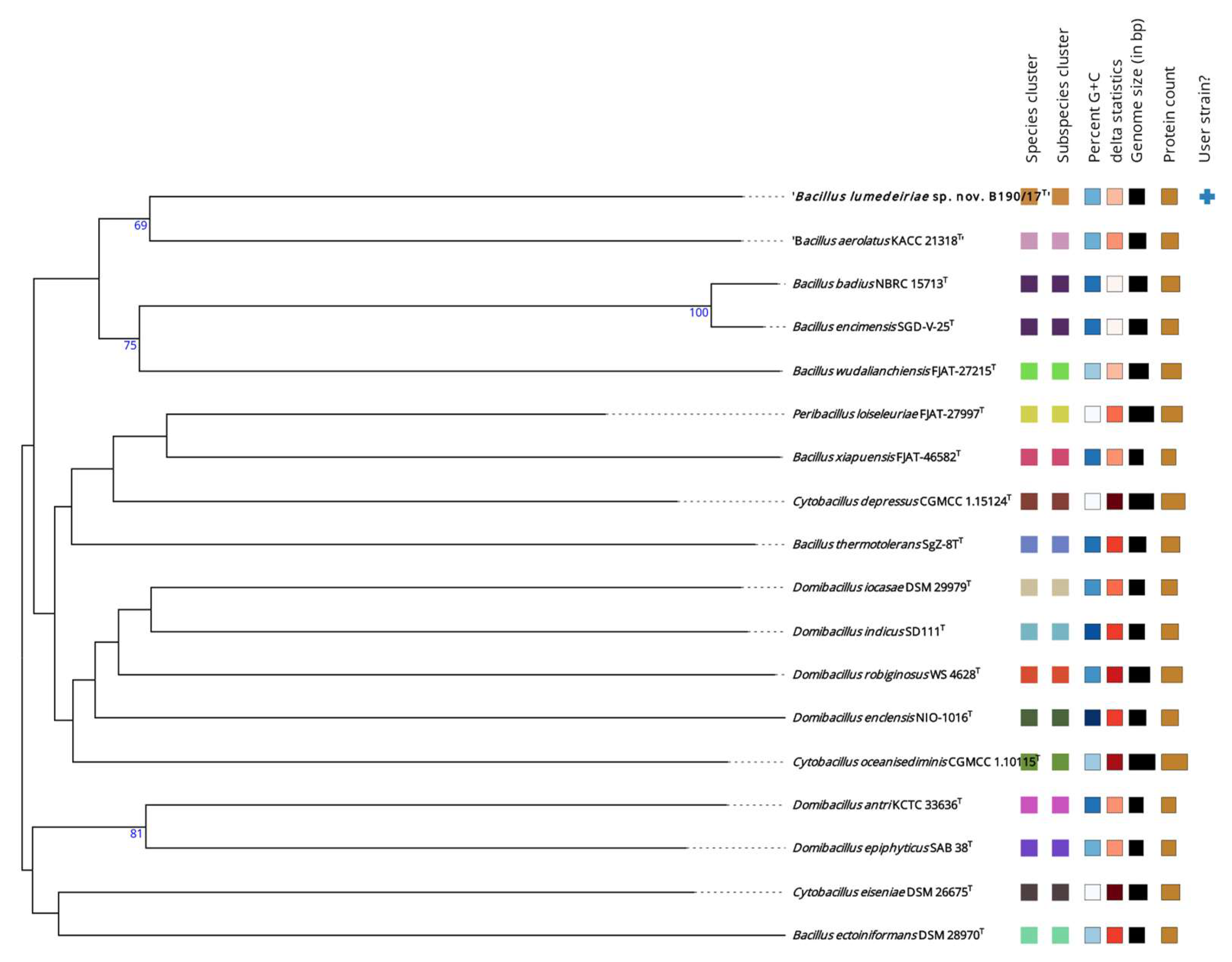
2.6. Genomic taxonomy analysis
2.7. Genome sequence deposit
2.8. Addition of B190/17 spectra to the VITEK® MS RUO and MALDI Biotyper® database
3. Results and Discussion
Description of Bacillus lumedeiriae sp. nov.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costa, L.V.D.; Miranda, R.V.D.S.L.; Reis, C.M.F.; Andrade, J.M.; Cruz, F.V.; Frazão, A.M.; Fonseca, E.L.; Ramos, J.N.; Brandão.
- M. L.L.; Vieira, V.V. MALDI-TOF MS database expansion for identification of Bacillus and related genera isolated from a pharmaceutical facility. J. Microbiol. Methods 2022, 203, 106625. [Google Scholar] [CrossRef]
- Song, M.; Li, Q.; Liu, C.; Wang, P.; Qin, F.; Zhang, L.; Fan, Y.; Shao, H.; Chen, G.; Yang, M. A comprehensive technology strategy for microbial identification and contamination investigation in the sterile drug manufacturing facility—a case study. Front. Microbiol. 2024, 15, 1327175. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency. European Medicines Agency. European Medicines Agency. European Union guidelines for good manufacturing practice for medicinal products for human and veterinary use. In The Rules Governing Medicinal Products in the European Union; Annex 1: Manufacture of Sterile Medicinal Products; European Medicines Agency: Brussels, Belgium, 2022; Volume 4. [Google Scholar]
- Miranda, R.V.D.S.L.; da Costa, L.V.; Albuquerque, L.S.; Dos Reis, C.M.F.; Braga, L.M.P.D.S.; de Andrade, J.M.; Ramos, J.N.; Mattoso,J. M.V.; Forsythe, S.J.; Brandão, M.L.L. Identification of Sutcliffiella horikoshii strains in an immunobiological pharmaceutical industry facility. Lett. Appl. Microbiol. 2023, 76, ovad056. [Google Scholar] [CrossRef]
- Mattoso, J.M.V.; Costa, L.V.C.; Vale, B.A.; Reis, C.M.F.; Andrade, J.M.; Braga, L.M.P.S.; Conceição, G.M.S.; Costa, P.B.M.; Silva, I.B.; Rodrigues, L.A.P.; Anjos, J.P.; Brandão, M.L.L. Quantitative and qualitative evaluation of microorganism profile identified in bioburden analysis in a biopharmaceutical facility in Brazil: Criteria for classification and management of results. PDA J. Pharm. Sci. Technol. 2024, 78, 3. [Google Scholar] [CrossRef] [PubMed]
- Stamatoski, B.; Ilievska, M.; Babunovska, H.; Sekulovski, N.; Panov, S. Optimized genotyping method for identification of bacterial contaminants in pharmaceutical industry. Acta Pharm. 2020, 2, 289–295. [Google Scholar] [CrossRef]
- Caldeira, N.G.S.; de Souza, M.L.S.; de Miranda, R.V.D.S.L.; da Costa, L.V.; Forsythe, S.J.; Zahner, V.; Brandão, M.L.L. Characterization by MALDI-TOF MS and 16S rRNA gene sequencing of aerobic endospore-forming bacteria isolated from pharmaceutical facility in Rio de Janeiro, Brazil. Microorganisms 2024, 12, 724. [Google Scholar] [CrossRef]
- Costa, L.V.D.; Miranda, R.V.D.S.L.; Fonseca, E.L.; Gonçalves, N.P.; Reis, C.M.F.; Frazão, A.M.; Cruz, F.V.; Brandão, M.L.L.; Ramos, J.N.; Vieira, V.V. Assessment of VITEK® 2, MALDI-TOF MS and full gene 16S rRNA sequencing for aerobic endospore-forming bacteria isolated from a pharmaceutical facility. J. Microbiol. Methods 2022, 194, 106419. [Google Scholar] [CrossRef]
- Husni, A.A.A.; Ismail, S.I.; Jaafar, N.M.; Zulperi, D. Current classification of the Bacillus pumilus group species, the rubber-pathogenic bacteria causing trunk bulges disease in Malaysia as assessed by MLSA and multi rep-PCR approaches. Plant. Pathol. J. 2021, 37, 243. [Google Scholar] [CrossRef]
- Qi, H.Y.; Wang, D.; Han, D.; Song, J.; Ali, M.; Dai, X.; Zhang, X.; Chen, J. Unlocking antagonistic potential of Bacillus amyloliquefaciens KRS005 to control gray mold. Front. Microbiol. 2023, 14, 1189354. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Patel, S.; Saini, N.; Chen, S. Robust demarcation of 17 distinct Bacillus species clades, proposed as novel Bacillaceae genera, by phylogenomics and comparative genomic analyses: Description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus Bacillus limiting it only to the members of the Subtilis and Cereus clades of species. Int. J. Syst. Evol. Microbiol. 2020, 70, 5753–5798. [Google Scholar] [CrossRef]
- Patel, S.; Gupta, R.S. A phylogenomic and comparative genomic framework for resolving the polyphyly of the genus Bacillus: Proposal for six new genera of Bacillus species, Peribacillus gen. nov., Cytobacillus gen. nov., Mesobacillus gen. nov., Neobacillus gen. nov., Metabacillus gen. nov. and Alkalihalobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 406–438. [CrossRef]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Teng, J.L.L.; Tse, H. Yuen, K-Y. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infec. 2008, 14, 908–934. [CrossRef]
- Xiang, C-Y.; Gao, F.; Jakovlic, I.; Lei, H-P.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G-T.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, e87. [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Res. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinformatics 2020, 70, e102. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; Meyer, F.; Olsen, G.J.; Olson, R.; Osterman, A.L.; Overbeek, R.A.; McNeil, L.K.; Paarmann, D.; Paczian, T.; Parrello, B.; Pusch, G.D.; Reich, C.; Stevens, R.; Vassieva, O.; Vonstein, V.; Wilke, A.; Zagnitko, O. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008, 9, 1–15. [Google Scholar] [CrossRef]
- Lee, I.; Kim, Y.O.; Park, S.C.; Chun, J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–3. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Chaumeil, P.A. , Mussig, A. J., Hugenholtz, P., Parks, D.H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2020, 6, 1925–1927. [Google Scholar] [CrossRef]
- bioMerieux. Guide for VITEK® 2 BCL card, v. bioMerieux. Guide for VITEK® 2 BCL card, v. 045519- 02 - 2019-03. 2019, 1-24.
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.; Meyer, S.D.; Trujillo, M.E. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Wang, D.; Ren, Q.; Wu, J.; Jiang, Y.; Wu, Z.; Pan, Y.; Zhong, Y.; Guan, Y.; Chen, K.; Zhang, G. Bacillus aerolatus sp. nov., a novel member of the genus Bacillus, isolated from bioaerosols in a school playground. Arch Microbiol 2020, 202, 2373–2378. [CrossRef]
- Verma, A.; Pal, Y.; Ojha, A.K.; Kumari, M.; Khatri, I.; Rameshkumar, N.; Schumann, P.; Dastager, S.G.; Mayilraj, S.; Subramanian, S.; Krishnamurthi, S. Taxonomic insights into the phylogeny of Bacillus badius and proposal for its reclassification to the genus Pseudobacillus as Pseudobacillus badius comb. nov. and reclassification of Bacillus wudalianchiensis Liu et al., 2017 as Pseudobacillus wudalianchiensis comb. nov. Syst. Appl. Microbiol. 2019, 42, 360–372. [CrossRef]
- Dhruw, C. Husain, K., Kumar, V., Sonawane, V. C.. Novel xylanase producing Bacillus strain X2: molecular phylogenetic analysis and its application for production of xylooligosaccharides. 3 Biotech 2020, 10, 1–15. [CrossRef]
- Husni, A.A.A. Ismail, S.I., Jaafar, N.M., Zulperi, D. Current classification of the Bacillus pumilus group species, the rubber-pathogenic bacteria causing trunk bulges disease in Malaysia as assessed by MLSA and multi rep-PCR approaches. Plant Pathol J 2021, 37, 243. [CrossRef]
- Ben Gharsa, H. , Bouri, M., Mougou Hamdane, A., Schuster, C., Leclerque, A., Rhouma, A. Bacillus velezensis strain MBY2, a potential agent for the management of crown gall disease. PLoS One 2021, 16, e0252823. [Google Scholar] [CrossRef]
- Cuellar-Gaviria, T.Z. García-Botero, C., Ju, K. S., Villegas-Escobar, V. The genome of Bacillus tequilensis EA-CB0015 sheds light into its epiphytic lifestyle and potential as a biocontrol agent. Front Microbiol 2023, 14, 1135487. [CrossRef]
- Yang, G. Zhou, X., Zhou, S., Yang, D., Wang, Y., Wang, D. Bacillus thermotolerans sp. nov., a thermophilic bacterium capable of reducing humus. IJSEM 2013, 63, 3672–3678. [CrossRef]
- Riesco, R.; Trujillo, M.E. Update on the proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2024, 74, 006300. [Google Scholar] [CrossRef] [PubMed]
- Bruker. Species/Entry List MBT Compass Library Revision K. 2022, 1-75.

| Biochemical test | Result | Biochemical test | Result | Biochemical test | Result | Biochemical test | Result | Biochemical test | Result | Biochemical test | Result |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BXYL | - | LysA | - | AspA | - | LeuA | + | PheA | + | ProA | - |
| BGAL | - | PyrA | - | AGAL | - | AlaA | + | TyrA | + | BNAG | - |
| APPA | + | CDEX | - | dGAL | - | GLYG | - | INO | - | MdG | - |
| ELLM | + | MdX | - | AMAN | - | MTE | - | GlyA | - | dMAN | - |
| dMNE | - | dMLZ | - | NAG | - | PLE | - | IRHA | - | BGLU | - |
| BMAN | - | PHC | - | PVATE | - | AGLU | - | dTAG | - | dTRE | - |
| INU | - | dGLU | - | dRIB | - | PSCNa | - | NaCI 6.5% | - | KAN | - |
| OLD | - | ESC | - | TTZ | - | POLYB_R | - |
| Characteristics | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Cell shape | rod | rod | rod | ovoid | rod | rod |
| Motility | - | + | + | - | + | + |
| Optimum temperature for growth (ºC) | 37 | 37 | 30 | 50 | 37 | 25 |
| Catalase | + | + | + | + | + | + |
| Starch | - | - | - | - | - | - |
| L-arabinose | - | - | - | - | - | - |
| L-rhamnose | - | - | - | - | - | ND |
| Lactose | - | - | - | - | - | - |
| Glucose | - | + | - | - | - | + |
| L-sorbose | - | - | - | + | - | ND |
| Mannitol | - | + | - | - | - | - |
| Sucrose | - | + | - | - | - | - |
| Amygdalin | - | + | - | - | - | ND |
| Inositol | - | + | - | - | - | - |
| G + C content (%) | 41.6 | 42.3 | 41.2 | 44.4 | 44.0 | 44.2 |
| Genera | Number of species/group of species in VITEK® 2 database | Number of species described |
|---|---|---|
| Alicyclobacillus | 1 | 29 |
| Aneurinibacillus | 1 | 9 |
| Bacillus | 21 | 111 |
| Brevibacillus | 8 | 33 |
| Geobacillus | 4 | 12 |
| Lysinibacillus | 1 | 22 |
| Paenibacillus | 14 | 310 |
| Virgibacillus | 2 | 34 |
| Strains | 16S rRNA (%) | rpoB (%) |
gyrB (%) |
Ortho ANI (%) | GGDC (%) | Mol GC distance (%) |
|---|---|---|---|---|---|---|
| ‘Bacillus aerolatus’ CX253T | 98.28 | 87.50 | 85.43 | 80.01 | 24.00 | 0.70 |
| Bacillus badius NBRC 15713T | 97.96 | 86.91 | 80.90 | 76.97 | 21.60 | 2.33 |
| Bacillus thermotolerans SGZ8T | 97.21 | 84.09 | NSSF | 73.73 | 20.10 | 2.81 |
| Bacillus wudalianchiensis FJAT 27215T | 98.51 | 87.38 | 81.17 | 78.22 | 22.50 | 0.39 |
| Bacillus xiapuensis FJAT 46582T | 97.63 | 82.91 | NSSF | 72.82 | 21.10 | 2.63 |
| Characteristics | B190/17 |
|---|---|
| Estimated genome size (bp) | 3,434,160 |
| Coverage | 73x |
| G+C content (%) | 41.6 |
| N50 | 219177 |
| L50 | 4 |
| Number of contigs | 89 |
| Number of Coding Sequences | 3544 |
| Number of RNA genes | 159 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





