1. Introduction
Banana (
Musa paradisiaca), a staple food in tropical regions and in many developing countries, plays an essential role in serving as a pillar for economic growth and social development of local communities, for its ability to maintain consistent production throughout the year [
1]. Ecuador’s banana production is vital for its economy and food security [
2]. The main producing provinces are Guayas, El Oro and Los Ríos, highlighting El Oro for its quality, while Guayas and Los Ríos for the major producers [
2,
3].
In production, various factors, both external and internal, affect the quality of the fruit during the post-harvest phase. External factors include environmental conditions such as relative humidity and temperature and among internal factors, metabolic changes and the presence of fungal pathogens are highlighted [
2,
4].
It is important to emphasize that fungal pathogens, in particular diseases associated with
Fusarium spp.
, Penicillium spp.
, Trichoderma spp.
, and
Aspergillus spp., represent the main causes of banana wilt [
5,
6]. These diseases manifest as a reduction in the firmness of the superficial tissues in the areas of the rachis and the banana crown, accompanied by changes in leaf coloration and cracks at the base of the pseudo stem [
7]. This causes internal rot in the fingers of the banana, eventually resulting in the crown rot and, in more severe cases, the death of the fruit [
8,
9].
Traditionally, these diseases have been managed using synthetic fungicides. However, the growing interest in organic farming and the concerns about toxic residues in food have prompted the search for more sustainable alternatives [
10]. In this context, EOs derived from plants such as
Origanum vulgare, Salvia rosmarinus, Syzygium aromaticum, Thymus vulgaris, and
Cinnamomum verum have emerged as promising alternatives [
11]. These oils not only possess antimicrobial properties but have also been shown to extend the shelf life of fruits by providing fungitoxic effects and enhancing resistance to postharvest diseases [
12,
13].
EOs contain bioactive compounds such as terpenes and phenols, which are documented for their antimicrobial effects. These compounds can inhibit the growth of fungal pathogens, including those responsible for postharvest rot in bananas [
14]. Previous studies have demonstrated the antifungal efficacy of EOs such as oregano, rosemary, and clove, although the results vary depending on the concentration, the type of fungus, and the storage conditions [
15,
16]. However, despite the promising results, there is limited research specifically evaluating the antifungal activity of these oils against
Fusarium spp.,
Penicillium spp.,
Trichoderma spp., and
Aspergillus spp. in bananas, especially under real postharvest conditions.
This investigation focuses on assessing the antifungal effectiveness of commercially available EOs against fungal pathogens isolated from banana peels. The research helps in the development of sustainable alternatives to synthetic fungicides, promoting eco-friendly agricultural practices and enhancing post-harvest management [
17]. Understanding the composition of the EOs used is crucial to identifying the bioactive compounds responsible for the observed antifungal effects. EOs of oregano and thyme share similar volatile compounds [
18], such as the carvacrol, thymol and p-cymene, the rosemary contains alpha-pinene, 1.8 cineol, camphor and verbenone [
19,
20]. Clove oil, is characterized by its content of eugenol, acetyl eugenol and α and β caryophyllenes and the basil contains estragole, linalool, eugenol and methyl cinnamate [
19], Cinnamon oil is rich in cinnamaldehyde and eugenol, which have demonstrated significant antifungal and bioactive properties [
21].
Although EOs from oregano, basil, thyme, clove, cinnamon, and rosemary have a potent antifungal effect, their phytotoxicity must be carefully monitored. Excessive or misdosed use of these oils can cause damage to plants, such as chlorosis, necrosis, and reduced root growth, especially at concentrations above 1%. It is recommended to use safe concentrations and conduct preliminary tolerance tests on specific plant species before large-scale application [
11,
22].
The objective is to evaluate the potential of EOs as an ecological alternative for controlling fungal diseases in bananas, promoting sustainable agriculture and organic production. Specifically, this study evaluates the antifungal efficacy of these oils against Trichoderma spp., Penicillium spp., Aspergillus spp., and Fusarium spp. isolated from Musa paradisiaca through of in vitro and ex vivo.
Fusarium a filamentous genus known for its ability to adapt to diverse environmental conditions, is a major cause of vascular wilt in bananas, blocking the transport of water and nutrients, and reducing both the quality and quantity of fruit production [
23]. In Ecuador is generally found the species
oxysporum, verticillioides and
solani that are studied to innovate strategies to avoid damage to the banana [
3,
24].
Penicillium distributed in various environments, some species have antagonistic activity against pathogens that can cause deterioration of the fruit in post-harvest [
25].
P. citrinum, P. expansum, and
P. digitatum are frequently isolated from banana surfaces, and EOs like mandarin have been shown to inhibit their growth [
26,
27].
Trichoderma is a genus of filamentous, cosmopolitan fungi is known for its biocontrol properties [
28,
29].
Trichoderma species produce antibiotics and secondary metabolites that inhibit phytopathogenic fungi [
30]. India distributes certified species of
Trichoderma asperellum, T. atroviride, T. gamsii, T. hamatum, T. harzianum, T. polysporum, T. virens, and
T. viride and with genetic engineering made significant improvements to apply in industrial processes [
28,
31].
Aspergillus spp. including species such as
A. niger,
A. flavus, and
A. parasiticus, are common contaminants in bananas during storage and transportation [
32]. These fungi produce a variety of enzymes that decompose plant tissues and can also produce harmful aflatoxins, which pose a risk to human health [
14,
32].
Aspergillus parasiticus is similar to
A. flavus, this species can also produce aflatoxin and affect the quality and safety of bananas [
33].
In light of the above, the objective of the current study was to evaluate the potential of oregano, rosemary, clove, cinnamon, and basil EOs as an ecological alternative for controlling fungal diseases in (
Musa paradisiaca) bananas, promoting sustainable agriculture and organic production [
34]. Specifically, this study evaluates the antifungal efficacy of these oils against
Trichoderma spp.,
Penicillium spp.,
Aspergillus spp., and
Fusarium spp. isolated from
Musa paradisiaca through in vitro and ex vivo analysis. By promoting sustainable agriculture and reducing reliance on chemical fungicides, this research supports the development of more eco-friendly, effective strategies for postharvest disease management.
2. Materials and Methods
In this research, we used
Musa paradisiaca (Ecuadorian bananas harvested with export considerations). A sample of bananas was taken and exposed to ambient conditions of approximately 25°C temperature until signs of rot were observed. Bananas showed at least 50% of the symptoms of the presence of fungi in the banana peels were selected [
35].
2.1. Isolation and Purification of Microorganisms
The isolation of pathogenic fungi from infected
Musa paradisiaca peels was carried out using Potato Dextrose Agar (PDA) medium (Difco™, Detroit, USA), widely recognized for its ability to promote fungal growth [
36]. To inhibit bacterial contamination, chloramphenicol (Merck, Ecuador) was added at a final concentration of 0.5 g/L following the agar diffusion method [
37]. The antibiotic was incorporated to prevent bacterial contamination during the fungal isolation process, considering the agar diffusion method. The prepared medium was then poured into sterile Petri dishes and allowed to solidify at room temperature.
The medium was prepared by dissolving 39 g of PDA in 1 L of distilled water, sterilized by autoclaving, poured into sterile Petri dishes, and left to solidify at room temperature [
38]. Thirty banana samples with visible symptoms were collected, extracting approximately 20 g of peel per fruit. The peels were washed twice with sterile distilled water to remove superficial contaminants. Subsequently, the fragments were transferred to Erlenmeyer flasks containing 200 mL of a 0.05% (v/v) Tween 80 aqueous solution (Merck, Ecuador), vortexed for 2 minutes to facilitate spore release.
Four serial dilutions (0.1%) were prepared from the suspension to ensure homogeneity [
39]. From each dilution, 0.1 mL was inoculated onto PDA plates supplemented with 0.05% chloramphenicol and incubated at 25 ± 2 °C, 75 ± 5% relative humidity, and a photoperiod of 12 h light / 12 h dark [
40]. Emerging colonies were monitored daily, individually isolated using a sterile loop, and subcultured weekly until pure cultures were obtained for macroscopic and microscopic morphological analyses [
41].
2.2. Morphological Identification
Once pure fungi were obtained, macroscopic analysis was conducted to assess the characteristics of the fungal colonies, including shape, color, and texture. For the identification of the causative agents affecting the banana, the upper and lower surfaces of the Petri dishes were observed macroscopically, considering the morphological similarity obtained through direct comparisons. The pure cultures were examined in triplicate weekly after inoculation with the isolated pathogens [
42]. Their macroscopic characteristics were recorded and compared with bibliographic information from books and descriptive guides of fungal morphology to identify the genus of the pathogen [
39,
43]. Aspects such as colony shape, elevation, edges and appearance of pure fungi were considered.
Microscopic examination was performed using conventional methods, to observe the fungal reproductive structures, including hyphae, conidia, and spores. To visualize the microscopic and specialized structures, a piece of adhesive tape was used to collect the aerial mycelium of the fungus and it was mounted on a microscope slide [
39]. The plate was examined using a compound microscope with a 40X and 60X objective lens, this analysis was performed in triplicate weekly. The evaluation was based on the observation of hyphae, mycelium, spores and other microscopic structures present.
2.3. Molecular Identification Through DNA Sequencing
Molecular identification of fungi isolated from
Musa paradisiaca peel was performed through DNA sequencing. Genomic DNA was extracted from pure fungal colonies using the commercial Invitrogen kit (Novogene, California, USA), following the manufacturer’s instructions. The quality and quantity of the extracted DNA were evaluated using spectrophotometry with a Nanodrop spectrophotometer (Thermo Fisher Scientific, Massachusetts, USA) and by performing 1% agarose gel electrophoresis to assess DNA integrity [
44].
The ribosomal DNA fragment corresponding to the Internal Transcribed Spacer (ITS) region was amplified using the universal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) [
45]. The PCR amplification was carried out under optimal conditions in a thermocycler, and the amplified products were visualized on a 1.5% agarose gel stained with ethidium bromide under UV light to confirm successful amplification of the ITS region [
37,
44].
Purified amplicons were sent to Macrogen Inc. (Seul, South Korea) for sequencing. The obtained sequences were analyzed using BioEdit software, version 7.0 [
46], for alignment and subsequent comparison with public databases, such as NCBI GenBank (
https://www.ncbi.nlm.nih.gov/genbank/), using the BLASTn algorithm. The fungal species were identified based on a ≥98% similarity to deposited sequences in the database.
Phylogenetic analysis was conducted by aligning ITS sequences using the ClustalW algorithm. Phylogenetic trees were constructed employing both the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) and the Neighbor-Joining approach, implemented in MEGA version 11 [
47]. Pairwise, identity matrices and genetic distances were calculated based on the Tamura-Nei substitution model. The reliability of clade groupings was assessed through 1,000 bootstrap replicates. This protocol is consistent with prior studies that support the effectiveness of ITS sequences for fungal identification and phylogenetic inference [
11,
48,
49].
2.4. Ex Vivo Fungal Activity
After completing the macroscopic and microscopic characterization of the pathogens, the ex vivo antifungal activity was assessed. The pathogens were isolated and purified from banana peel samples and included
Trichoderma spp.
, Aspergillus spp.
, Penicillium spp.
, and
Fusarium spp. [
50]. These fungi were selected from a total of 15 due to their relevance in postharvest diseases of bananas during storage
The growth index was calculated based on the development of these funguses on fruit, with 20 samples used for each fungus with four replicates for each treatment. The inoculum concentration was adjusted to 10
6 conidia/mL to maintain a constant infection [
14]. A volume of 100 µL of the standardized fungal inoculum was applied, and the samples were maintained at approximately 13 ± 1 °C and 92 ± 3% relative humidity. Fungal growth diameter was measured weekly for up to 5 weeks post-inoculation, providing a comprehensive timeline of the pathogen’s progression and activity. The severity of infection was classified based on the growth diameter of the fungal colonies, with all the purified pathogens considered in the analysis [
43]. This classification provides valuable data on their interaction with the banana peel under controlled conditions [
50]. A one-way ANOVA was applied to identify statistically significant differences in species growth, followed by Tukey’s HSD post-hoc test.
2.5. In Vitro Antifungal Activity with Essential Oils
The antifungal efficacy of EOs derived from six species: oregano (
Origanum vulgare), rosemary (
Salvia rosmarinus), clove (
Syzygium aromaticum), thyme (
Thymus vulgaris), cinnamon (
Cinnamomum verum), and basil (
Ocimum basilicum) was assessed in vitro using a method of incorporation into the medium [
50,
51,
52]. Antifungal activity was assessed under controlled laboratory conditions at 25 ± 2 °C and 75 ± 5% relative humidity, with a photo period of 12 hours of light and 12 hours of darkness and monitored daily. Essential oils (batch 20230516) were used, supplied by Green Harmony, a company specialized in the production and distribution of pure and natural oils.
EOs were obtained using steam distillation, a widely accepted method for isolating plant-derived bioactive compounds. Oregano oil was extracted from dried leaves, cinnamon oil from dried bark, and clove oil from dried flower buds. For rosemary, basil, and thyme, EOs were derived from fresh leaves and flowers. All oils were purchased from certified commercial suppliers. Specifically, oregano EO originated from Turkey, cinnamon from Sri Lanka, clove from Indonesia, thyme from Japan, basil from India, and rosemary from Spain. It is worth noting that the geographic origin of EOs plays a critical role in determining their chemical composition and biological activity [
53,
54].
Cinnamon EOs consist mainly of cinnamaldehyde (60-70%) and eugenol (5-15%), both known for their antimicrobial and antioxidant effects. Basil EOs are characterized by their high eugenol content (50-70%), linalool (10-15%), and methyl chavicol (5-15%), which are compounds with antimicrobial, anti-inflammatory, and antioxidant properties. Oregano EOs are predominantly composed of carvacrol (60-80%), a phenolic compound recognized for its antimicrobial and antioxidant properties. It also contains thymol (5-10%), p-cymene (5-10%), and γ-terpinene (2-5%), which act together to inhibit microbial growth.
Clove EOs is distinguished by their high eugenol content (70-85%), followed by acetyl eugenol (5-15%) and β-caryophyllene (5-12%), all of which contribute to their antifungal and antibacterial effectiveness. The EOs of thyme contain thymol (30-50%) and carvacrol (10-20%), along with p-cymene (15-25%) and γ-terpinene (10-15%), which are responsible for their antifungal properties. The EOs of rosemary are mainly made up of 1,8-cineole (20-50%), carnosol (5-15%), rosmarinic acid (2-5%), and α-pinene (10-20%), compounds known for their antioxidant, antimicrobial, and potential neuroprotective effects.
To prepare the solutions, each EOs was diluted in a 0.05% Tween 80 solution to form a homogeneous emulsion, concentrations of 200, 400, 600, 800, and 1000 ppm [
55]. These emulsions were added to cooled PDA medium before solidification. The pathogens were inoculated to determine the most effective concentration. Visual assessments were carried out every 24 hours, considering three independent replicates and four subsamples per replicate for each microorganism. Mycelial growth inhibition percentages were calculated to assess the antifungal efficacy of the applied essential oils.
A negative control was included, consisting of PDA medium supplemented with 0.05% Tween 80 but without essential oils (EOs), allowing for the establishment of a baseline fungal growth reference in the absence of treatment. This facilitated the evaluation of the antifungal efficacy of the tested EOs. The experimental design followed a 6 × 5 mixed-factor model, where the independent variables were six different types of EOs and five concentration levels, while the dependent variable was the percentage of mycelial growth inhibition. Statistical analyses were performed to identify the optimal concentration of each EO capable of significantly inhibiting fungal growth. The findings provide meaningful insights into the potential of plant-derived essential oils as natural antifungal agents, supporting their future use in biocontrol strategies and sustainable agriculture.
3. Results
3.1. Morphological Identification
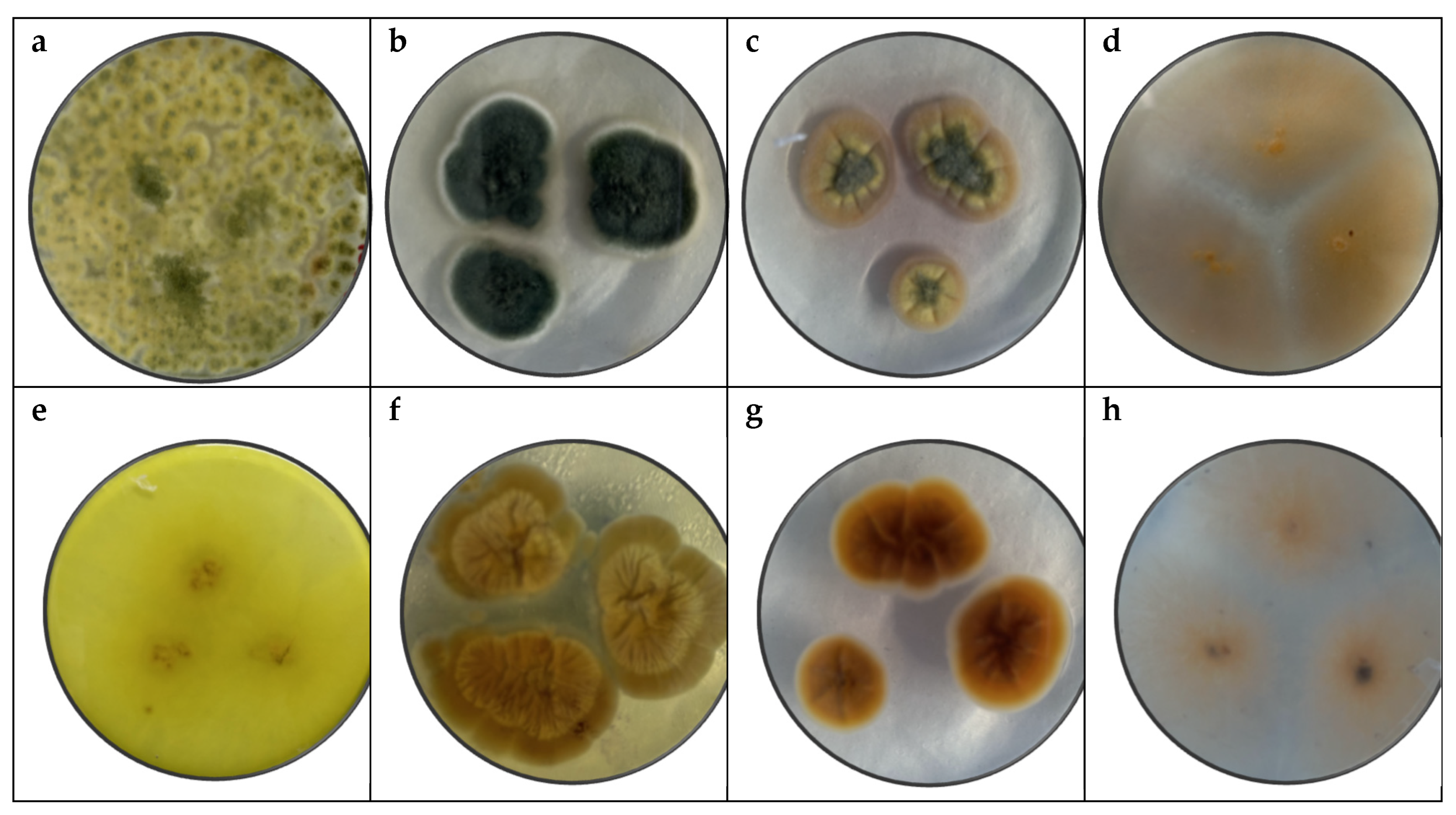
Figure 1 shows the pure fungi isolated from banana peel rot on selective medium (PDA + Chloramphenicol) and stored in PDA at approximately 25°C, with macroscopic images of the front and reverse sides of
Trichoderma spp.,
Penicillium spp.,
Aspergillus spp., and
Fusarium spp.
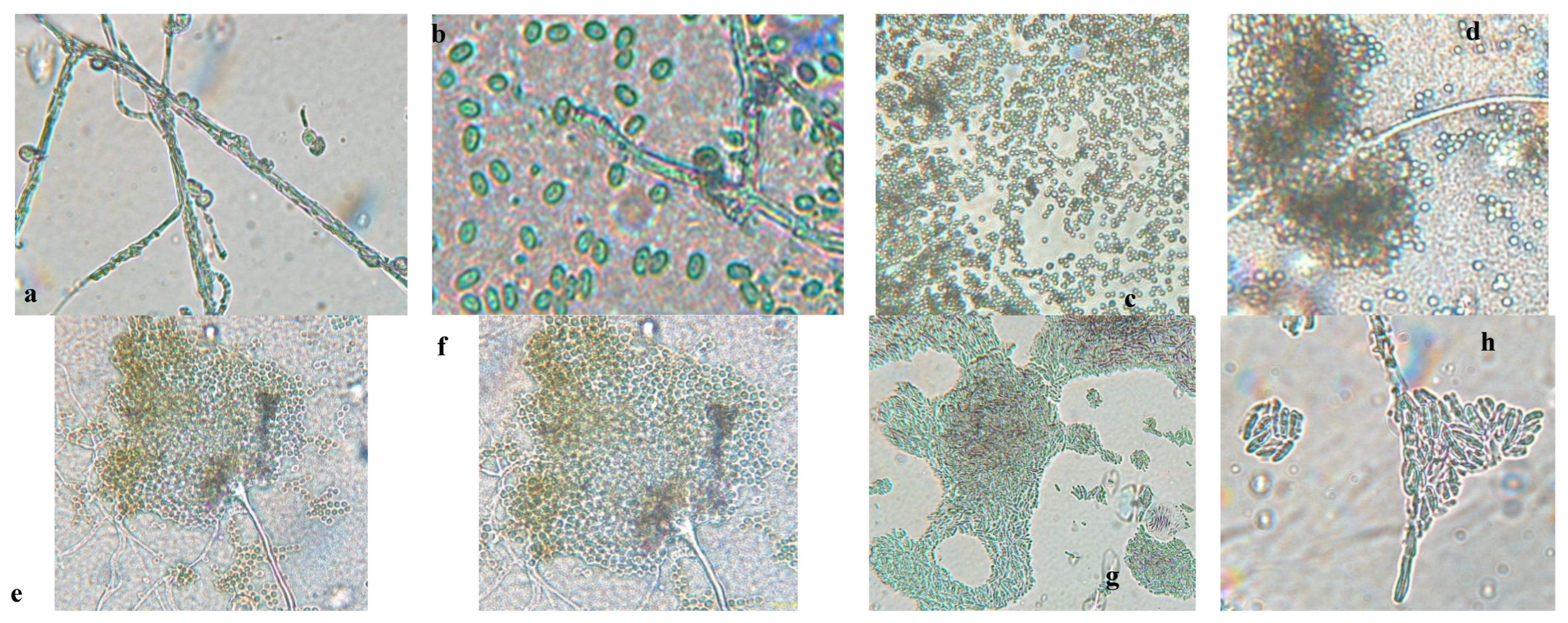
Figure 2 presents the aerial mycelium of the fungi. These observations provide visual information that complements the morphological analysis and microscopic images observed under 40X and 60X magnifications.
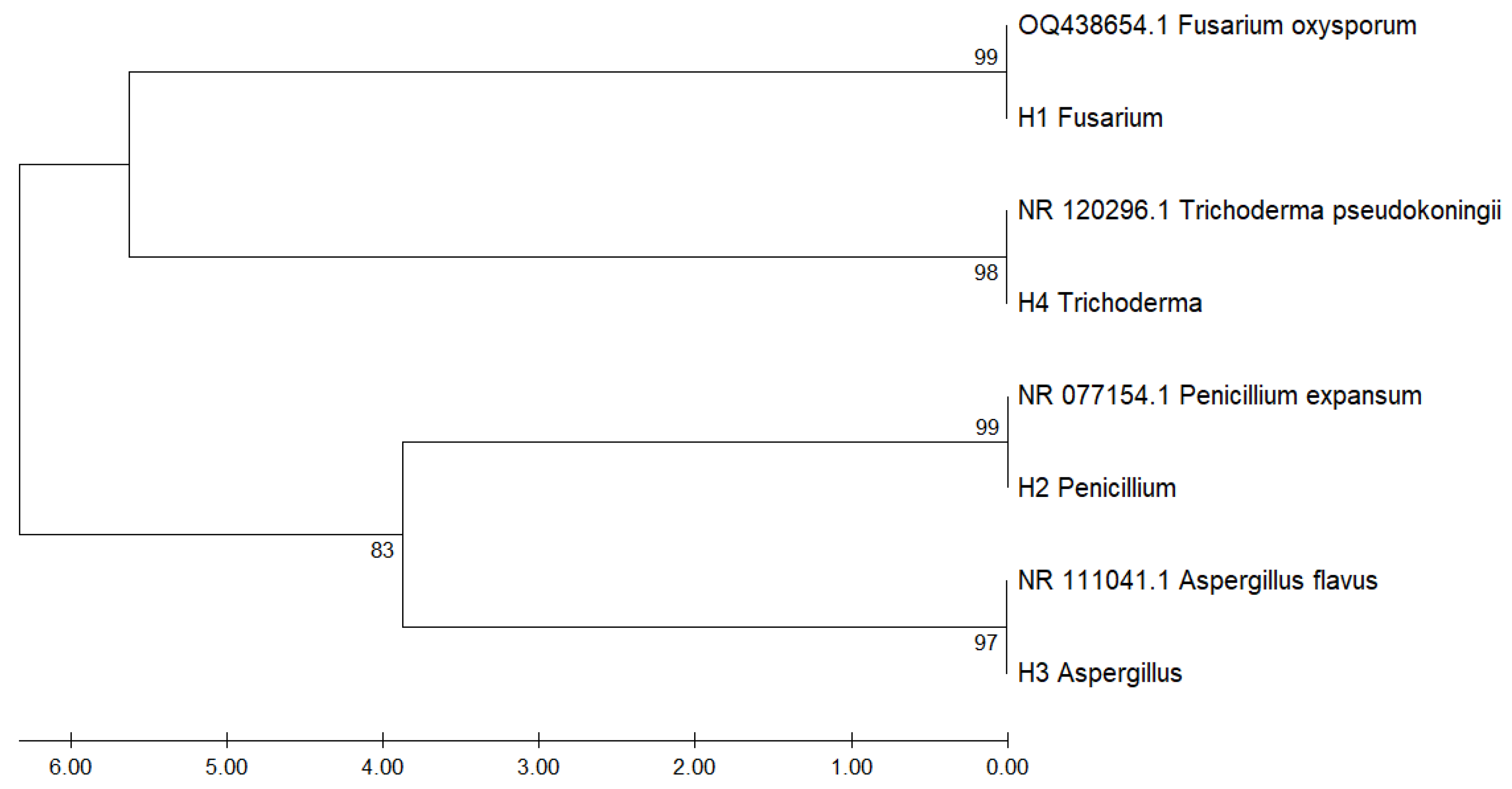
Figure 1 shows the phylogenetic tree constructed from ITS region sequences of fungal isolates recovered from
Musa paradisiaca, aligned against GenBank reference sequences. The analysis revealed consistent clustering by genus and species, supported by high bootstrap values confirming the molecular identification of the isolates.
Isolate H1 clustered closely with Fusarium oxysporum (OQ438654.1) with 99% bootstrap support, while H4 grouped with Trichoderma pseudokoningii (NR_120296.1) with 98%. Similarly, H2 aligned with Penicillium expansum (NR_077154.1) and H3 with Aspergillus flavus (NR_111041.1), with bootstrap values of 99% and 97%, respectively.
The phylogenetic tree also revealed two main clades: one comprising Fusarium and Trichoderma, and the other comprising Penicillium and Aspergillus, supported by a bootstrap value of 83%. These findings highlight the utility of ITS sequencing in accurately discriminating fungal pathogens in tropical crops such as banana.
Table 1 presents a summary of the results from sequencing the ITS region of fungi isolated from banana peels. It lists the identified fungal organisms, the genetic fragments utilized for sequencing, and the percentage similarity derived from comparison with the database. The sequences exhibited a high degree of similarity (≥98%), which supports the reliability of the fungal genera and species identification in the analyzed samples.
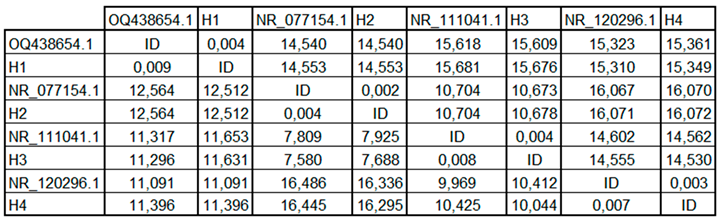
The pairwise identity matrix derived from ITS sequence alignment (
Table 2) revealed high similarity levels between fungal isolates and GenBank reference sequences. Isolate H1 exhibited 99.6% identity with
Fusarium oxysporum (OQ438654.1), while H4 showed 99.7% identity with
Trichoderma pseudokoningii (NR_120296.1). Likewise, H2 matched
Penicillium expansum (NR_077154.1) with 99.8% similarity, and H3 shared 99.2% identity with
Aspergillus flavus (NR_111041.1).
Figure 2.
Microscopy images of (a) Trichoderma spp, (c) Penicillium spp., (e) Aspergillus spp., (g) Fusarium spp. to 40 X and (b) Trichoderma spp., (d) Penicillium spp., (f) Aspergillus spp. (h) Fusarium spp. to 60 X.
Figure 2.
Microscopy images of (a) Trichoderma spp, (c) Penicillium spp., (e) Aspergillus spp., (g) Fusarium spp. to 40 X and (b) Trichoderma spp., (d) Penicillium spp., (f) Aspergillus spp. (h) Fusarium spp. to 60 X.
Figure 3.
Phylogenetic tree based on ITS region sequences of fungal isolates associated with Musa paradisiaca.
Figure 3.
Phylogenetic tree based on ITS region sequences of fungal isolates associated with Musa paradisiaca.
Table 1.
Sequencing results for fungi isolated from the samples of Musa paradisiaca.
Table 1.
Sequencing results for fungi isolated from the samples of Musa paradisiaca.
| Organism |
Fragment |
NCBI |
% identity |
| Fusarium oxysporum |
ITS |
OQ438654.1 |
99.26% |
| Penicillium expansum |
ITS |
NR_077154.1 |
99.68% |
| Trichoderma pseudokoringii |
ITS |
NR_111041.1 |
99.24% |
| Aspergillus flavus |
ITS |
NR_120296.1 |
99.34% |
The lowest identity values were observed between different genera, with genetic distances exceeding 10,000 units, highlighting clear taxonomic differentiation. This matrix quantitatively supports the phylogenetic proximity observed in the dendrogram and confirms molecular identification at the species level.
Table 2.
Pairwise genetic distance matrix of ITS sequences among fungal isolates and GenBank reference strains.
Table 2.
Pairwise genetic distance matrix of ITS sequences among fungal isolates and GenBank reference strains.
3.3. Fungal Activity Ex Vivo
The severity of fungal infection in banana samples was assessed through ex vivo analysis, which 20 banana samples were monitored over a 6-week period. Fungal growth was evaluated for various species, including
Trichoderma spp.,
Penicillium spp.,
Aspergillus spp., and
Fusarium spp., with
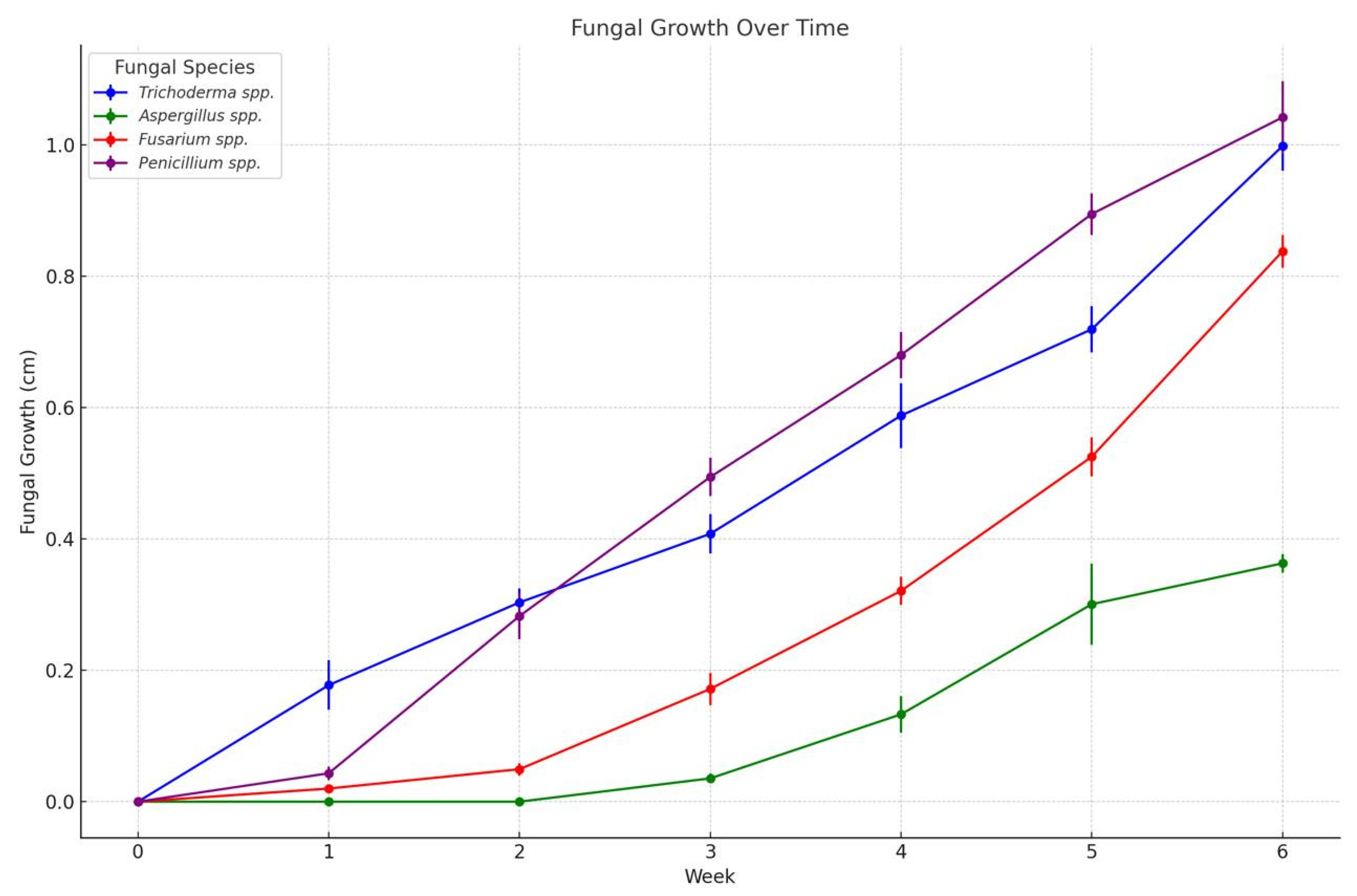
Figure 4 providing a visual representation of this growth over time.
The progression of fungal growth on Musa paradisiaca peel samples over a six-week storage period. The analysis utilized an ANOVA approach to examine the impact of different treatments and concentrations on fungal development. Statistically significant differences were observed among treatments (p < 0.05), indicating that both the type of treatment and its concentration had a notable effect on fungal growth.
In addition, a one-way ANOVA revealed highly significant differences in growth among the four fungal species evaluated (Penicillium, Trichoderma, Fusarium, and Aspergillus) (F = 48.25, p < 0.001). These differences underscore the varying capacity of these fungi to colonize postharvest banana tissue.
A Tukey’s Honest Significant Difference (HSD) post hoc test (α = 0.05) was conducted to identify pairwise differences. The results showed that Penicillium and Trichoderma exhibited the highest growth rates, with no significant difference between them (p > 0.05), while both displayed significantly greater growth compared to Fusarium and Aspergillus. Among all, Aspergillus spp. showed the lowest growth, differing significantly from the other species (p < 0.001). These findings highlight the variability in fungal infections in bananas and emphasize the importance of effective treatment strategies to control postharvest decay.
Assumptions of normality and homogeneity of variances were verified through residual diagnostic plots and statistical tests. The Shapiro–Wilk test confirmed the normal distribution of residuals (p > 0.05), while Levene’s test indicated homogeneity of variances across groups (p > 0.05). Additionally, visual inspection of standard deviations revealed consistent dispersion among treatment groups, supporting the validity of the ANOVA results.
3.4. In Vitro Antifungal Activity with Essential Oils
Figure 5,
Figure 6,
Figure 7 and
Figure 8 show the in vitro growth of
Trichoderma spp.,
Penicillium spp.,
Aspergillus spp., and
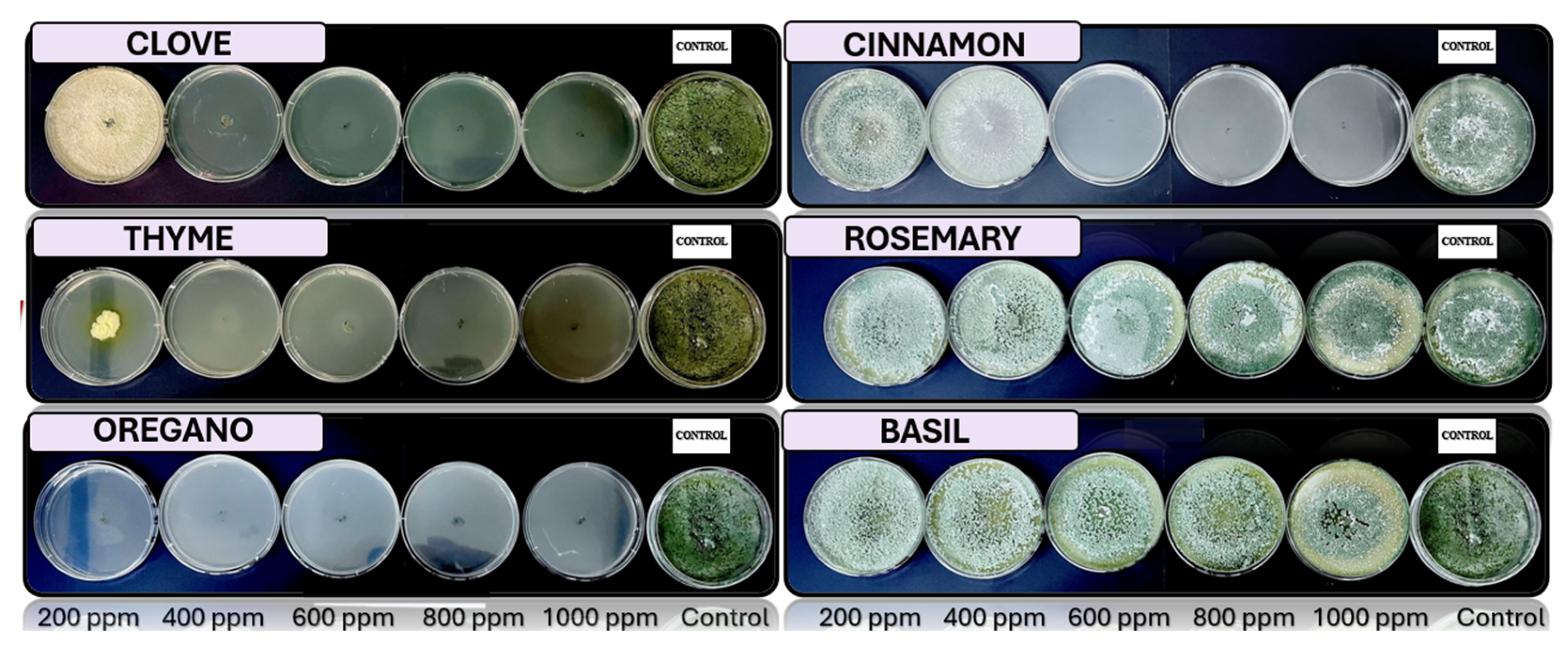
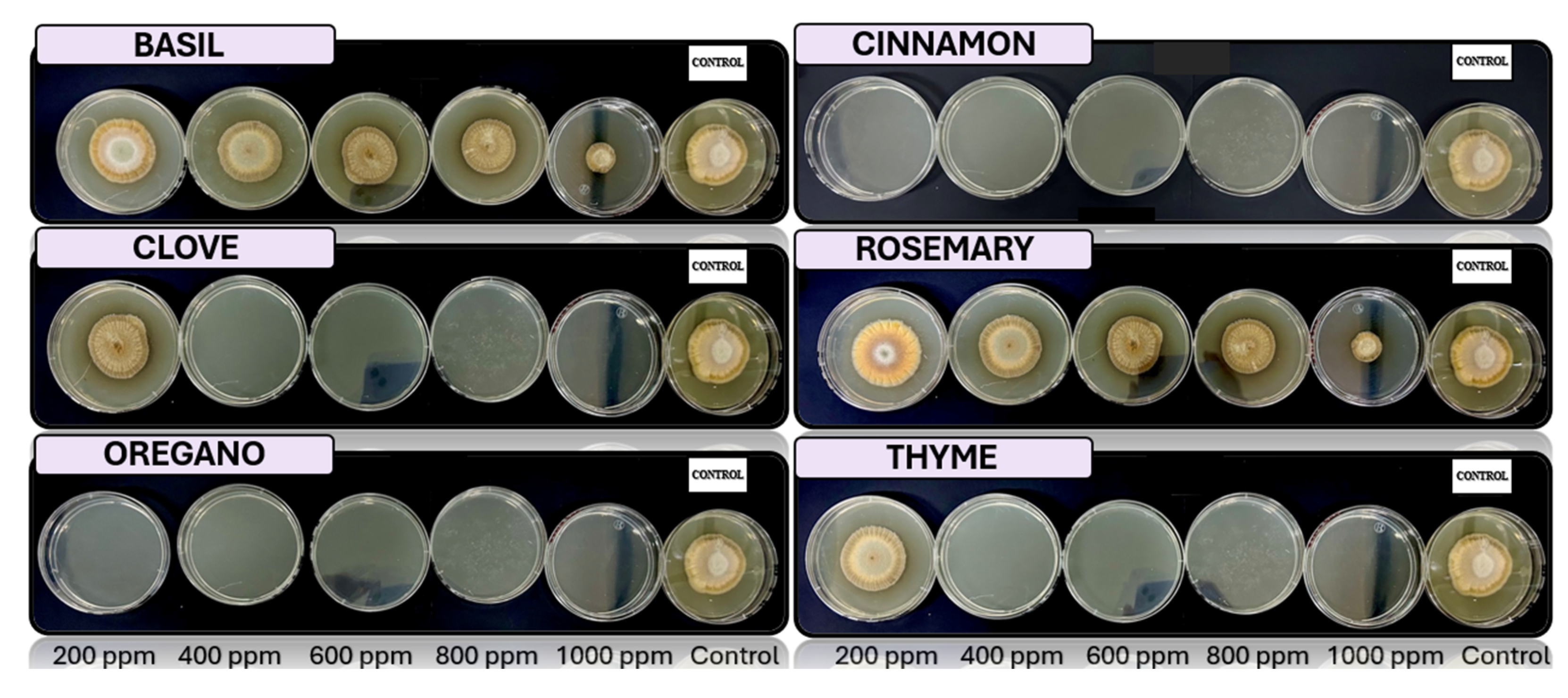
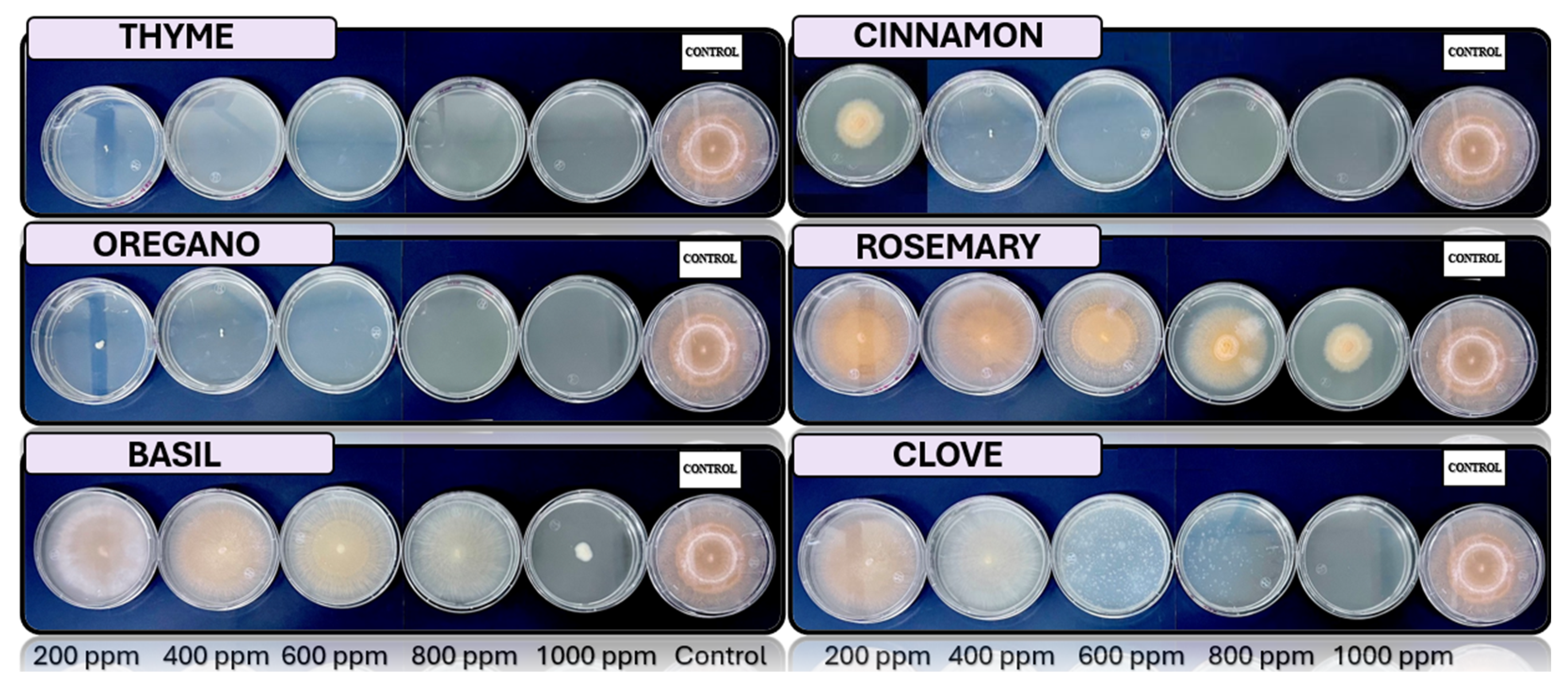
Fusarium spp., respectively, in PDA medium with 200 ppm, 400 ppm, 600 ppm, 800 ppm, and 1000 ppm of oregano, basil, cinnamon, rosemary, thyme, and clove EOs.
In this study, the antifungal efficacy of six essential oils (EOs) was evaluated in vitro against
Trichoderma spp.,
Penicillium spp.,
Aspergillus spp., and
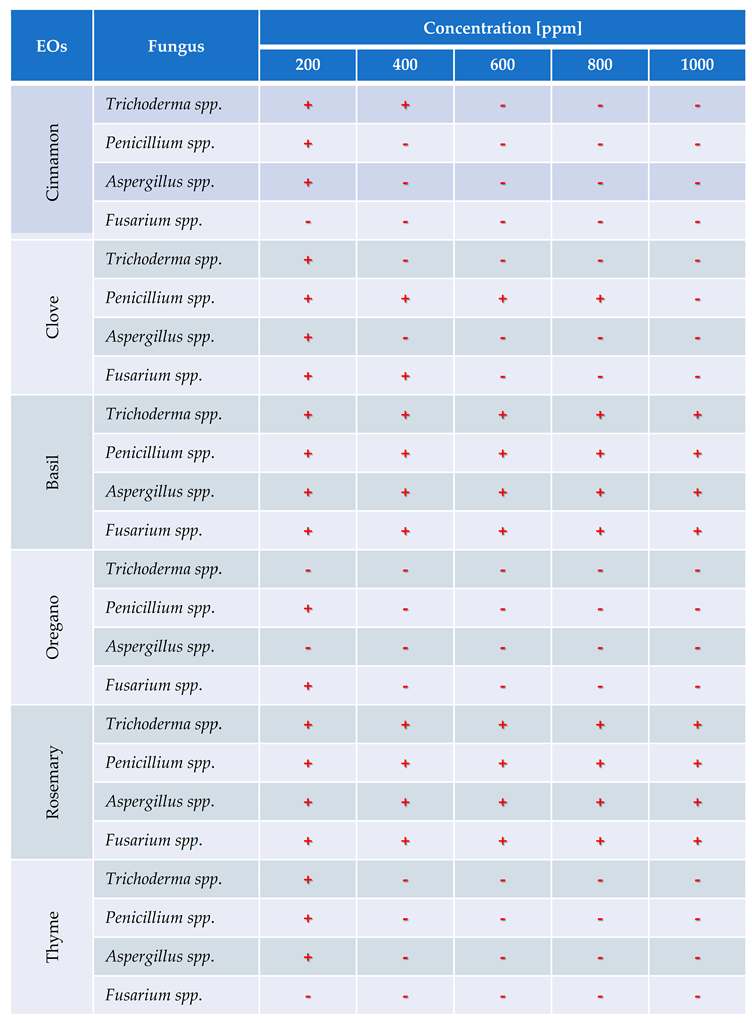
Fusarium spp. at five concentration levels ranging from 200 to 1000 ppm. Fungal inhibition was recorded as the absence of visible mycelial growth and denoted by “−”, whereas growth was indicated by “+”.
Table 3 presents a detailed overview of these inhibitory effects.
The results exhibit a clear concentration-dependent inhibition pattern. Cinnamon, oregano, and thyme oils demonstrated the highest antifungal efficacy, each achieving 23 out of 25 possible inhibitions across all tested fungi and concentrations. Conversely, basil oil displayed the weakest performance, with only 16 inhibitory responses.
Among the fungal genera, Fusarium spp. was the most sensitive, exhibiting consistent inhibition by most oils at concentrations of 400 ppm and above. Trichoderma spp. and Penicillium spp. showed moderate susceptibility, while Aspergillus spp. appeared comparatively more resistant, maintaining growth at lower concentrations for several oils.
These findings underscore the potential of cinnamon, oregano, and thyme essential oils as effective natural alternatives for the control of fungal pathogens in Musa paradisiaca, supporting their possible application in postharvest disease management strategies.
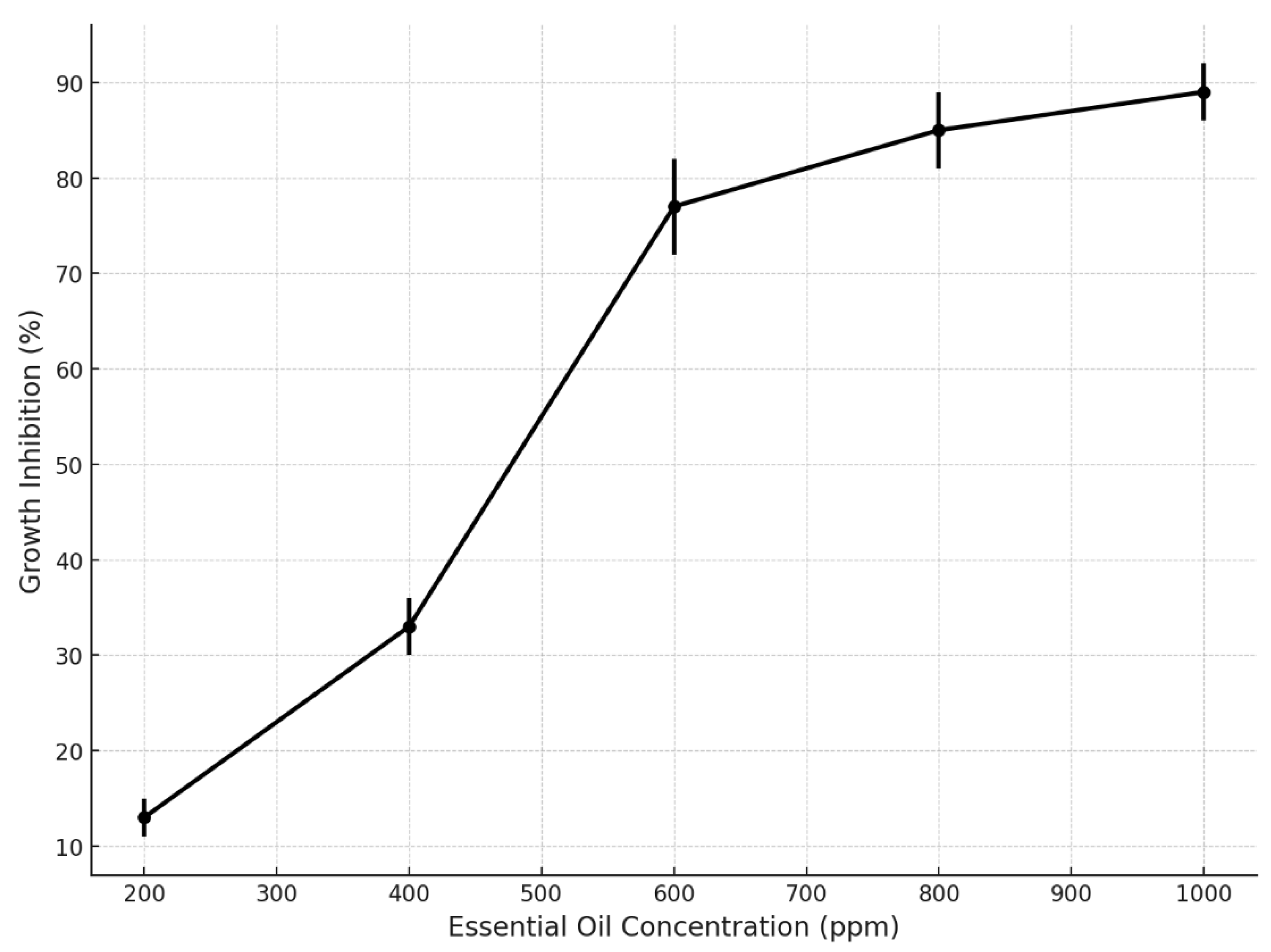
The analysis of fungal growth inhibition revealed a clear concentration-dependent response to the application of essential oils (EOs). As shown in
Figure 9, inhibition percentages increased progressively with EO concentration, reaching statistically relevant thresholds. At the lowest tested concentration (200 ppm), the average inhibition was approximately 13%, while at 400 ppm it increased to 33%. A marked rise in antifungal activity was observed at 600 ppm, achieving 77% inhibition, with subsequent increases at 800 ppm (85%) and 1000 ppm (89%).
4. Discussion
4.1. Morphological Identification
In
Figure 1 and 2, the results of the macroscopic and microscopic analysis are presented and detailed below.
Trichoderma spp. grows rapidly, with colonies that vary in color (white, green, yellow, or orange) and texture (cottony, velvety, or granular) depending on growth conditions. Microscopically, it produces branched conidiophores with small, oval or cylindrical conidia that form in chains or clusters. A characteristic odor may be present but is not a reliable feature [
56].
Fusarium spp. has networks of filaments (hyphae), conidia (asexual and sexual spores or ascospores). In the microscope, the phialide is generally thin, with a bottle – shaped that can be simple or branched; short or long, they can have different characteristics depending on their species [
57].
Aspergillus spp. is characterized by a prominent columella and highly organized plumose structures, where conidia emerge symmetrically. This distinct arrangement differentiates from
Penicillium and
Fusarium, which have simpler conidiophore structures.
Penicillium spp. can cause stains and discoloration on the peel of the banana, affecting its appearance and quality,
expansum can affect the bananas during storage, causing rotting and loss of firmness, while
digitatum is more common in citrus fruits but can affect bananas if are stored in wet and warm storage conditions, causing rotting and stains on the peel [
58]. To prevent the presence of this fungus should be handled properly during the storage and transport of the fruit.
Trichoderma spp., including T. asperellum, T. harzianum, and T. koningii, are characterized by ellipsoidal conidia grouped together and thin, septate hyphae [
59]. The colonies exhibit rapid growth and are typically green, with a circular shape, rough surface, regular edges, and slightly elevated contours [
60]. The texture varies from fuzzy to cottony, reflecting their adaptability to different growth conditions.
Penicillium spp. displays septate hyphae of variable length, forming structures that resemble plumes. The colonies range in color from white to green or blue, with a velvety surface and well-defined edges. The characteristic brush-like arrangement of conidia is a defining feature of this genus, enabling clear morphological differentiation from other fungi [
25].
Aspergillus spp. is distinguished by septate hyphae and conidiophores bearing long, ellipsoidal conidia arranged in characteristic plumose structures. The colonies exhibit a range of colors, typically yellow to green, with a rugged surface and a texture resembling tufts. Edges are regular or slightly wavy, and the colony elevation often shows a cratiform pattern [
33].
Fusarium spp. is characterized by fusiform, cylindrical conidia arranged in chains or clusters, supported by septate hyphae. The colonies appear pink, red, or orange, with a cottony or velvety texture, diffuse edges, and a flat or slightly elevated surface. These macroscopic and microscopic features distinguish
Fusarium spp. from other genera [
24].
The macroscopic and microscopic analysis of the fungal colonies, in combination with DNA sequencing, revealed distinct characteristics that facilitated accurate identification of the species. The following discussion outlines the macroscopic features observed for Fusarium oxysporum, Penicillium expansum, Trichoderma pseudokoringii, and Aspergillus flavus.
Fusarium oxysporum exhibited typical characteristics of the
Fusarium genus, including pink to orange colonies with a cottony texture and regular edges. The colony surface was flat or slightly elevated, which is consistent with the morphological described [
61]. The conidia, arranged in chains, were observable under the microscope. The macroscopic features, including the color and texture, provided reliable indicators for the identification of
Fusarium oxysporum when compared to other species.
Penicillium expansum showed green colonies with white edges and a powdery, flat surface, which are characteristic of many
Penicillium species, particularly those associated with decaying fruits and vegetables [
62]. The white edge and powdery surface were key distinguishing features that aligned with the macroscopic description of
Penicillium expansum. The appearance supports its identification, which may have similar macroscopic features but differ in detail such as colony color or edge definition.
Trichoderma pseudokoringii include green colonies with a rough, granular surface and slightly elevated, regular edges. These features, common in many
Trichoderma species [
63], were consistent with the macroscopic description of
T. pseudokoringii. The growing of the colonies and the distinctive texture helped confirm its identification. The rough surface and elevated edges serve as distinguishing features when compared to other [
30].
Aspergillus flavus exhibited yellow-green colonies with a rough surface and cratiform elevation, consistent with the known morphological traits of
A. flavus [
33,
64]. The coloration and rough texture of the colonies made it easy to differentiate
A. flavus from other
Aspergillus species. The cratiform elevation, a distinct feature, further facilitated the identification. The yellow-green color and texture are particularly indicative of the species.
4.2. Molecular Identification Through DNA Sequencing
The DNA sequencing of fungi isolated from
Musa paradisiaca samples provided highly reliable identification of the species through the ITS region, with all isolates showing a percentage identity above 98% [
44]. These results validate the initial morphological analysis and emphasize the precision of molecular techniques in fungal taxonomy.
Fusarium oxysporum was identified with a 99.26% identity in the ITS region, confirming its classification within the Fusarium genus. This high similarity aligns with the macroscopic features previously observed, such as the pink to orange colonies with cottony texture and regular edges. The combined findings provide strong evidence of its presence in the samples analyzed.
Penicillium expansum showed the highest percentage identity (99.68%) among the isolates, reflecting its genetic distinction. The molecular data corroborates its characteristic macroscopic traits, such as green colonies with white edges and a powdery, flat surface. The DNA sequencing and morphological analysis solidifies its identification and highlights its significance of Musa paradisiaca samples, association with fruit decay.
Trichoderma pseudokoringii exhibited a 99.24% identity, supporting its classification aligns with its observed rapid growth and green colonies with a granular texture. The genetic match enhances confidence in identification, particularly given the importance of Trichoderma species in biological control and agricultural systems.
Aspergillus flavus was identified with a 99.34% identity, confirming its classification. The sequencing results are consistent with their distinctive morphologies, such as yellow-green colonies with a rough surface and cratiform elevation. This high genetic identity underscores the reliability of ITS sequencing for distinguishing A. flavus, a species of economic and health significance due to its ability to produce aflatoxins.
The pairwise distance matrix derived from ITS sequences (
Table 1 and
Table 2) revealed significant genetic divergence among the fungal isolates analyzed. The lowest genetic distance was recorded between
Fusarium oxysporum (OQ438654.1) and its corresponding local isolate H1 (0.004), indicating a high degree of sequence similarity. In contrast, the highest divergence was observed between
Penicillium expansum (NR_077154.1) and
Trichoderma pseudokoningii (NR_120296.1), with values exceeding 16.4 units, reflecting distinct phylogenetic separation.
These findings align with the clustering patterns observed in the phylogenetic tree (
Figure 3), further validating the internal transcribed spacer (ITS) region as a robust molecular marker for delineating taxonomic relationships among filamentous fungi. Moreover, the distance matrix (
Table 2) reinforces the bootstrap-supported clade assignments, particularly between
Penicillium and
Aspergillus species, which exhibited moderate genetic distances.
4.3. Ex Vivo Fungal Activity
The inhibition rate was evaluated by measuring fungal growth from 20 inoculations of each fungus to assess the severity of the pathogens. The ex vivo growth of fungal species isolated from
Musa paradisiaca was monitored over six weeks, as shown in
Figure 4. The data demonstrates distinct growth patterns among the fungal species (
Trichoderma pseudokoringii,
Aspergillus flavus,
Fusarium oxysporum, and
Penicillium expansum), highlighting their varying rates of colonization and adaptation.
Trichoderma pseudokoringii exhibited the slowest growth among the species, with minimal increases in fungal colony size during the first three weeks. By week 6, the colony size reached approximately 0.4 cm. The initial slow growth may reflect the competitive nature of
Trichoderma spp. as a biological control agent, requiring time to adapt and establish in the substrate [
63]. However, its accelerated growth in later weeks suggests its ability to utilize the available nutrients efficiently once adapted.
Aspergillus flavus displayed moderate growth, showing a consistent increase in colony size throughout the six weeks and reaching approximately 0.8 cm by the end of the observation period. This growth pattern aligns with its documented ability to colonize plant materials, particularly in nutrient-rich environments such as stored grains and fruits [
65,
66]. The steady growth indicates
A. flavus’s adaptability and efficiency in utilizing
Musa paradisiaca as a substrate.
Fusarium oxysporum demonstrated a growth rate between that of
Trichoderma pseudokoringii and
Aspergillus flavus. By week 6, the colony size was approximately 0.6 cm. The intermediate growth rate is consistent with the ecological behavior of
Fusarium spp., which thrives under specific conditions but does not dominate rapidly in environments with competing organisms [
56,
61].
Penicillium expansum showed the highest growth rate among the species, reaching over 1 cm by week 6. This rapid growth reflects its opportunistic nature and strong ability to colonize decaying organic matter, including fruits like
Musa paradisiaca [
26,
62,
67]. The powdery texture and efficiency in nutrient utilization are characteristic of
P. expansum, enabling its dominance in the substrate.
4.4. In Vitro Antifungal Activity with Essential Oils
The in vivo experiments with EOs at concentrations (200, 400, 600, 800, and 1000 ppm) demonstrated distinct antifungal efficacy across fungal species (Trichoderma pseudokoringii, Penicillium expansum, Aspergillus flavus, and Fusarium oxysporum). The results emphasize the importance of EOs as natural antifungal agents in agricultural and food preservation applications.
Clove and cinnamon EOs contain active compounds such as eugenol and cinnamaldehyde, that have demonstrated significant antimicrobial properties. These compounds interact with the fungal cell membranes, increasing their permeability and causing the leakage of essential components, which leads to fungal cell death [
14].
Furthermore, cinnamaldehyde in cinnamon oil inhibits the production of intracellular enzymes such as amylases and proteases, resulting in the degradation of the cell wall and a high degree of cellular lysis. This action contributes to the efficacy of cinnamon oil inhibiting the growth of various pathogens.
EOs also affect fungal spore germination. For instance, studies have shown that clove oil inhibits the germination of
F. oxysporum and
F. solani spores at concentrations of 125 µL/L, suggesting its potential as a preventive biofungicide. Likewise, thyme oil has been shown to inhibit the germination of
Colletotrichum acutatum spores, significantly reducing the spread of this pathogen [
68]. These findings support the efficacy of clove and cinnamon EOs as natural antimicrobial agents, offering sustainable alternatives for controlling fungal pathogens in the postharvest management of
Musa paradisiaca.
Trichoderma pseudokoringii displayed a response to cinnamon and clove oils, were most effective at higher concentrations (800–1000 ppm), showing significant inhibition of fungal growth, probably due to the phenolic compounds such as cinnamaldehyde and eugenol that disrupt fungal cell membranes [
63]. Basil and rosemary oils exhibited moderate inhibition at all concentrations, while oregano and thyme oils showed minimal effects. These findings align with previous studies highlighting the potential of
Trichoderma pseudokoringii as a biological control agent when combined with natural compounds [
69].
The growth of
Penicillium expansum was highly sensitive to cinnamon and clove oils, with complete inhibition observed at 800–1000 ppm. Basil and rosemary oils provided moderate inhibition across all concentrations, while thyme and oregano oils had limited effects. The pronounced sensitivity of
Penicillium expansum to phenolic compounds suggests their potential use in controlling post-harvest diseases in fruits and vegetables [
70].
Aspergillus flavus showed significant growth inhibition when treated with cinnamon and clove oils, particularly at higher concentrations (800–1000 ppm). Thyme and oregano oils provided moderate inhibition, while basil and rosemary oils were less effective. The susceptibility of
Aspergillus flavus to these oils underscores their utility in controlling fungal contamination and reducing aflatoxin production [
71].
Fusarium oxysporum demonstrated varied responses to EOs. Cinnamon and clove oils showed moderate inhibition at concentrations above 600 ppm, while basil and rosemary oils exhibited consistent but limited effects across all concentrations. Oregano and thyme oils had minimal inhibitory activity. These results suggest that
Fusarium oxysporum may require combined treatments, such as the integration of EOs with chemical fungicides or other biological agents, to achieve effective control [
72].
Figure 9 shows the inhibition curve reveals a clear concentration-dependent antifungal effect of essential oils, with inhibition rates increasing notably from 200 to 1000 ppm. A significant threshold was observed at 600 ppm, where the inhibition exceeded 75%, reaching nearly 90% at the highest concentration. These results highlight the effectiveness of EOs in fungal suppression, with reduced variability at higher doses, confirming their potential as natural antifungal agents for postharvest protection in
Musa paradisiaca.
Cinnamon and clove oils exhibited the highest antifungal activity across all species, particularly at higher concentrations. Their effectiveness is attributed to the high phenolic content, which disrupts fungal cell structures and inhibits growth [
73]. Basil and rosemary oils demonstrated moderate antifungal activity, suggesting their potential for combined use with other antifungal agents. Thyme and oregano oils exhibited limited antifungal effects [
74,
75], particularly against
Fusarium oxysporum and
Trichoderma pseudokoringii, highlighting the need for higher concentrations or combinations with other treatments.
5. Conclusions
Our findings showed that the characterization of the fungus by order of severity identified Penicillium expansum, Trichoderma pseudokoringii, Fusarium oxysporum, and Aspergillus flavus as the fungus affecting Musa paradisiaca during postharvest. The in vitro analysis of essential oil efficacy revealed that Penicillium expansum was controlled by 400 ppm of cinnamon, oregano, and thyme essential oils. Trichoderma pseudokoringii was inhibited by 200 ppm of oregano, 400 ppm of clove and thyme, and 600 ppm of cinnamon essential oils. Fusarium oxysporum was most effectively managed with 200 ppm of cinnamon and thyme and 400 ppm of oregano essential oils. Aspergillus flavus was controlled by 200 ppm of oregano, and 400 ppm of cinnamon, clove, and thyme essential oils.
These findings suggest that essential oils, from cinnamon, clove, oregano, and thyme, are promising natural alternatives to synthetic fungicides. They offer an eco-friendly solution for managing fungal infections in bananas. Future research should focus on optimizing their application in agricultural systems and investigating their synergistic effects with other control methods to enhance their antifungal efficacy
Figure 1.
Macroscopy images of (a) Trichoderma spp, (b) Penicillium spp., (c) Aspergillus spp., (d) Fusarium spp., considering the appearance of the front side and (e) Trichoderma spp., (f) Penicillium spp., (g) Aspergillus spp. (h) Fusarium spp. of reverse side.
Figure 1.
Macroscopy images of (a) Trichoderma spp, (b) Penicillium spp., (c) Aspergillus spp., (d) Fusarium spp., considering the appearance of the front side and (e) Trichoderma spp., (f) Penicillium spp., (g) Aspergillus spp. (h) Fusarium spp. of reverse side.
Figure 4.
Fungal growth (cm) during 6 weeks in 20 banana samples inoculated with Trichoderma spp, Penicillium spp., Aspergillus spp., and Fusarium spp., stored at 13°C and 95% HR approximately.
Figure 4.
Fungal growth (cm) during 6 weeks in 20 banana samples inoculated with Trichoderma spp, Penicillium spp., Aspergillus spp., and Fusarium spp., stored at 13°C and 95% HR approximately.
Figure 5.
In vitro analysis of Trichoderma spp. in PDA medium supplied with basil, cinnamon, clove, oregano, rosemary, and thyme essential oils at various concentrations of 200, 400, 600, 800, and 1000 ppm (n=12).
Figure 5.
In vitro analysis of Trichoderma spp. in PDA medium supplied with basil, cinnamon, clove, oregano, rosemary, and thyme essential oils at various concentrations of 200, 400, 600, 800, and 1000 ppm (n=12).
Figure 6.
In vitro analysis of Penicillium spp. in PDA medium supplied with basil, cinnamon, clove, oregano, rosemary, and thyme essential oils at various concentrations of 200, 400, 600, 800, and 1000 ppm (n=12).
Figure 6.
In vitro analysis of Penicillium spp. in PDA medium supplied with basil, cinnamon, clove, oregano, rosemary, and thyme essential oils at various concentrations of 200, 400, 600, 800, and 1000 ppm (n=12).
Figure 7.
In vitro analysis of Aspergillus spp. in PDA medium supplied with basil, cinnamon, clove, oregano, rosemary, and thyme essential oils at various concentrations of 200, 400, 600, 800, and 1000 ppm (n=12).
Figure 7.
In vitro analysis of Aspergillus spp. in PDA medium supplied with basil, cinnamon, clove, oregano, rosemary, and thyme essential oils at various concentrations of 200, 400, 600, 800, and 1000 ppm (n=12).
Figure 8.
In vitro analysis of Fusarium spp. in PDA medium supplied with basil, cinnamon, clove, oregano, rosemary, and thyme essential oils at various concentrations of 200, 400, 600, 800, and 1000 ppm (n=12).
Figure 8.
In vitro analysis of Fusarium spp. in PDA medium supplied with basil, cinnamon, clove, oregano, rosemary, and thyme essential oils at various concentrations of 200, 400, 600, 800, and 1000 ppm (n=12).
Figure 9.
Fungal growth inhibition according to Essential Oils concentration.
Figure 9.
Fungal growth inhibition according to Essential Oils concentration.
Table 3.
Evaluation of antifungal activity in vitro of Trichoderma spp., Penicillium spp., Aspergillus spp. and Fusarium spp. use essential oil of oregano, rosemary, clove, thyme, cinnamon and basil.
Table 3.
Evaluation of antifungal activity in vitro of Trichoderma spp., Penicillium spp., Aspergillus spp. and Fusarium spp. use essential oil of oregano, rosemary, clove, thyme, cinnamon and basil.