1. Introduction
Selecting the right journal for a scientific manuscript remains a critical step for researchers, regardless of their experience level [
1]. A misaligned journal choice can lead to delayed publication, immediate rejection, and reduced visibility within the most relevant research communities [
2]. Even seasoned authors sometimes grapple with overlapping scopes and emerging journal niches, while early-career researchers or those working on cross-disciplinary topics often lack clear criteria for identifying the most suitable outlets. The continuous creation of new journals and the evolution of established ones make it even more difficult for authors to keep pace with this shifting array of publication venues [
3]. These issues make the case for data-driven systems that can automate and streamline the journal selection process, thereby reducing guesswork, saving time, and potentially improving the likelihood of a favorable peer-review outcome [
4].
Historically, automated journal recommendation systems have relied on traditional information retrieval and machine learning techniques [
5]. Early approaches, such as bag-of-words models and term frequency–inverse document frequency (TF–IDF [
6]), primarily matched manuscripts to journal scopes based on term co-occurrences and document similarity. For instance, Wang et al. [
7] applied softmax regression with TF–IDF-based feature selection to recommend computer science journals, demonstrating modest accuracy. Other studies employed n-gram classification for textual analysis [
8] or combined stylometric features with collaborative filtering [
9] to identify suitable journals. However, these methods often struggled to capture the semantic richness of scientific texts, especially in fields with rapidly evolving jargon or interdisciplinary overlap.
Various commercial platforms, including those offered by well-known publishing companies such as Springer and Elsevier, aim to assist authors in selecting journals. These tools typically rely on keyword-based matching or limited datasets, which can restrict their applicability across diverse research areas. Beel et al. [
10] highlighted key limitations of these platforms, such as their reliance on proprietary data, lack of transparency in recommendation algorithms, and inability to effectively handle nuanced linguistic variations or semantic relationships.
The advent of Transformer-based language models, such as BERT and its variants [
11,
12], has revolutionized natural language processing by capturing more nuanced, context-sensitive relationships among words. Sentence Transformer models, specifically designed for semantic similarity tasks, have emerged as powerful tools for comparing scientific texts [
13]. These models generate dense semantic embeddings that preserve meaningful relationships in a continuous vector space, enabling robust similarity matching even when different terms describe the same concept [
14]. Applications of such embeddings range from semantic search and clustering to question answering and recommender systems (Arora et al., 2020).
In journal recommendation tasks, Transformer embeddings offer significant advantages over traditional methods. Recent studies, including Michail et al. [
15], have shown that models such as Sentence Transformers can outperform earlier techniques by encoding article abstracts into high-dimensional vectors and comparing them via cosine similarity. These approaches have proven effective across a variety of domains, from computer science to biomedicine, where capturing the thematic nuances of a manuscript is essential for identifying appropriate journals [
14].
Despite this potential and all the pragmatic hurdles described above, a key question remains: which Sentence Transformer model best captures domain-specific semantic nuances for journal recommendation [
16]? Existing models differ in training objectives, corpora, and parameter settings [
17]. Domain-general models—such as all-mpnet-base-v2—often excel at broad semantic similarity, although they are not specifically tuned to scientific literature. Alternatively, compact models like all-MiniLM-L6-v2 promise faster runtime but may sacrifice some representational depth. Given these trade-offs, a comparative study of multiple models is needed to determine which embedding approach yields the most accurate, context-sensitive journal recommendations in a biomedical setting.
To address this issue, we propose an automated journal recommendation pipeline for the Life Sciences and systematically evaluate five Sentence Transformer models: all-mpnet-base-v2, all-MiniLM-L6-v2, all-MiniLM-L12-v2, multi-qa-distilbert-cos-v1, and all-distilroberta-v1. We started with test articles, which we used to extract keywords through KeyBERT [
18]. We then queried PubMed [
19] with these KeyBERT-derived keywords to retrieve custom corpora of potentially related articles. We then encoded the titles+abstracts of these articles and the test manuscripts into embeddings, calculate their cosine similarity scores, and generate a data-driven list of recommended journals. In addition to comparing the models’ semantic alignment, we assessed their computational efficiency, domain coverage, and alignment with expert-driven expectations.
2. Materials and Methods
2.1. Data Collection
We created a pool corpus of scientific articles by querying PubMed using NCBI E-utilities. To retrieve a representative dataset, we issued queries based on selected keywords derived from test articles (e.g., “clostridium AND infection AND difficile”). For each query, we retrieved article PMIDs. We then randomly sampled the retrieved PMIDs to create a manageable pool size of up to 5,000 articles per test case. Metadata for each article, including PMID, Title, Abstract, and Journal, were retrieved using Entrez.efetch and stored in a pandas DataFrame [
20]. To avoid data leakage, the test article’s PMID was explicitly excluded from the pool corpus.
2.2. Preprocessing and Keyword Extraction
For each test article, we extracted its Title and Abstract. A keyword extraction step was performed using KeyBERT to identify three salient keywords representing the article’s core content. These keywords guided PubMed queries to build a dynamically tailored pool corpus for each test article. The number of keywords (n=3) was empirically chosen to balance corpus size and thematic relevance. This procedure was repeated for multiple test articles (50 in total) to assess model performance on a broader scale.
2.3. Embedding Models
We employed five Sentence Transformer models to encode textual data into high-dimensional vector embeddings: all-mpnet-base-v2 (mpnet), all-MiniLM-L6-v2 (minilm-l6), all-MiniLM-L12-v2 (minilm-l12), multi-qa-distilbert-cos-v1 (multi-qa-distilbert), and all-distilroberta-v1 (roberta). For each article, we concatenated its Title and Abstract into a single string and used the model.encode() method to generate embeddings. All embeddings were computed on Google Colab GPUs.
2.4. Similarity Computation
Cosine similarity was calculated between the test article embedding and all embeddings in the pool corpus. Using PyTorch’s torch.topk [
21], we identified the top 10 most similar articles for each model, recording their associated PMIDs and journals.
Alternatively, articles were grouped by journal, and a mean embedding was calculated for each journal by averaging the embeddings of all articles belonging to that journal. The cosine similarity between the test article embedding and each journal’s mean embedding was then computed, yielding a ranked list of journals by average thematic alignment.
2.5. Evaluation Metrics
We further quantified the diversity and concentration of recommendation scores using two complementary metrics: Shannon entropy and the Gini coefficient.
2.5.1. Shannon Entropy
Shannon entropy is widely used in information theory to measure the diversity or “spread” of a distribution [
22]. We applied Shannon entropy to the cosine similarity scores of the top recommended articles for each test case, aiming to capture how evenly the model distributed its top similarity scores among different articles or journals. Formally, for a given model M and a test article t, let {p
i} be the normalized similarity scores (so that ∑
ip
i=1) of the top N recommended items, where p
i=
and s
i is the raw similarity for item i. Shannon entropy was calculated using the following formula [
23]:
H(X)=p(xi)log2 p(xi)
A higher entropy indicates a more even distribution of similarity scores—i.e., no single article or journal dominates—while a lower entropy suggests one or a few items have much higher similarity scores than the rest.
2.5.2. Gini coefficient
The Gini coefficient is commonly used in economics to assess inequality within a distribution. We employed it here to measure how concentrated the similarity scores are among top-ranked articles or journals. Following the procedure in Dorfman [
24], the Gini coefficient G ranges from 0 (perfect equality in similarity scores) to 1 (perfect concentration in one or a few items). Conceptually, a low Gini coefficient implies that the model assigns relatively similar similarity scores across its top recommendations, whereas a higher Gini coefficient indicates that only a small subset of articles or journals receive distinctly higher similarity scores.
2.6. Software and Environment
All code was implemented in Python (version 3.10) within a Google Colab environment [
25]. The following primary libraries were used:
All code was executed under Google Colab T4 GPU runtime configurations, respecting NCBI E-utilities usage policies (including Entrez.email specification and rate-limiting queries).
3. Results
3.1. Case study
In this study, we implemented a pipeline designed to recommend journals for a single test article by dynamically generating a corpus of potentially relevant PubMed articles. Our approach comprised four major steps: (1) extracting keywords from the test article’s title and abstract using KeyBERT, (2) issuing an ad hoc PubMed query to gather a custom pool of papers, (3) encoding the resulting pool with Sentence Transformers to create high-dimensional embeddings, and (4) computing similarity between the test article embedding and the pool embeddings.
To this purpose, we set out to evaluate and compare the recommendations produced by five Sentence Transformer models— all-mpnet-base-v2, all-MiniLM-L6-v2, all-MiniLM-L12-v2, multi-qa-distilbert-cos-v1, and all-distilroberta-v1—.
To get a general overview of the performance of these models, we randomly chose a test article from a corpus of 10000 scientific articles randomly retrieved from PubMed: “
Problems of Clostridium difficile infection (CDI) in Polish healthcare units”, published on Annals of agricultural and environmental medicine: AAEM [
31].
We began by applying KeyBERT, an unsupervised keyword extraction tool, on the article’s title and abstract. We thus identified the three most representative keywords describing the article’s core content.
Based on these three keywords—clostridium, infection, and difficile—we conducted a PubMed query to locate potentially relevant articles, by joining the keywords with the AND operator. The number of keywords (n=3) was empirically determined to retrieve a sufficiently large corpus, without excessively restricting the query. PubMed returned 15,466 candidate articles matching the query “clostridium AND infection AND difficile”. For computational feasibility, we randomly selected 5,000 articles from among those 15,466. We then ensured that our test article’s own PMID was not included in this pool, so as not to artificially inflate similarity scores.
Once we generated the 5,000-article corpus, we extracted the title, abstract, and journal fields from each article using Biopython’s Entrez utilities. The goal was to measure two distinct but complementary aspects of recommendations:
Article-Level Similarity: For each model, we sought the top ten articles (out of those 5,000) most similar to the test article and identified the journals where they were published.
Journal-Level Similarity: We computed the average similarity of each journal’s articles to the test article, then ranked the journals accordingly.
To ensure consistency, each model was applied systematically. First, the model encoded all 5,000 articles, concatenating their titles and abstracts into a single text input per article and transformed the text into embeddings. Next, the same model encoded the test article’s title and abstract. By comparing each article’s embedding with the test article’s embedding using cosine similarity, we obtained a comprehensive list of similarity scores. The top ten articles with the highest scores were treated as the
closest articles and their journal can be found in
Table 1 (in the Top Article column). Simultaneously, we grouped articles by their journals and computed a mean embedding per journal—by averaging the embeddings of all articles belonging to that journal. A second round of similarity checks identified the top ten journals with the highest average similarity to the test article (column Top Avg Journal in
Table 1).
A preliminary comparison of computation time shows the great disparity between mpnet, which required a minute and a half to encode all the 5000 titles and abstracts of the dataset, and the minilm-l6 and minilm-l12 models, which only took 9 seconds to encode all 5000 titles and abstracts.
3.1.1. Article-Level Findings
Each model produced a list of ten articles deemed most semantically similar to the test article. A list of the journals where these articles appeared can be found as Supplementary
Table 1. In general, all models identified articles published in journals that focused on infectious diseases, hospital-acquired infections, or microbial pathogens, confirming that the basic textual content was indeed guiding the retrieval. As shown in
Table S1, mpnet consistently produced the highest similarity scores across its top ten recommendations, with articles often exceeding 0.85, and multiple entries from journals such as the
International Journal of Environmental Research and Public Health and the
Journal of Preventive Medicine and Hygiene. “Hospital management of Clostridium difficile infection: a review of the literature”[
32] was mpnet’s suggestion as closest article, with similarity=90.62. The overall data suggest mpnet's robustness in identifying journals strongly aligned with the test article’s thematic content, particularly those related to public health and infection control. Minilm-l6 and minilm-l12 exhibited overlapping trends in their recommendations, with journals such as the
Journal of Hospital Infection and the
International Journal of Environmental Research and Public Health frequently appearing in their top lists. However, minilm-l12 (top choice: “Clostridium difficile infection perceptions and practices: a multicenter qualitative study in South Africa.”[
33], similarity=86.5) often returned higher maximum similarity scores than minilm-l6 (top choice: “The Burden of Clostridium Difficile (CDI) Infection in Hospitals, in Denmark, Finland, Norway And Sweden.”[
34], similarity=82.3) for the same journal, suggesting that the additional model complexity of minilm-l12 might provide a marginally better thematic alignment.
The multi-qa-distilbert model mostly aligned with the results by the other models (top choice: “Ten-year review of Clostridium difficile infection in acute care hospitals in the USA, 2005-2014.”[
35]), although also diverged from them by identifying broader healthcare-related journals, such as the
European Review for Medical and Pharmacological Sciences and the
Bulletin of Mathematical Biology. Despite this, multi-qa-distilbert consistently included journals like the
Journal of Hospital Infection, indicating its capacity to recognize core infectious disease topics within its broader thematic spectrum. The roberta model demonstrated a strong performance, particularly with journals addressing clinical microbiology and infection control, such as
The Journal of Hospital Infection (87.07 similarity) and
International Journal of Environmental Research and Public Health. Noticeably, roberta identified the same top choice article as mpnet: “Hospital management of Clostridium difficile infection: a review of the literature”[
32].
3.1.2. Journal-Level Suggestions
To get a broader view on the dataset, we calculated the average similarity of each journal’s articles to the test article. For every journal in the 5,000-article pool, we averaged all embeddings belonging to that journal, then measured how close that average embedding was to the test article’s embedding. The results can be found in Supplementary
Table 2. Mpnet consistently identified journals strongly aligned with infectious diseases, hospital-acquired infections, and antimicrobial resistance. Its top-ranked journals, such as the
International Journal of Environmental Research and Public Health and the
Journal of Preventive Medicine and Hygiene, demonstrated high average similarity scores, with values exceeding 92% for the leading journal, and over 86% similarity for
Infection, disease & health, which ranks 10
th and last for this model. This pattern confirms mpnet's capability to prioritize thematically congruent journals with precise alignment.
Similarly, minilm-l12 showed a strong (but lower than mpnet’s and always lower than 86%) affinity for journals relevant to infection control and epidemiology, with its highest-ranked journal also being the International Journal of Environmental Research and Public Health with 85.2% similarity. Notable overlap was observed between mpnet and minilm-l12, particularly in their top journal selections, which included titles such as the International Journal of Environmental Research and Public Health, Journal of preventive medicine and hygiene, American Journal of Infection Control and The Journal of Hospital Infection. This confirms that these models share a nuanced ability to identify core journals within the test article's domain.
Minilm-l6, while maintaining a comparable thematic focus, included a broader range of journals such as Emerging Infectious Diseases and Frontiers in Public Health. Its slightly lower average similarity scores compared to mpnet and minilm-l12 (always lower than 85%) highlight a broader distribution of relevance, potentially indicating a greater inclusion of interdisciplinary journals. Similarly, multi-qa-distilbert displayed a tendency toward multidisciplinary recommendations. Journals such as Risk Management and Healthcare Policy and Journal of Global Health appeared prominently in its rankings. Finally, roberta showcased a balanced profile, with its top-ranked journal being the International Journal of Environmental Research and Public Health. While there was some overlap with the other models, such as its inclusion of Przeglad Epidemiologiczny and The Journal of Hospital Infection, roberta also included journals like Journal of Global Health and Minerva Medica.
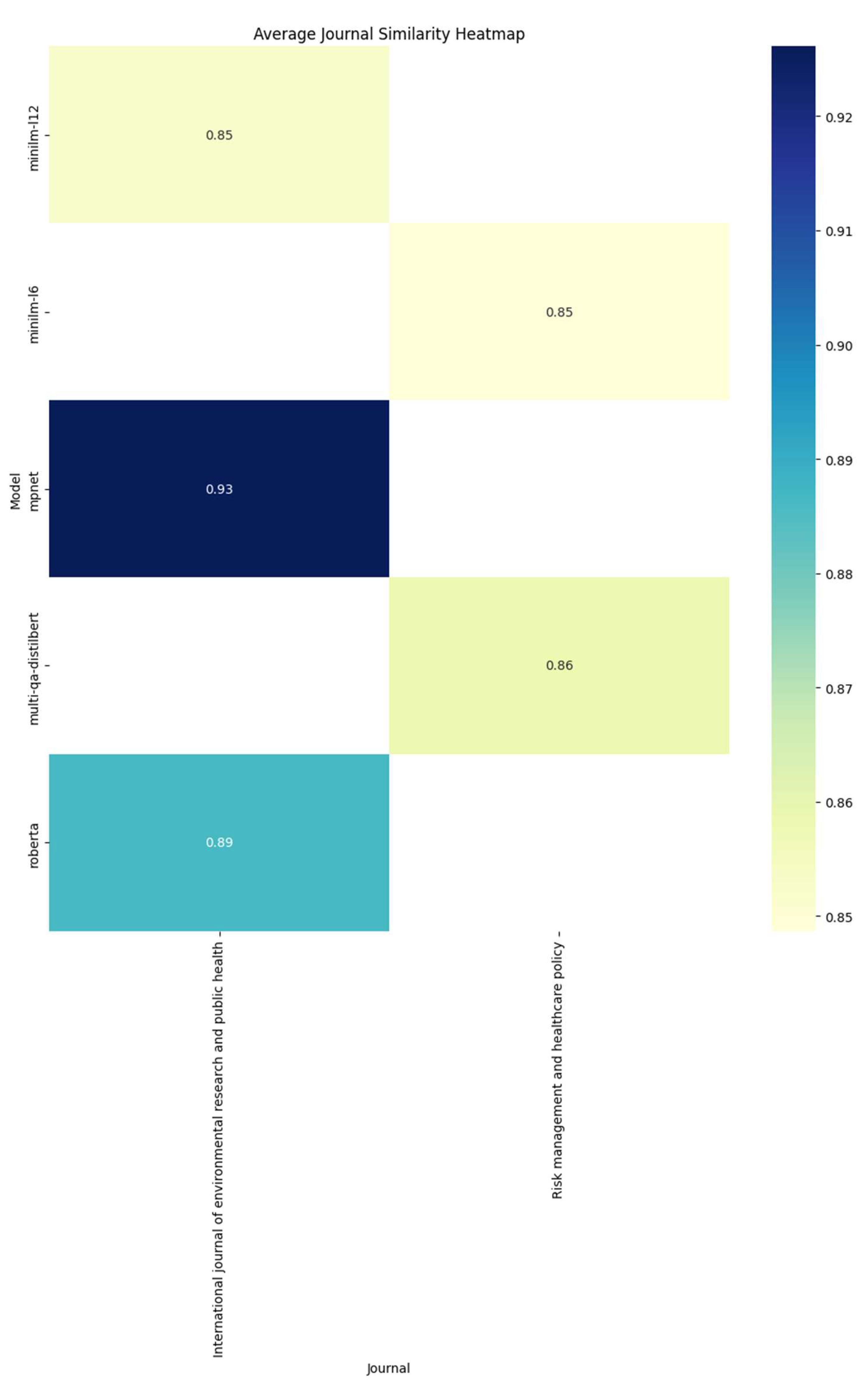
Figure 1 summarizes these findings, highlighting how the models identify 2 journals as their most probable candidate, the
International Journal of Environmental Research and Public Health for mpnet, minilm-l12 and roberta, and
Risk Management and Healthcare Policy for minilm-l6 and multi-qa-distilbert. Interestingly, none of the models correctly identified the actual journal where the test article was published.
3.1.3. Overlap Among Model Recommendations
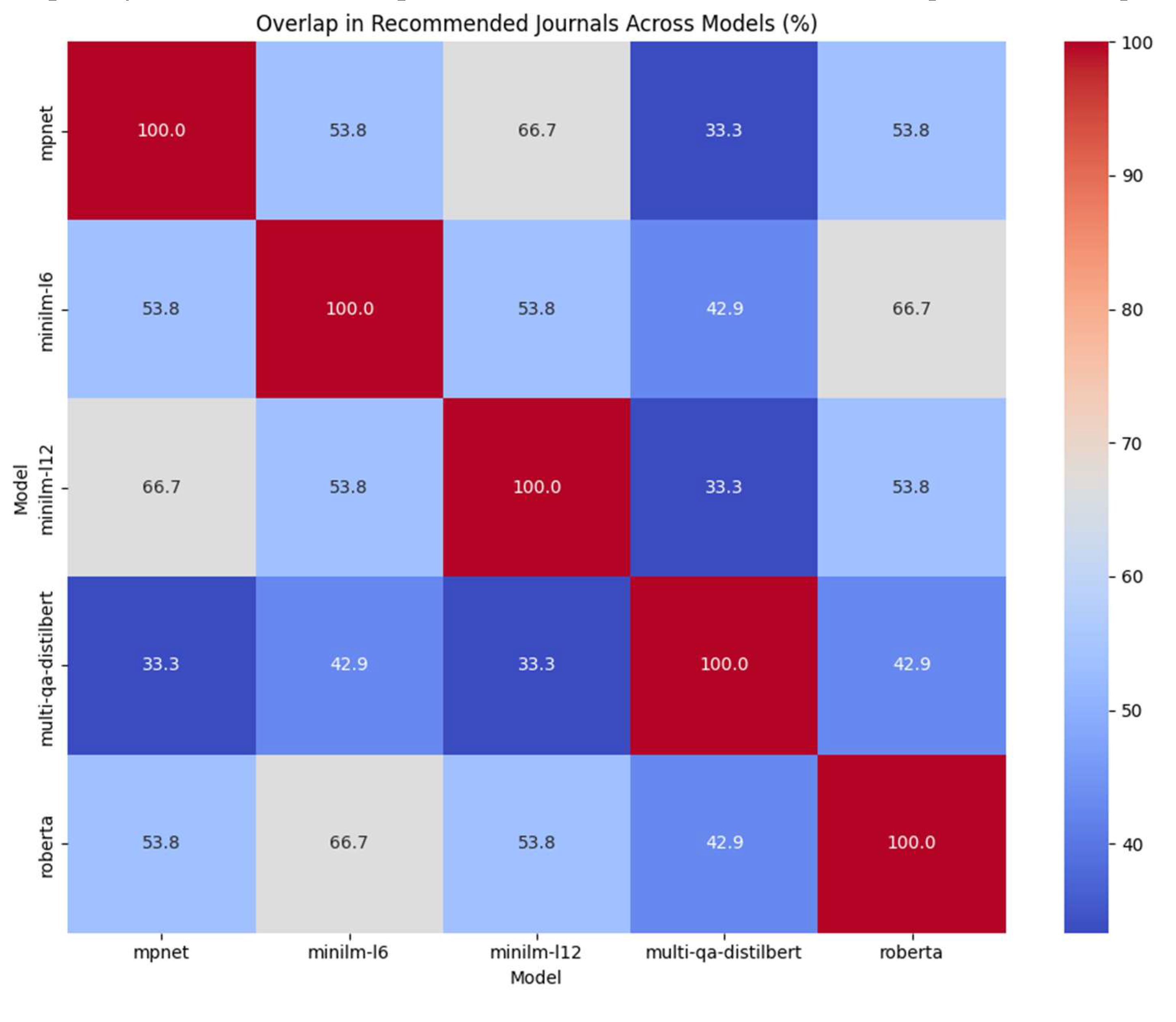
To quantify the overlap in recommended journals, we calculated the set intersection and union of the top ten journals from each pair of models, as calculated according to the average similarity.
A heatmap displayed the percentage overlap, where the color scale ranged from minimal overlap (~30%) to substantial overlap (~60–70%) for certain model pairs; mpnet and minilm-l12 exhibited notably higher overlap, with 66.7% of their recommended journals in common. Multi-qa-distilbert frequently diverged, sharing less than 35% with minilm-12 and mpnet, suggesting it captures text semantics or research topics differently. Minilm-l6 yielded intermediate high overlap percentages with roberta (66.7%)— with also a relatively high alignment with mpnet or minilm-l12 (53.8%).
3.2. Quantitative comparison
We then extended our initial analysis of a single test article to encompass a broader evaluation involving 50 distinct test articles (
Table S3), each randomly selected from the initial test dataset. This expansion aimed to provide a more generalized understanding of how various Sentence Transformer models performed in identifying thematically similar articles and recommending appropriate journals for publication. For each test article, we employed KeyBERT to extract the top three keywords, which were subsequently used to query PubMed, retrieving up to 20,000 PMIDs per query. To maintain computational efficiency and ensure manageable data processing, we randomly sampled 5,000 PMIDs (if available) from each pool, carefully excluding the test article’s own PMID if present. The extracted articles were then consolidated into a DataFrame, ensuring the removal of duplicate entries based on PMID, resulting in a final pool size of maximum 5,000 unique articles per test case.
Each of the five Sentence Transformer models was systematically applied to encode both the pooled articles and the respective test articles. For every model, we identified the top 10 most similar articles based on these scores. Additionally, we aggregated the similarity scores at the journal level by computing the average similarity of articles within each journal to the test article, thereby ranking journals according to their thematic alignment with the test case.
The analysis yielded a comprehensive set of metrics for each model across all 50 test articles. Shannon entropy was calculated to assess the diversity of similarity scores among the top-ranked articles, with higher entropy values indicating a more even distribution of similarities and lower values suggesting dominance by one or a few highly similar articles. The maximum similarity score within the top 10 was recorded to measure the peak alignment each model achieved with the most similar article. Furthermore, we evaluated whether the actual journal of the test article was included within the top 10 recommended journals based on average similarity, providing a binary measure of each model’s effectiveness in correctly identifying the appropriate publication venue.
Aggregating the results across all test articles (
Table 2), we observed that mpnet consistently demonstrated the highest mean similarity scores, averaging 0.71 ± 0.04, indicating a strong ability to identify journals closely aligned with the test articles’ themes. This was followed by roberta, with an average mean similarity of 0.68 ± 0.04, further showing its proficiency in aligning with relevant journals. The minilm-l12 and minilm-l6 models displayed comparable performance, achieving mean similarity scores of 0.67 ± 0.04 and 0.66 ± 0.04, respectively, reflecting a solid alignment with test articles but slightly lower than that of mpnet and roberta. Conversely, multi-qa-distilbert exhibited the lowest mean similarity score of 0.63 ± 0.04, indicating a more moderate alignment with the test articles.
Additionally, mpnet achieved the highest Top Article similarity score of 0.83, emphasizing its capacity to match individual articles very closely to the test articles’ themes. Roberta followed with a similarity of 0.80, while minilm-l12, minilm-l6, and multi-qa-distilbert achieved Top Article similarity scores of 0.79, 0.77, and 0.76, respectively.
The JournalInTop10 metric revealed that mpnet and minilm-l6 were the most proficient in correctly identifying the actual journal of the test article within their top 10 recommendations, achieving inclusion rates of 14.5%. This was closely followed by minilm-l12, roberta, and multi-qa-distilbert, all of which had inclusion rates of 12.5%.
All models exhibited identical Shannon entropy values of approximately 3.24, suggesting that the similarity scores were evenly distributed across their recommendations. This low variability reflects a tendency to concentrate recommendations among a few top-ranked journals rather than distributing them broadly. The Gini coefficients were uniformly low across all models, with minilm-l12 showing the highest value of 0.0361, reflecting slightly higher concentration of similarity scores among its top recommendations. Mpnet and roberta followed closely, with Gini values of 0.03 and 0.0336, respectively, indicating a relatively even distribution of similarity scores across their top journals.
4. Discussion
The present study used a simple pipeline for automated journal recommendation as a tool to evaluate the performance of 5 freely available sentence transformer models. The study tested all-mpnet-base-v2, all-MiniLM-L6-v2, all-MiniLM-L12-v2, multi-qa-distilbert-cos-v1, and all-distilroberta-v1 against one randomly chosen test article or across a dataset of equally random 50 test articles, examining the ability of these models to match manuscripts with thematically aligned journals. Our findings support the notion that Transformer-based text embeddings can outperform earlier approaches such as bag-of-words or TF–IDF in capturing nuanced thematic overlaps between manuscripts and journals.
Nevertheless, this approach rests on a key assumption: namely, that a potential publication venue for an article would be chosen primarily based on thematic alignment, which is clearly not always the case. The final decision on where to submit a manuscript often involves additional considerations. Factors such as journal impact factor, acceptance rate, publication costs, and the expected turnaround time for reviews frequently influence authors’ choices [
36]. Moreover, authors might initially aim for more prestigious journals and use automated recommendations only after facing rejection from their preferred venues [
37]. For the sake of our investigation, however, we only considered the scenario where the scholar needs to identify journals where similar articles have been recently published.
Our pipeline relied on Transformer-based embeddings, which are numerical representation of sentence semantics. Embeddings allow for easy comparison between the meaning of 2 or more sentences [
38,
39,
40,
41]. Our two-level evaluation (article-level versus journal-level) underscores how embedding-based similarity can help authors in different scenarios. By highlighting specific articles within a journal, the pipeline enables researchers to assess how closely their manuscript aligns with previously published work. When aggregating at the journal level, the system offers a broader view of which venues consistently publish content resembling the manuscript’s themes.
Consistently with our previous findings [
16], mpnet consistently demonstrated the highest performance among the models, while requiring the longest time to encode the articles. When tested against 50 randomly chosen articles, mpnet achieved the highest average similarity score (MeanSim: 0.71 ± 0.04) and the highest maximum similarity (MaxSimilarity: 0.83) across test articles, underscoring its strong ability to identify journals closely aligned with thematic content. Additionally, mpnet displayed a relatively higher JournalInTop10 inclusion rate (14.5%) as compared to its competitors, indicating its focused recommendations sometimes aligned with the actual test article’s journal.
Minilm-l12 also performed strongly, achieving a MeanSim score of 0.67 ± 0.04 and a MaxSimilarity of 0.79, with a much faster processing time than mpnet. While slightly lower than mpnet, its recommendations demonstrated notable thematic alignment, particularly with journals related to infectious diseases and epidemiology. Similarly, minilm-l6 achieved a MeanSim score of 0.66 ± 0.04 and matched mpnet in JournalInTop10 inclusion rate (14.5%), despite its smaller architecture. This model (just like minilm-l6) creates 328-long embeddings, which are computationally lighter and require less computational power. Its lightweight design is therefore well-suited for scenarios requiring real-time processing or computational efficiency, without substantial sacrifices in recommendation quality.
While mpnet yielded the highest domain-specific alignment, roberta also scored highly (MeanSim: 0.68), and slightly above minilm-l12 (0.67). However, roberta included more interdisciplinary journals in its top recommendations, suggesting it might strike a balance between precision and broader thematic coverage. By contrast, multi-qa-distilbert displayed the lowest average similarity (0.63), reflecting its generalist nature and possibly broader, policy-related scope. This diversity might be advantageous for interdisciplinary manuscripts or exploratory research, where thematic breadth is prioritized over narrow precision, and future research should address this issue, to optimize the use of this model in the biomedical field.
The metrics we used provided additional insights into the recommendation dynamics. Minilm-l12 exhibited slightly higher Gini coefficients (0.036), indicating a greater concentration of similarity scores among top-ranked journals. In contrast, mpnet had lower Gini coefficients (0.029), reflecting a more equitable distribution of similarity scores across broader journal recommendations. These patterns highlight the trade-offs between precision and diversity among the models.
One intriguing result is that none of the models in our case study identified the actual journal in which the test article was published. Although the recommended titles were thematically relevant, they did not match the manuscript’s real publication venue. This outcome may reflect the granularity of journal scopes or the sheer variability in editorial decisions, or even the very simple absence of the journal from the dynamically generated dataset. The mismatch, however, highlights the importance of considering factors beyond semantic alignment, including a journal’s editorial preferences, the manuscript’s novelty or methodological approach, and even the presence of special issues or calls for papers. It also underscores a possible avenue for future improvement: incorporating editorial or reviewer information, acceptance rates, or historical publication patterns might increase the likelihood of recommending the journal that ultimately published the manuscript.
From a methodological standpoint, our use of KeyBERT for keyword extraction and random sampling of up to 5,000 articles per test case demonstrates how a scalable NLP pipeline can be easily assembled with open-source tools. However, future work might expand upon this by integrating advanced query expansion techniques or alternative keyword extraction models that leverage domain-specific ontologies (e.g., MeSH terms for biomedical research).
Overall, the study demonstrates the strengths and limitations of Transformer-based models for automated journal recommendation. While mpnet excels in thematic precision, multi-qa-distilbert and roberta offer broader recommendations potentially suited for interdisciplinary research with competitive computational speed.
5. Conclusions
Our pipeline for automated journal recommendation, based on Sentence Transformer embeddings and PubMed metadata, explores the potential of advanced NLP techniques in addressing a well-known pain in academic publishing. By evaluating multiple models, we showed that no single embedding architecture is universally superior; rather, performance varies based on factors such as domain specificity, computational efficiency, and the breadth of recommendations. Mpnet consistently outperformed the other models in similarity, with roberta close behind in mean similarity. Both minilm-l12 and minilm-l6 offered solid performance, while excelling in more limited computational demands. Meanwhile, multi-qa-distilbert displayed the broadest range of journal recommendations, potentially benefiting interdisciplinary research. These differences underscore the importance of model selection according to the specific needs of authors and institutions.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Table S1: This table provides the list of article-level journal recommendations generated by the five different Sentence Transformer models tested when applied to the test article; Table S2: This table provides the list of journal-level journal recommendations generated by the five different Sentence Transformer models tested when applied to the test article; Table S3: This table contains the list of the 50 test articles used for quantitative appraisal of the model performance.
Author Contributions
Conceptualization, C.G., M.M. and E.C..; methodology, C.G.; software, C.G.; formal analysis, C.G. and M.T.C.; data curation, S.G. and M.T.C.; writing—original draft preparation, C.G. and M.M.; writing—review and editing, S.G. and E.C.; All the authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available on request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Welch, S.J. Selecting the Right Journal for Your Submission. J Thorac Dis 2012, 4, 336–338. [Google Scholar] [CrossRef]
- Nicholas, D.; Herman, E.; Clark, D.; Boukacem-Zeghmouri, C.; Rodríguez-Bravo, B.; Abrizah, A.; Watkinson, A.; Xu, J.; Sims, D.; Serbina, G.; et al. Choosing the ‘Right’ Journal for Publication: Per-ceptions and Practices of Pandemic-era Early Career Researchers. Learned Publishing 2022, 35, 605–616. [Google Scholar] [CrossRef]
- Larsen, P.O.; von Ins, M. The Rate of Growth in Scientific Publication and the Decline in Coverage Provided by Science Citation Index. Scientometrics 2010, 84, 575–603. [Google Scholar] [CrossRef] [PubMed]
- Kreutz, C.K.; Schenkel, R. Scientific Paper Recommendation Systems: A Literature Review of Recent Publications. International Journal on Digital Libraries 2022, 23, 335–369. [Google Scholar] [CrossRef] [PubMed]
- Park, D.H.; Kim, H.K.; Choi, I.Y.; Kim, J.K. A Literature Review and Classification of Recommender Systems Research. Expert Syst Appl 2012, 39, 10059–10072. [Google Scholar] [CrossRef]
- Qaiser, S.; Ali, R. Text Mining: Use of TF-IDF to Examine the Relevance of Words to Documents. Int J Comput Appl 2018, 181, 25–29. [Google Scholar] [CrossRef]
- Wang, D.; Liang, Y.; Xu, D.; Feng, X.; Guan, R. A Content-Based Recommender System for Computer Science Publications. Knowl Based Syst 2018, 157, 1–9. [Google Scholar] [CrossRef]
- Medvet, E.; Bartoli, A.; Piccinin, G. Publication Venue Recommendation Based on Paper Abstract. In Proceedings of the 2014 IEEE 26th International Conference on Tools with Artificial Intelligence; IEEE, November 2014; pp. 1004–1010. [Google Scholar]
- Yang, Z.; Davison, B.D. Venue Recommendation: Submitting Your Paper with Style. In Proceedings of the 2012 11th International Conference on Machine Learning and Applications; IEEE, December 2012; pp. 681–686. [Google Scholar]
- Beel, J.; Gipp, B.; Langer, S.; Breitinger, C. Research-Paper Recommender Systems: A Literature Survey. International Journal on Digital Libraries 2016, 17, 305–338. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. Bert: Pre-Training of Deep Bidirectional Transformers for Language Understanding. arXiv preprint arXiv:, arXiv:1810.04805 2018.
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. Adv Neural Inf Process Syst 2017, 30. [Google Scholar]
- Liu, Q.; Kusner, M.J.; Blunsom, P. A Survey on Contextual Embeddings. arXiv preprint arXiv:, arXiv:2003.07278 2020.
- Noh, J.; Kavuluru, R. Improved Biomedical Word Embeddings in the Transformer Era. J Biomed Inform 2021, 120, 103867. [Google Scholar] [CrossRef] [PubMed]
- Michail, S.; Ledet, J.W.; Alkan, T.Y.; İnce, M.N.; Günay, M. A Journal Recommender for Article Submission Using Transformers. Scientometrics 2023, 128, 1321–1336. [Google Scholar] [CrossRef]
- Galli, C.; Donos, N.; Calciolari, E. Performance of 4 Pre-Trained Sentence Transformer Models in the Semantic Query of a Systematic Review Dataset on Peri-Implantitis. Information 2024, 15, 68. [Google Scholar] [CrossRef]
- Stankevičius, L.; Lukoševičius, M. Extracting Sentence Embeddings from Pretrained Transformer Models. Applied Sciences 2024, 14, 8887. [Google Scholar] [CrossRef]
- Issa, B.; Jasser, M.B.; Chua, H.N.; Hamzah, M. A Comparative Study on Embedding Models for Keyword Extraction Using KeyBERT Method. In Proceedings of the 2023 IEEE 13th International Conference on System Engineering and Technology (ICSET); IEEE; 2023; pp. 40–45. [Google Scholar]
- Jin, Q.; Leaman, R.; Lu, Z. PubMed and beyond: Biomedical Literature Search in the Age of Artificial Intelligence. EBioMedicine 2024, 100. [Google Scholar] [CrossRef]
- Mckinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the Proceedings of the 9th Python in Science Conference; van der Walt, S., Millman, J., Eds.; 2010; pp. 51–56.
- Paszke, A.; Gross, S.; Chintala, S.; Chanan, G.; Yang, E.; Facebook, Z.D.; Research, A.I.; Lin, Z.; Desmaison, A.; Antiga, L.; et al. Automatic Differentiation in PyTorch. In Proceedings of the 31st Conference on Neural Information Processing Systems; 2017. [Google Scholar]
- Godden, J.W.; Bajorath, J. Analysis of Chemical Information Content Using Shannon Entropy. Reviews in computational chemistry 2007, 23, 263–289. [Google Scholar]
- Vajapeyam, S. Understanding Shannon’s Entropy Metric for Information. arXiv preprint arXiv:, arXiv:1405.2061 2014.
- Dorfman, R. A Formula for the Gini Coefficient. Rev Econ Stat 1979, 61, 146. [Google Scholar] [CrossRef]
- Bisong, E. Google Colaboratory. In Building Machine Learning and Deep Learning Models on Google Cloud Platform: A Comprehensive Guide for Beginners; Bisong, E., Ed.; Apress: Berkeley, CA, 2019; ISBN 978-1-4842-4470-8. [Google Scholar]
- Chapman, B.; Chang, J. Biopython: Python Tools for Computational Biology. ACM Sigbio Newsletter 2000, 20, 15–19. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array Programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Chintala, S.; Chanan, G.; Yang, E.; DeVito, Z.; Lin, Z.; Desmaison, A.; Antiga, L.; Lerer, A. Automatic Differentiation in Pytorch. 2017.
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput Sci Eng 2007, 9. [Google Scholar] [CrossRef]
- Waskom, M. Seaborn: Statistical Data Visualization. J Open Source Softw 2021, 6. [Google Scholar] [CrossRef]
- Kiersnowska, Z.; Lemiech-Mirowska, E.; Ginter-Kramarczyk, D.; Kruszelnicka, I.; Michałkiewicz, M.; Marczak, M. Problems of Clostridium Difficile Infection (CDI) in Polish Healthcare Units. Annals of Agricultural and Environmental Medicine 2021, 28, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Khanafer, N.; Voirin, N.; Barbut, F.; Kuijper, E.; Vanhems, P. Hospital Management of Clostridium Difficile Infection: A Review of the Literature. Journal of Hospital Infection 2015, 90, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Legenza, L.; Barnett, S.; Rose, W.; Safdar, N.; Emmerling, T.; Peh, K.H.; Coetzee, R. Clostridium Difficile Infection Perceptions and Practices: A Multicenter Qualitative Study in South Africa. Anti-microb Resist Infect Control 2018, 7, 125. [Google Scholar] [CrossRef]
- Nordling, S.; Anttila, V.J.; Norén, T.; Cockburn, E. The Burden of Clostridium Difficile (CDI) Infection in Hospitals, in Denmark, Finland, Norway And Sweden. Value in Health 2014, 17, A670. [Google Scholar] [CrossRef]
- Luo, R.; Barlam, T.F. Ten-Year Review of Clostridium Difficile Infection in Acute Care Hospitals in the USA, 2005–2014. Journal of Hospital Infection 2018, 98, 40–43. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xie, J.; Sun, J.; Cheng, Y. Factors Affecting Authors’ Manuscript Submission Behaviour: A Systematic Review. Learned Publishing 2023, 36, 285–298. [Google Scholar] [CrossRef]
- Gaston, T.E.; Ounsworth, F.; Senders, T.; Ritchie, S.; Jones, E. Factors Affecting Journal Submission Numbers: Impact Factor and Peer Review Reputation. Learned Publishing 2020, 33, 154–162. [Google Scholar] [CrossRef]
- Worth, P.J. Word Embeddings and Semantic Spaces in Natural Language Processing. Int J Intell Sci 2023, 13, 1–21. [Google Scholar] [CrossRef]
- Yao, Z.; Sun, Y.; Ding, W.; Rao, N.; Xiong, H. Dynamic Word Embeddings for Evolving Semantic Discovery. WSDM, Proceedings of the 11th ACM International Conference on Web Search and Data Mining 2018, 2018-Febua, 673–681. [CrossRef]
- Si, Y.; Wang, J.; Xu, H.; Roberts, K. Enhancing Clinical Concept Extraction with Contextual Embed-dings. Journal of the American Medical Informatics Association 2019, 26, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, L.; Keith, B. A Systematic Literature Review on Word Embeddings. Proceedings of the Trends and Applications in Software Engineering: Proceedings of the 7th International Conference on Software Process Improvement (CIMPS 2018) 7Springer, 2019; pp. 132–141.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).