Submitted:
06 January 2025
Posted:
07 January 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Fungus Isolation
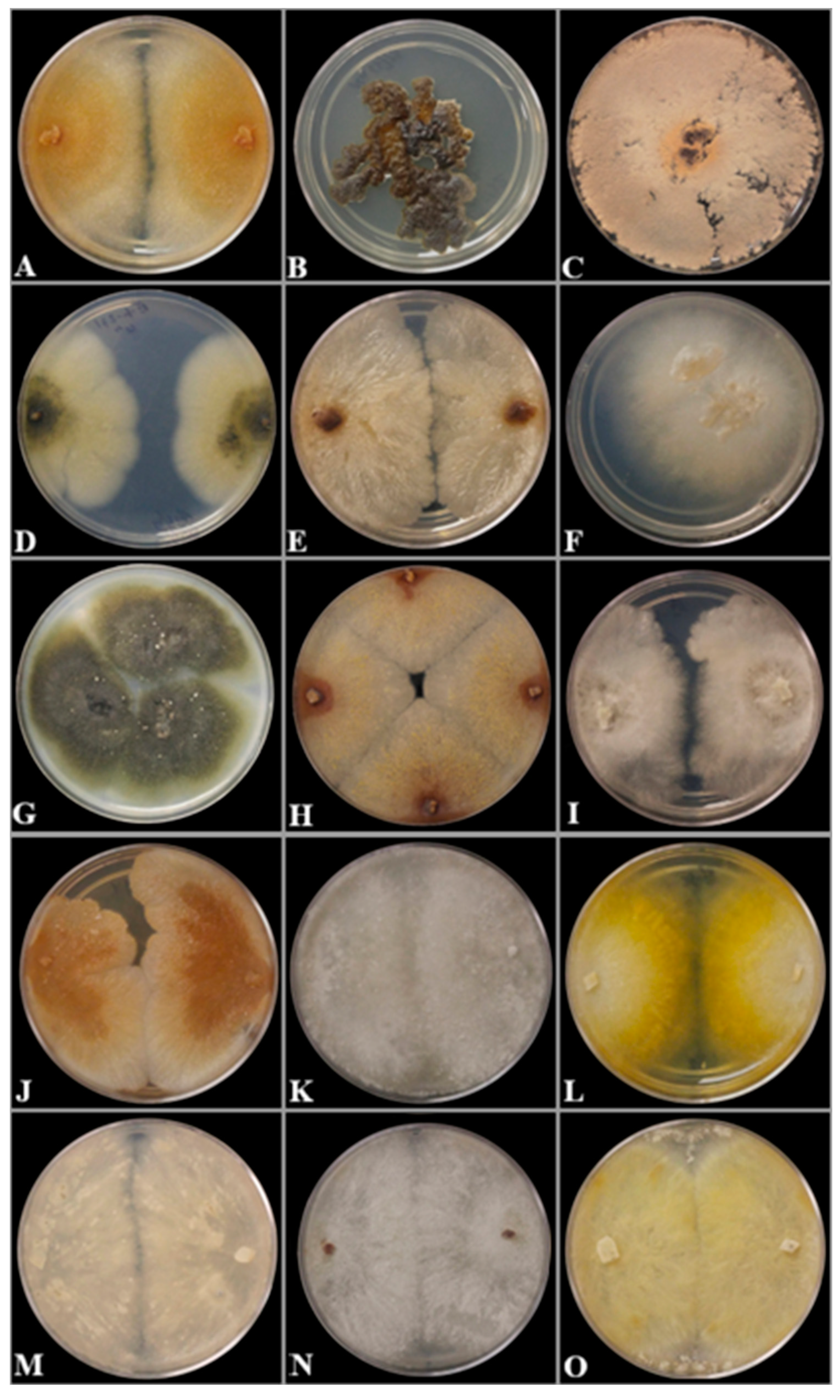
2.2. Morphological Characterization
2.3. DNA Extraction, PCR Amplification and Sequencing
2.4. Sequence Alignment and Phylogenetic Analysis
3. Results
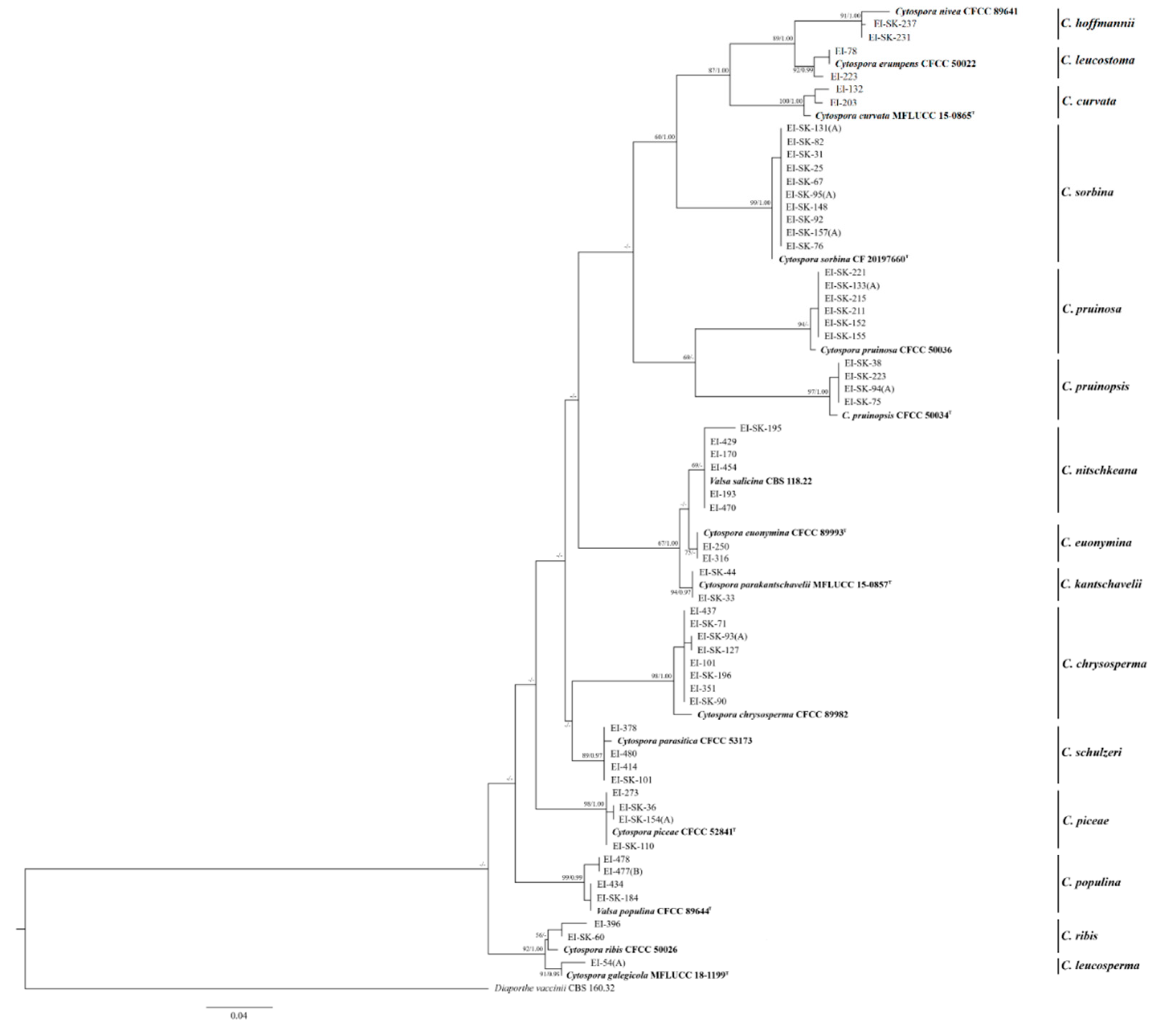
3.1. Phylogenetic Analysis
3.2. Taxonomy
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Casiraghi, M.; Labra, M.; Ferri E, Galimberti, A.; De Mattia, F. DNA Barcoding: A Six-Question Tour to Improve Users' Awareness about the Method. Brief Bioinform. 2010, 11, 440–453.
- Ćelepirović, N.; Novak Agbaba, S.; Karija Vlahović, M. DNA Barcoding of Fungi in the Forest Ecosystem of the Psunj and Papuk Mountains in Croatia. SEEFOR 2020, 11, 145–152.
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W. Fungal Barcoding Consortium; Fungal Barcoding Consortium Author List. Nuclear Ribosomal Internal Transcribed Spacer (ITS) Region as a Universal DNA Barcode Marker for Fungi. Proc Natl Acad Sci U S A. 2012, 109, 6241–6246.
- Pan, M.; Zhu, H.Y.; Tian, C.M.; Huang, M.R.; Fan, X.L. Assessment of Cytospora Isolates from Conifer Cankers in China, with the Descriptions of Four New Cytospora species. Front. Plant Sci. 2021, 12, 636460.
- Lin, L.; Pan, M.; Tian, C.; Fan, X. Fungal Richness of Cytospora Species Associated with Willow Canker Disease in China. J. Fungi 2022, 8, 377.
- Lin, L.; Fan, XL.; Groenewald, J.Z.; Jami, F.; Wingfield, M.J.; Voglmayer, H., Jaklitsch, W., Castlebury, L.A.; Tian, C.M.; Crous, P.W. Cytospora: an Important Genus of Canker Pathogens. Studies in Mycology 2024, 109, 323–401.
- Adams, G.C.; Wingfield, M.J.; Common, R.; Roux, J. Phylogenetic Relationships and Morphology of Cytospora Species and Related Teleomorphs (Ascomycota, Diaporthales, Valsaceae) from Eucalyptus. Stud. Mycol. 2005, 52, 1–147.
- Biggs, A.R. Integrated Approach to Controlling Leucostoma Canker of Peach in Ontario. Plant Dis. 1989, 73, 869–874.
- Adams, G.C.; Surve-Iyer, R.S.; Iezzoni, A.F. Ribosomal DNA Sequence Divergence and Group I Introns within the Leucostoma species L. cinctum, L. persoonii, and L. parapersoonii sp. nov., Ascomycetes that Cause Cytospora Canker of Fruit Trees. Mycologia 2002, 94, 947–967.
- Fan, X.L.; Tian, C.M.; Yang, Q.; Liang, Y.M.; You, C.J.; Zhang, Y.B. Cytospora from Salix in Northern China. Mycotaxon 2015, 2, 303–315.
- Adams, G.C.; Roux, J.; Wingfield, M.J. Cytospora species (Ascomycota, Diaporthales, Valsaceae): Introduced and Native Pathogens of Trees in South Africa. Australasian Plant Pathology 2006, 35, 521–548.
- Hanifeh, S.; Zafari, D.; Soleimani, M.J.; Arzanlou, M. Multigene Phylogeny, Morphology, and Pathogenicity Trials Reveal Novel Cytospora Species Involved in Perennial Canker Disease of Apple Trees in Iran. Fungal Biol. 2022, 126, 707–726.
- Wang, S.; Jiang, N.; Ma, R. Morphology and Phylogeny Reveal Three New Species of Cytospora Associated with Tree Cankers in China. J. Fungi 2024, 10, 139.
- He, Z.; Abeywickrama, P.D.; Wu, L.; Zhou, Y.; Zhang, W.; Yan, J.; Shang, Q.; Zhou, Y.; Li, S. Diversity of Cytospora Species Associated with Trunk Diseases of Prunus persica (Peach) in Northern China. J. Fungi 2024, 10, 843.
- Li, J.-T.; Li, J.-R.; Jiang, N. Identification of Cytospora Species Isolated from Branch Canker Diseases of Woody Plants in Tibet, China. Forests 2024, 15, 121.
- Phookamsak, R.; Norphanphoun, C.; Tanaka, K.; Dai, D.-Q.; Luo, Z.-L.; Liu, J.-K.; Su, H.-Y.; Bhat, D.J.; Bahkali, A.H.; Mortimer, P.E.; et al. Towards a Natural Classification of Astrosphaeriella-like Species; Introducing Astrosphaeriellaceae and Pseudoastrosphaeriellaceae fam. nov. and Astrosphaeriellopsis, gen. nov. Fungal Divers. 2015, 74, 143–197.
- White, T.J.; Bruns, T.D.; Lee, S.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 312–322.
- Carbone, I.; Kohn, L.M. A Method for Designing Primer Sets for Speciation Studies in Filamentous Ascomycetes. Mycologia 1999, 3, 553–556.
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position–Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680.
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729.
- Stamatakis, S. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313.
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian Inference of Phylogenetic Trees. Bioinformatics 2001, 17, 754–755.
- Mishra, B.; Ploch, S.; Weiland, C.; Thines, M. The TrEase Web Service: Inferring Phylogenetic Trees with Ease. Mycological Progress 2023, 22, 84.
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest–NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294.
- Hillis, D.M.; Bull, J.J. An Empirical Test of Bootstrapping as a Method for Assessing Confidence in Phylogenetic Analysis. Syst. Biol. 1993, 42, 182–192.
- Rambaut, A. FigTree, version 1.4.4; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2018.
- Norphanphoun, C.; Wen, T.C.; Hyde, K.D.; Doilom, M.; Daranagama, D.A.; Phookamsak, R.; Bulgakov, T.S. Revisiting the Genus Cytospora and Allied Species. Mycosphere 2017, 8, 51–97.
- Fan, X.L.; Bezerra, J.D.P.; Tian, C.M.; Crous, P.W. Cytospora (Diaporthales) in China. Persoonia 2020, 45, 1–45.
- Shang, Q.J.; Hyde, K.D.; Camporesi, E.; Maharachchikumbura, S.S.N.; Norphanphoun, C.; Brooks, S.; Liu, J.K. Additions to the Genus Cytospora with Sexual Morph in Cytosporaceae. Mycosphere 2020, 11, 189–224.
- Pan, M.; Zhu, H.Y.; Tian, C.M.; Alvarez, L.V.; Fan, X.L. Cytospora piceae sp. nov. Associated with Canker Disease of Picea crassifolia in China. Phytotaxa 2018, 383, 181–196.
- Li, J.; Li, J.; Jiang, N. Morphology and Phylogeny of Cytospora (Cytosporaceae, Diaporthales) Species Associated with Plant Cankers in Tibet, China. MycoKeys 2024, 104, 51-70.
- Yang, Q.; Fan, X.L.; Crous, P.W.; Liang, Y.M.; Tian, C.M. Cytospora from Ulmus pumila in Northern China. Mycol. Prog. 2015, 14, 74.
- Ariyawansa, H.A.; Hyde, K.D.; Jayasiri, S.C.; Buyck, B.; Chethana, K.T.; Dai, D.Q.; Dai, Y.C.; Daranagama, D.A.; Jayawardena, R.S.; Lücking, R.; et al. Fungal Diversity Notes 111–252-Taxonomic and Phylogenetic Contributions to Fungal Taxa. Fungal Divers. 2015, 75, 27–274.
- Pan, M.; Zhu, H.; Bonthond, G.; Tian, C.M.; Fan, X.L. High Diversity of Cytospora Associated with Canker and Dieback of Rosaceae in China, with 10 New Species Described. Front. Plant Sci. 2020, 11, 690.
- Úrbez-Torres, J.R.; Lawrence, D.P.; Hand, F.P.; Trouillas, F.P. Olive Twig and Branch Dieback in California Caused by Cytospora oleicola and the Newly Described Species Cytospora olivarum sp. nov. Plant Dis. 2020, 104, 1908–1917.
- Nouri, M.T.; Li, S.; Travadon, R.; Holtz, B.A.; Maguvu, T.E.; Trouillas, F. First Report of Cytospora azerbaijanica Causing Cytospora Canker and Shoot Dieback on Peach (Prunus persica) in California, U.S.A. Plant Dis. 2023. [CrossRef]
- Sijin, B.; Fenghui, S.; Yannan, Z.; Yanjiang, S.; Hongwei, H.; Jianyu, B. Cytospora chrysosperma Causing Branch Dieback and Canker of Black Walnut in China. Canadian Journal of Plant Pathology, 2020, 42, 203–209.
- Madar, Z.; Solel, Z.; Kimchi, M.; First Report of Cytospora Canker Caused by Cytospora chrysosperma on White Poplar in Israel. Plant Dis. 2004, 88, 220.
- Petrović, E.; Vrandečić, K.; Ivić, D.; Ćosić, J.; Godena S. First Report of Olive Branch Dieback in Croatia Caused by Cytospora pruinosa Défago. Microorganisms, 2023, 11, 1679.
- Dudley, M.M.; Tisserat, N.A.; Jacobi, W.R.; Negron, J.; Stewart, J.E. Pathogenicity and Distribution of Two Species of Cytospora on Populus tremuloides in Portions of the Rocky Mountains and Midwest in the United States. For. Ecol. Manag. 468, 118168.
- Lin, L.; Pan, M.; Bezerra, J.D.P.; Tian, C.; Fan, X. Re-evaluation of the Fungal Diversity and Pathogenicity of Cytospora Species from Populus in China. Plant Dis. 2023, 107, 83–96.

| GenBank accession numbers | |||||
|---|---|---|---|---|---|
| Species | Strain | Country | Host | ITS | act1 |
| Cytospora chrysosperma | CFCC 89982 | China | Ulmus pumila | KP281261 | KP310835 |
| EI-101 | Canada | Unknown tree | PQ356607 | PQ728056 | |
| EI-351 | Canada | Rhus sp. | PQ385601 | NA | |
| EI-437 | Canada | Populus tremuloides | PQ678997 | NA | |
| EI-SK-71 | Canada | Populus deltoides | PQ678998 | NA | |
| EI-SK-90 | Canada | Salix bebbiana | PQ679047 | NA | |
| EI-SK-93(A) | Canada | Populus tremula | PQ679032 | NA | |
| EI-SK-127 | Canada | Populus sp. | PQ679040 | NA | |
| EI-SK-196 | Canada | Salix alba | PQ679042 | NA | |
| Cytospora curvata | MFLUCC 15-0865T | Russia | Salix alba | KY417728 | KY417694 |
| EI-132 | Canada | Aronia sp. | PQ677273 | PQ728060 | |
| EI-203 | Canada | Syringa vulgaris | PQ677316 | PQ728061 | |
| Cytospora euonymina | CFCC 89993T | China | Euonymus kiautschovicus | MH933630 | MH933537 |
| EI-250 | Canada | Salix sp. | PQ678925 | NA | |
| EI-316 | Canada | Berberis vulgaris | PQ383411 | NA | |
| Cytospora hoffmannii | CFCC 89641 | China | Elaeagnus angustifolia | KF765683 | KU711006 |
| EI-SK-231 | Canada | Salix bebbiana | PQ677104 | PQ728057 | |
| EI-SK-237 | Canada | Salix sp. | PQ677112 | PQ728058 | |
| Cytospora kantschavelii | MFLUCC 15-0857T | Russia | Populus × sibirica | KY417738 | KY417704 |
| EI-SK-33 | Canada | Ulmus glabra | PQ678995 | PQ728067 | |
| EI-SK-44 | Canada | Acer spicatum | PQ678996 | PQ728068 | |
| Cytospora leucosperma | MFLUCC 18-1199T | Russia | Galega officinalis | MK912128 | MN685810 |
| EI-54(A) | Canada | Berberis vulgaris | PQ281438 | NA | |
| Cytospora leucostoma | CFCC 50022 | China | Prunus padus | MH933627 | MH933534 |
| EI-78 | Canada | Malus sp. | PP751512 | NA | |
| EI-223 | Canada | Vaccinium sp. | PQ368601 | PQ728059 | |
| Cytospora nitschkeana | CBS. 118.22 | Netherlands | Salix alba | MH854712 | KX964746 |
| EI-170 | Canada | Salix babylonica | PQ356735 | NA | |
| EI-193 | Canada | Berberis vulgaris | PQ362651 | NA | |
| EI-429 | Canada | Fraxinus americana | PQ421750 | NA | |
| EI-454 | Canada | Unknown tree | PQ425077 | NA | |
| EI-470 | Canada | Fraxinus nigra | PQ678921 | NA | |
| EI-SK-195 | Canada | Salix alba | PQ678915 | NA | |
| Cytospora piceae | CFCC 52841T | China | Picea crassifolia | MH820398 | MH820406 |
| EI-273 | Canada | Picea pungens | ON352565 | NA | |
| EI-SK-36 | Canada | Rhus sp. | PQ671332 | NA | |
| EI-SK-110 | Canada | Picea sp. | PQ671333 | NA | |
| EI-SK-154(A) | Canada | Fraxinus sp. | PQ666762 | NA | |
| Cytospora populina | CFCC 89644T | China | Salix psammophila | KF765686 | KU711007 |
| EI-434 | Canada | Magnolia sp. | PQ422182 | NA | |
| EI-477(B) | Canada | Sorbus sp. | PQ683229 | PQ728055 | |
| EI-478 | Canada | Acer platanoides | PQ425482 | NA | |
| EI-SK-184 | Canada | Aronia sp. | PQ683280 | NA | |
| Cytospora pruinopsis | CFCC 50034T | China | Ulmus pumila | KP281259 | KP310836 |
| EI-SK-38 | Canada | Populus deltoides | PQ678306 | PQ728066 | |
| EI-SK-75 | Canada | Ulmus americana | PQ678839 | NA | |
| EI-SK-94(A) | Canada | Malus sp. | PQ678815 | NA | |
| EI-SK-223 | Canada | Unknown shrubs | PQ678307 | NA | |
| Cytospora pruinosa | CFCC 50036 | China | Syringa oblata | KP310800 | KP310832 |
| EI-SK-133(A) | Canada | Aronia sp. | PQ677817 | PQ728065 | |
| EI-SK-152 | Canada | Syringa reticulata | PQ677975 | NA | |
| EI-SK-155 | Canada | Unknown shrubs | PQ680075 | NA | |
| EI-SK-211 | Canada | Syringa sp. | PQ677872 | NA | |
| EI-SK-215 | Canada | Fraxinus sp. | PQ677818 | NA | |
| EI-SK-221 | Canada | Unknown shrubs | PQ680079 | NA | |
| Cytospora ribis | CFCC 50026 | China | Ulmus pumila | KP281267 | KP310843 |
| EI-396 | Canada | Fraxinus americana | PQ393077 | PQ728070 | |
| EI-SK-60 | Canada | Syringa vulgaris | PQ683333 | NA | |
| Cytospora schulzeri | CFCC 53173 | China | Berberissp. | MK673070 | MK673040 |
| EI-378 | Canada | Sorbaria sorbifolia | PQ392014 | NA | |
| EI-414 | Canada | Malus sp. | PQ421083 | NA | |
| EI-480 | Canada | Acer platanoides | PQ432426 | PQ728069 | |
| EI-SK-101 | Canada | Malus sp. | PQ728069 | PQ683213 | |
| Cytospora sorbina | CF 20197660T | China | Sorbus tianschanica | MK673052 | MK673022 |
| EI-SK-25 | Canada | Malus sp. | PQ677620 | PQ728063 | |
| EI-SK-31 | Canada | Aronia sp. | PQ677471 | PQ728062 | |
| EI-SK-67 | Canada | Aronia sp. | PQ677674 | PQ728064 | |
| EI-SK-76 | Canada | Prunus sp. | PQ736325 | NA | |
| EI-SK-82 | Canada | Sorbus aucuparia | PQ677675 | NA | |
| EI-SK-92 | Canada | Prunus padus | PQ677676 | NA | |
| EI-SK-95(A) | Canada | Aronia sp. | PQ680078 | NA | |
| EI-SK-131(A) | Canada | Unknown shrubs | PQ677688 | NA | |
| EI-SK-148 | Canada | Sorbus sp. | PQ680076 | NA | |
| EI-SK-157(A) | Canada | Viburnum cf. trilobum | PQ680077 | NA | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





