Submitted:
30 December 2024
Posted:
31 December 2024
You are already at the latest version
Abstract
Background/Objectives: Ovarian cancer is a leading cause of gynecological cancer mortality worldwide, often diagnosed at advanced stages due to vague symptoms and lack of effective early detection methods. Long non-coding RNAs (lncRNAs) have emerged as key regulators in cancer biology, influencing cellular processes such as proliferation, apoptosis, and chemoresistance. This review explores the multifaceted roles of lncRNAs in ovarian cancer pathogenesis and their potential as biomarkers and therapeutic targets. Methods: A comprehensive literature review was conducted to analyze the structural and functional characteristics of lncRNAs and their contributions to ovarian cancer biology. This includes their regulatory mechanisms, interactions with signaling pathways, and implications for therapeutic resistance. Advanced bioinformatics and omics approaches were also evaluated for their potential in lncRNA research. Results: The review highlights the dual role of lncRNAs as oncogenes and tumor sup-pressors, modulating processes such as cell proliferation, invasion, and angiogenesis. Specific lncRNAs, such as HOTAIR and GAS5, demonstrate significant potential as di-agnostic biomarkers and therapeutic targets. Emerging technologies, such as single-cell sequencing, provide valuable insights into the tumor microenvironment and the het-erogeneity of lncRNA expression. Conclusions: LncRNAs hold transformative potential in advancing ovarian cancer di-agnosis, prognosis, and treatment. Targeting lncRNAs or their associated pathways offers promising strategies to overcome therapy resistance and enhance personalized medicine. Continued research integrating omics and bioinformatics will be essential to unlock the full clinical potential of lncRNAs in ovarian cancer management.
Keywords:
1. Introduction
2. LncRNAs: Structure and Function
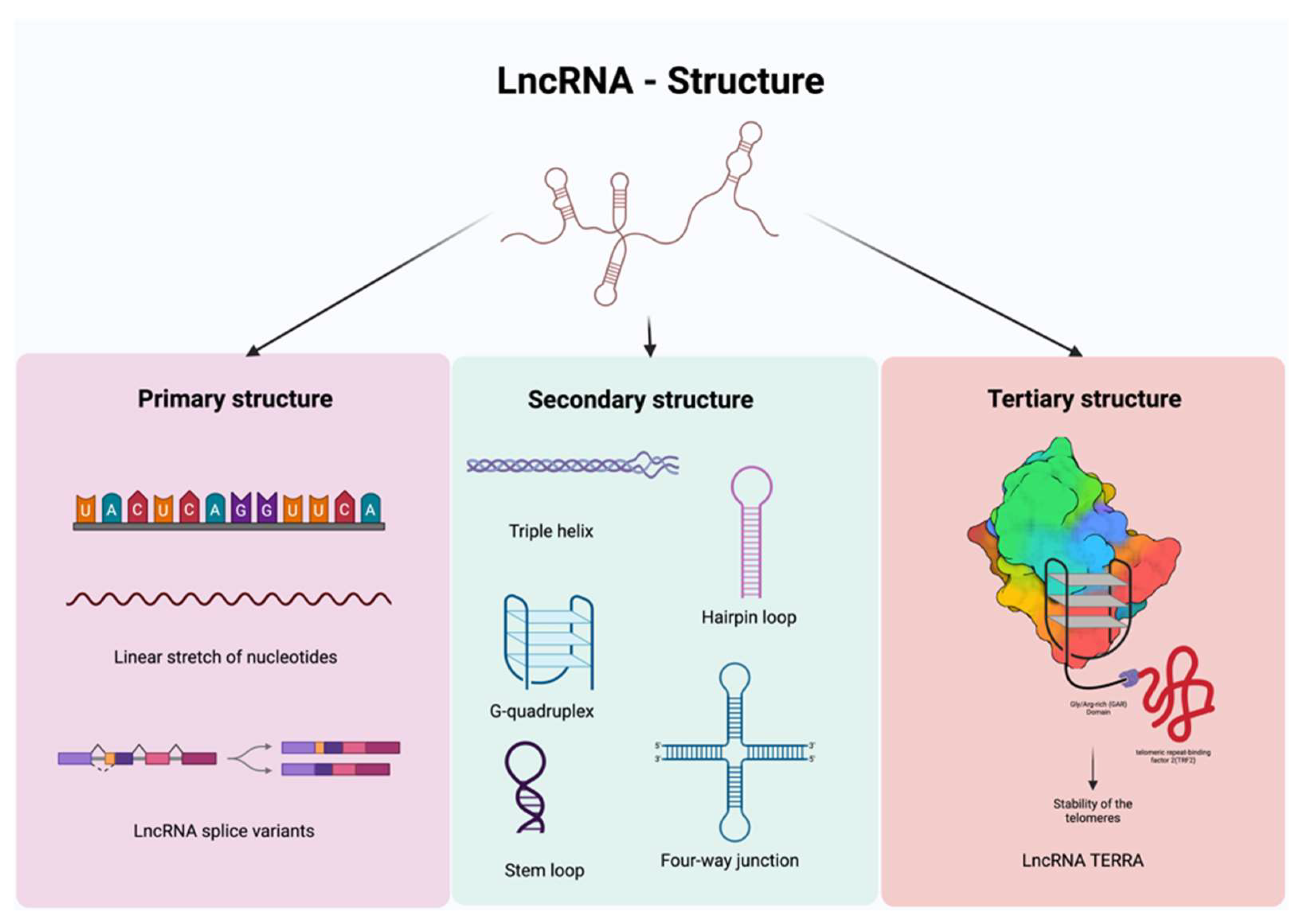
2.1. Primary, Secondary, and Tertiary Structures of lncRNAs
2.1.1. Primary Structure
2.1.2. Secondary Structure
2.1.3. Tertiary Structure
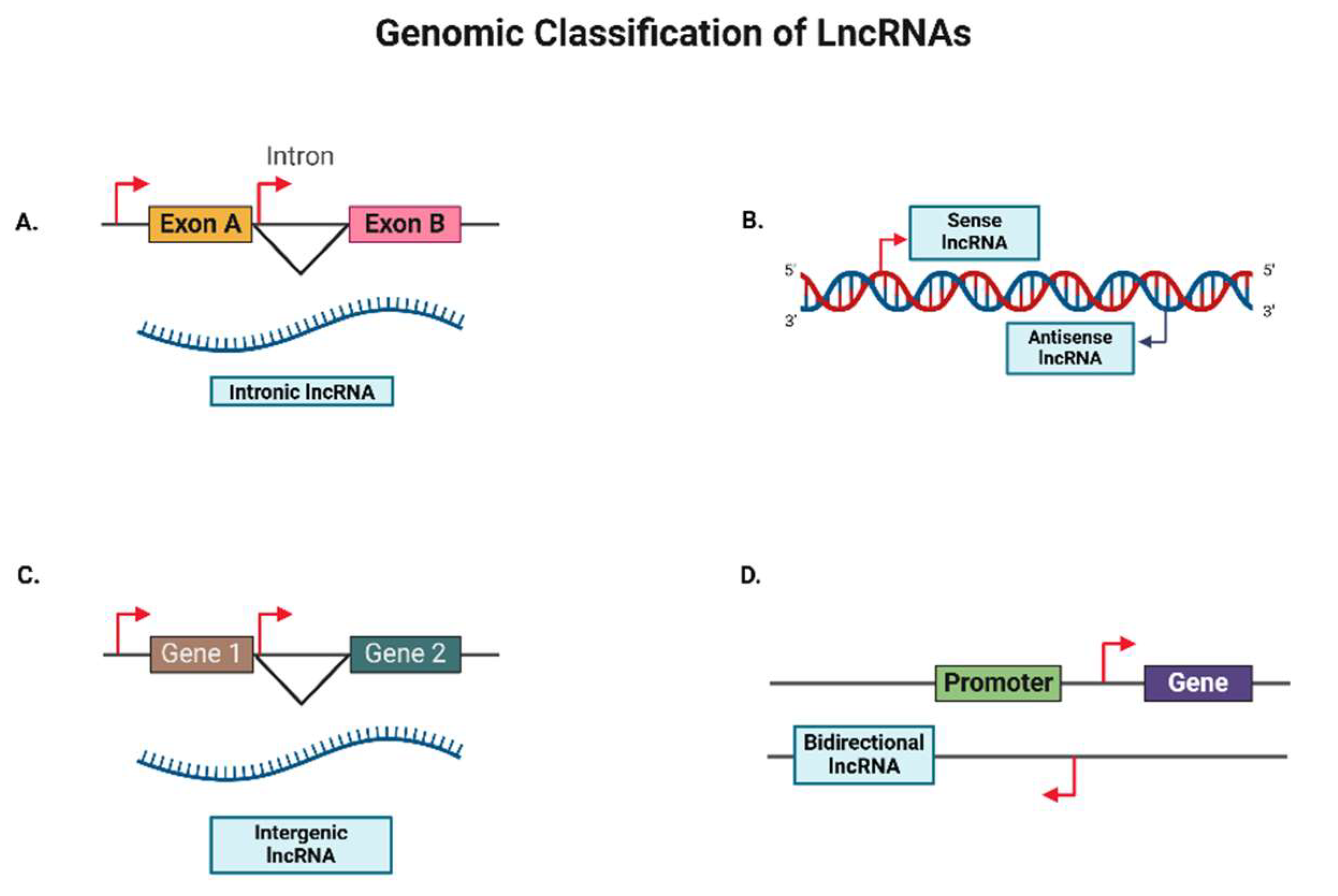
2.2. Positional Diversity of lncRNAs
2.2.1. Intronic LncRNAs
2.2.2. Antisense LncRNAs
2.2.3. Intergenic LncRNAs
2.2.4. Bidirectional LncRNAs
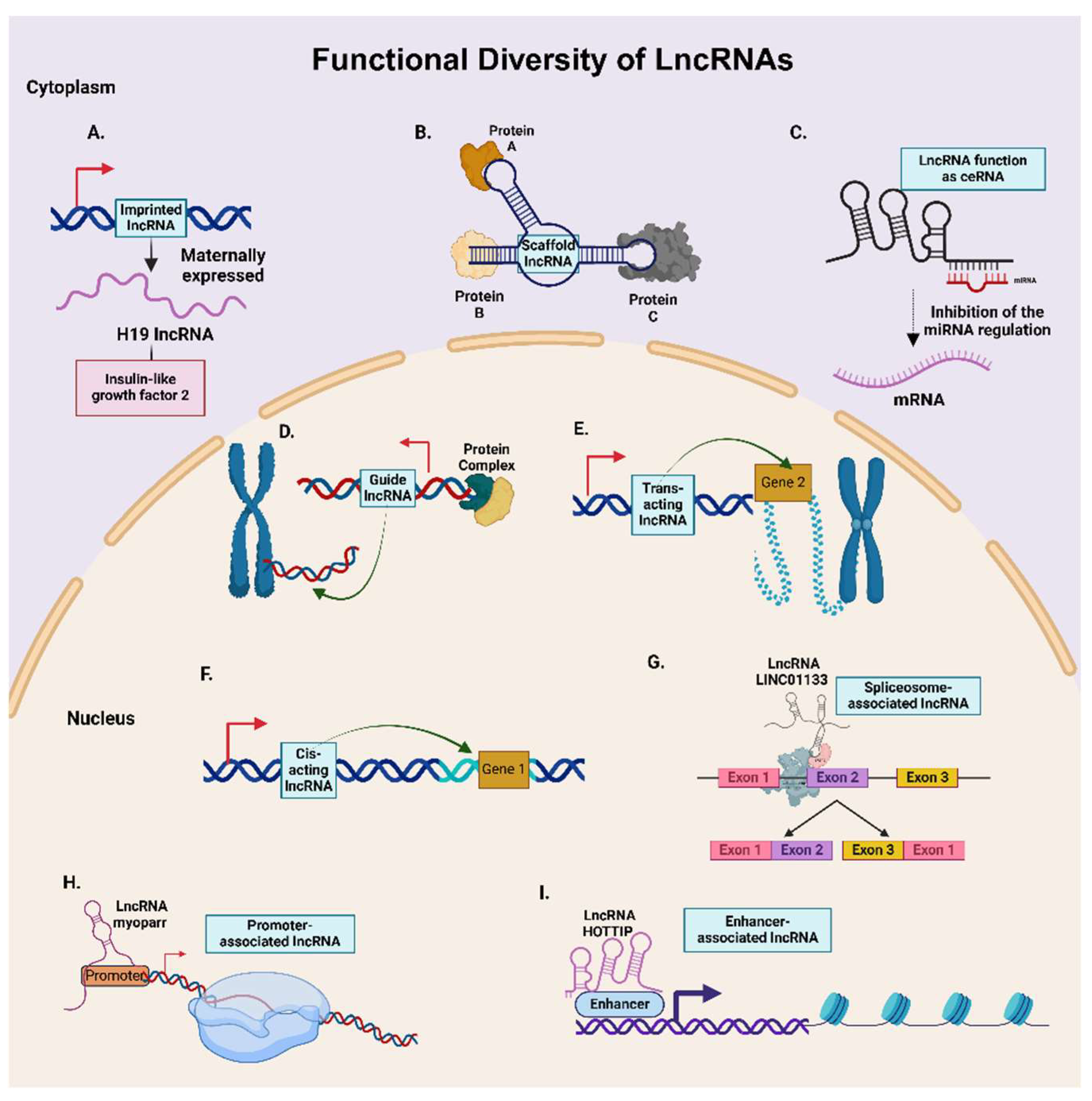
2.3. Functional Diversity of LncRNAs
2.3.1. Cis- and Trans-acting LncRNAs
2.3.2. Enhancer and Promoter-associated LncRNAs
2.3.3. Competing Endogenous RNAs (ceRNAs)
2.3.4. Guide and Scaffold LncRNAs
2.3.5. Imprinted LncRNAs
2.3.6. Spliceosome-associated LncRNAs
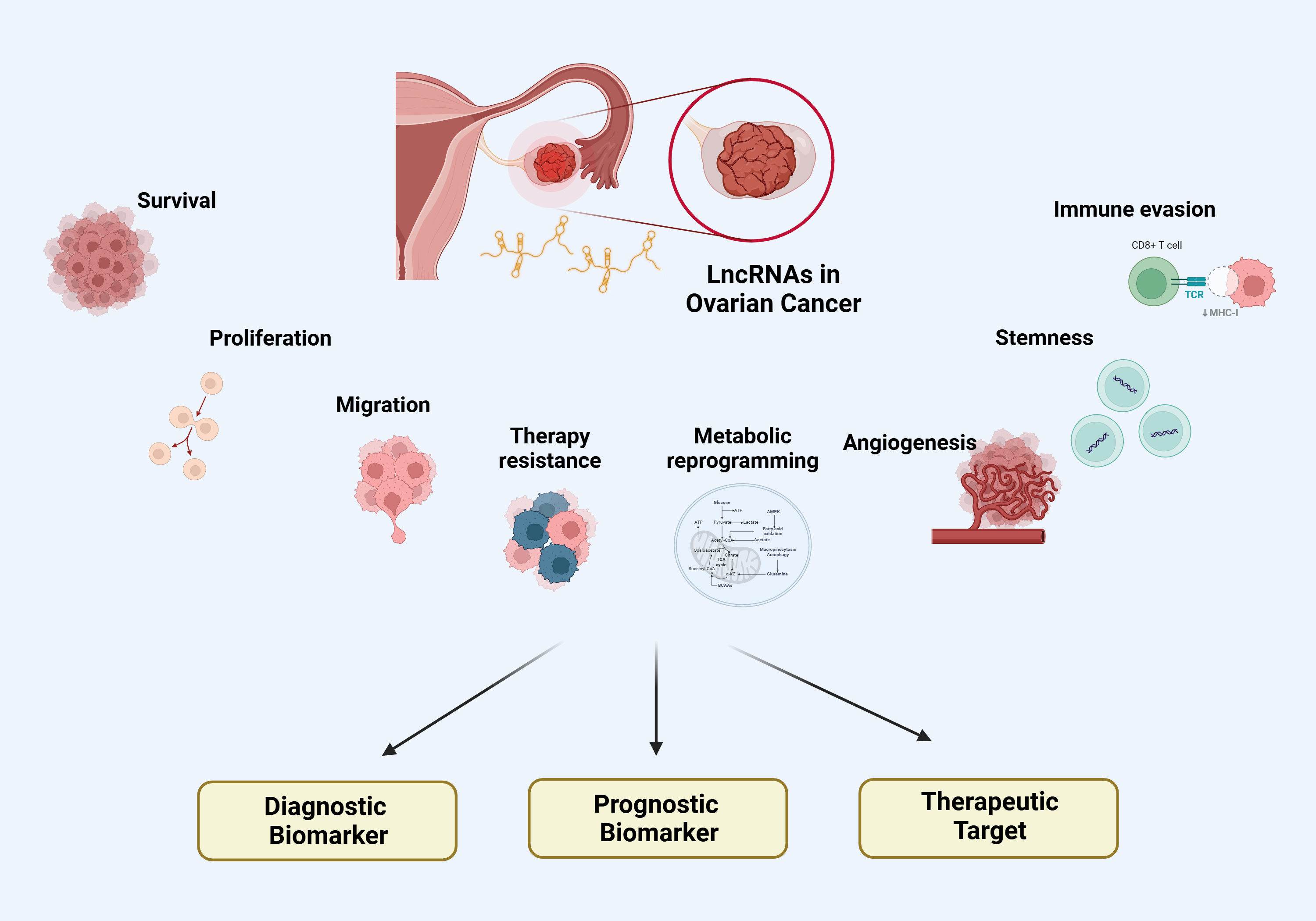
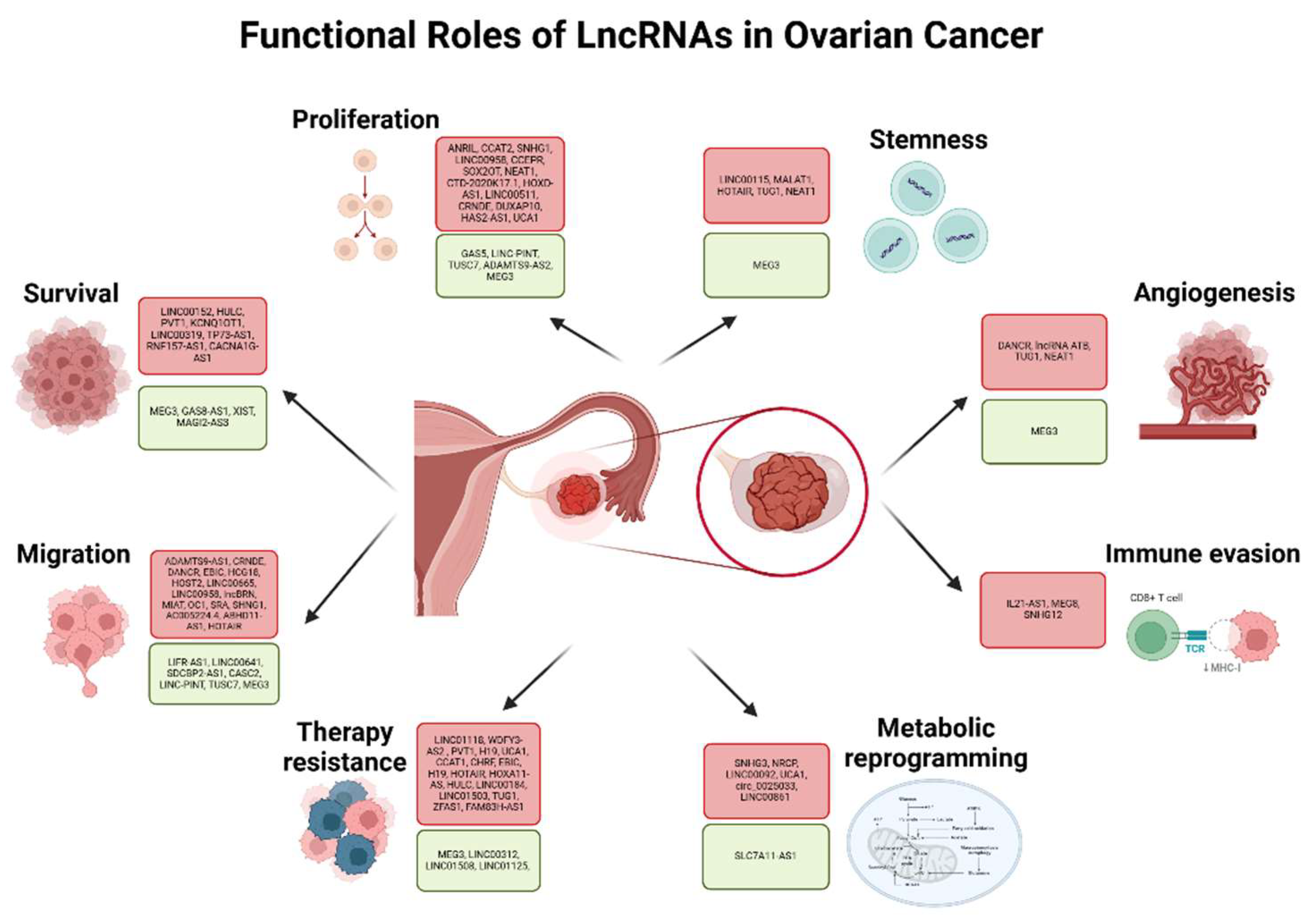
3. LncRNAs in Ovarian Cancer
3.1. LncRNAs in Ovarian Cancer Cell Proliferation
3.1.1. Regulation of Cell Proliferation
3.1.2. Regulation of Cell Cycle Progression
3.1.3. Tumor Suppression and Proliferation Inhibition
3.2. LncRNAs in Ovarian Cancer Cell Survival
3.2.1. Evasion of Apoptosis
3.2.2. Regulation of Ferroptosis
3.2.3. Modulation of Autophagy
3.3. LncRNAs in Ovarian Cancer Metabolic Reprogramming
3.3.1. Regulation of Glucose Metabolism
3.3.2. Regulation of Fatty Acid Metabolism
3.3.3. Regulation of Amino Acid Metabolism
3.3.4. Intercellular Metabolic Signaling in the Tumor Microenvironment
3.4. LncRNAs in Ovarian Cancer Cell Migration
3.4.1. Promotion of EMT by Oncogenic LncRNAs
3.4.2. Inhibition of EMT by Tumor Suppressor LncRNAs
3.4.3. Direct Regulation of Cell Migration
3.5. LncRNAs in Ovarian Cancer Angiogenesis
3.5.1. Promotion of Angiogenesis by Oncogenic LncRNAs
3.5.2. Inhibition of Angiogenesis by Tumor Suppressor LncRNAs
3.6. LncRNAs in Ovarian Cancer Stemness
3.6.1. Transcriptional Regulation of Stemness by LncRNAs
3.6.2. Epigenetic Modulation of Stemness by LncRNAs
3.6.3. Regulation of CSC Signaling Pathways by LncRNAs
3.7. LncRNAs in Ovarian Cancer associated with Immune Evasion
3.7.1. Modulation of Innate Immunity
3.7.2. Modulation of Adaptive Immunity
3.8. LncRNAs in Ovarian Cancer Therapy Resistance
3.8.1. Modulation of Drug Efflux and Uptake
3.8.2. Alteration of Drug Metabolism
3.8.3. Role of Tumor Suppressor LncRNAs in Therapy Sensitivity
3.8.4. LncRNAs and Radiotherapy Resistance
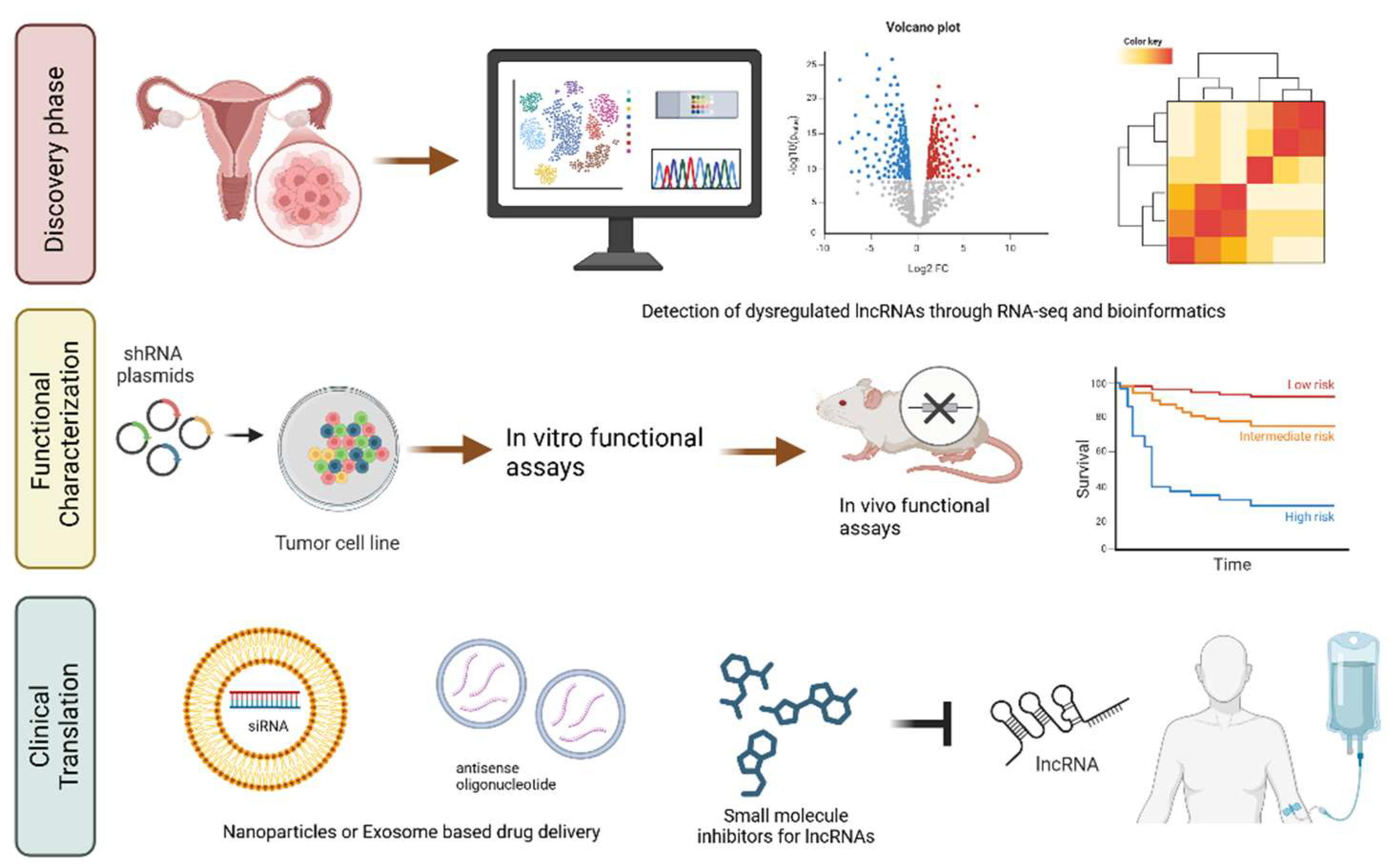
4. Clinical Implications
4.1. LncRNAs as diagnostic biomarkers in Ovarian Cancer
4.2. LncRNAs as prognostic indicators and predictors of treatment response
4.3. LncRNAs as Therapeutic Targets
4.4. Therapeutic potential of targeting lncRNAs in Ovarian Cancer
5. Summary and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OC | Ovarian Cancer |
| lncRNA | Long Non-Coding RNA |
| lincRNA | Long Intergenic Non-Coding RNA |
| ceRNA | Competing Endogenous RNA |
| miRNA | MicroRNA |
| piRNA | Piwi-Interacting RNA |
| snRNA | Small Nuclear RNA |
| circRNA | Circular RNA |
| siRNAs | Small Interfering RNAs |
| shRNA | Short Hairpin RNA |
| ASO | Antisense Oligonucleotide |
| RNA-seq | RNA Sequencing |
| CAF | Cancer Associated Fibroblasts |
| CSC | Cancer Stem Cells |
| TME | Tumor Micro-Environment |
| EMT | Epithelial-to-Mesenchymal Transition |
References
- Webb, P.M.; Jordan, S.J. Global epidemiology of epithelial ovarian cancer. Nat Rev Clin Oncol 2024. [Google Scholar] [CrossRef] [PubMed]
- Tavares, V.; Marques, I.S.; de Melo, I.G.; Assis, J.; Pereira, D.; Medeiros, R. Paradigm Shift: A Comprehensive Review of Ovarian Cancer Management in an Era of Advancements. Int J Mol Sci 2024, 25, 1845. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA: A Cancer Journal for Clinicians 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Lheureux, S.; Braunstein, M.; Oza, A.M. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA: A Cancer Journal for Clinicians 2019, 69, 280–304. [Google Scholar] [CrossRef]
- Chan, J.K.; Tian, C.; Kesterson, J.P.; Monk, B.J.; Kapp, D.S.; Davidson, B.; Robertson, S.; Copeland, L.J.; Walker, J.L.; Wenham, R. , et al. Symptoms of Women With High-Risk, Early-Stage Ovarian Cancer. Obstet Gynecol 2022, 139, 157–162. [Google Scholar] [CrossRef]
- Lõhmussaar, K.; Kopper, O.; Korving, J.; Begthel, H.; Vreuls, C.P.H.; van Es, J.H.; Clevers, H. Assessing the origin of high-grade serous ovarian cancer using CRISPR-modification of mouse organoids. Nat Commun 2020, 11, 2660. [Google Scholar] [CrossRef]
- Marth, C.; Abreu, M.H.; Andersen, K.K.; Aro, K.M.; De Lurdes Batarda, M.; Boll, D.; Ekmann-Gade, A.W.; Haltia, U.M.; Hansen, J.; Haug, A.J. , et al. Real-life data on treatment and outcomes in advanced ovarian cancer: An observational, multinational cohort study (<scp>RESPONSE</scp> trial). Cancer 2022, 128, 3080–3089. [Google Scholar] [CrossRef]
- Alatise, K.L.; Gardner, S.; Alexander-Bryant, A. Mechanisms of Drug Resistance in Ovarian Cancer and Associated Gene Targets. Cancers (Basel) 2022, 14, 6246. [Google Scholar] [CrossRef]
- Algethami, M.; Kulkarni, S.; Sadiq, M.T.; Tang, H.K.C.; Brownlie, J.; Jeyapalan, J.N.; Mongan, N.P.; Rakha, E.A.; Madhusudan, S. Towards Personalized Management of Ovarian Cancer. Cancer Manag Res 2022, 14, 3469–3483. [Google Scholar] [CrossRef]
- Nadhan, R.; Isidoro, C.; Song, Y.S.; Dhanasekaran, D.N. Long Non-coding RNAs in Cancer. In Handbook of Oncobiology: From Basic to Clinical Sciences, Sobti, R.C., Ganguly, N.K., Kumar, R., Eds. Springer Nature Singapore: Singapore, 2023; pp. 1-45. [CrossRef]
- Nadhan, R.; Isidoro, C.; Song, Y.S.; Dhanasekaran, D.N. Signaling by LncRNAs: Structure, Cellular Homeostasis, and Disease Pathology. Cells 2022, 11, 2517. [Google Scholar] [CrossRef]
- Nadhan, R.; Dhanasekaran, D.N. Decoding the Oncogenic Signals from the Long Non-Coding RNAs. Onco 2021, 1, 176–206. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nature Reviews Molecular Cell Biology 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Oo, J.A.; Brandes, R.P.; Leisegang, M.S. Long non-coding RNAs: novel regulators of cellular physiology and function. Pflugers Arch 2022, 474, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.H.; Kim, K.M.; McClusky, W.; Abdelmohsen, K.; Gorospe, M. Cytoplasmic functions of lncRNAs. Wiley Interdiscip Rev RNA 2018, 9, e1471. [Google Scholar] [CrossRef]
- Nadhan, R.; Dhanasekaran, D.N. Regulation of Tumor Metabolome by Long Non-Coding RNAs. 2022, 16, 1. [CrossRef]
- Nikpayam, E.; Tasharrofi, B.; Sarrafzadeh, S.; Ghafouri-Fard, S. The Role of Long Non-Coding RNAs in Ovarian Cancer. Iran Biomed J 2017, 21, 3–15. [Google Scholar] [CrossRef]
- Zheng, M.; Hu, Y.; Gou, R.; Nie, X.; Li, X.; Liu, J.; Lin, B. Identification three LncRNA prognostic signature of ovarian cancer based on genome-wide copy number variation. Biomedicine & Pharmacotherapy 2020, 124, 109810. [Google Scholar] [CrossRef]
- Loganathan, T.; Doss C, G.P. Non-coding RNAs in human health and disease: potential function as biomarkers and therapeutic targets. Funct Integr Genomics 2023, 23, 33. [Google Scholar] [CrossRef]
- Qian, Y.; Shi, L.; Luo, Z. Long Non-coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front Med (Lausanne) 2020, 7, 612393. [Google Scholar] [CrossRef]
- Karakas, D.; Ozpolat, B. The Role of LncRNAs in Translation. Noncoding RNA 2021, 7, 16. [Google Scholar] [CrossRef]
- Gao, N.; Li, Y.; Li, J.; Gao, Z.; Yang, Z.; Li, Y.; Liu, H.; Fan, T. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front Oncol 2020, 10, 598817. [Google Scholar] [CrossRef]
- Taniue, K.; Akimitsu, N. The Functions and Unique Features of LncRNAs in Cancer Development and Tumorigenesis. Int J Mol Sci 2021, 22, 632. [Google Scholar] [CrossRef] [PubMed]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J Cell Biol 2021, 220, e202009045. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Xu, Y.; Shen, Q.; Li, R.; Xiao, X.; Saw, P.E.; Xu, X. Role of long non-coding RNAs in cancer: From subcellular localization to nanoparticle-mediated targeted regulation. Molecular Therapy - Nucleic Acids 2023, 33, 774–793. [Google Scholar] [CrossRef] [PubMed]
- Kazimierczyk, M.; Kasprowicz, M.K.; Kasprzyk, M.E.; Wrzesinski, J. Human Long Noncoding RNA Interactome: Detection, Characterization and Function. Int J Mol Sci 2020, 21, 1027. [Google Scholar] [CrossRef]
- Ruan, X.; Li, P.; Chen, Y.; Shi, Y.; Pirooznia, M.; Seifuddin, F.; Suemizu, H.; Ohnishi, Y.; Yoneda, N.; Nishiwaki, M. , et al. In vivo functional analysis of non-conserved human lncRNAs associated with cardiometabolic traits. Nat Commun 2020, 11. [Google Scholar] [CrossRef]
- Huang, W.; Xiong, T.; Zhao, Y.; Heng, J.; Han, G.; Wang, P.; Zhao, Z.; Shi, M.; Li, J.; Wang, J. , et al. Computational prediction and experimental validation identify functionally conserved lncRNAs from zebrafish to human. Nature Genetics 2024, 56, 124–135. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G. , et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Cruz, J.A.; Westhof, E. The Dynamic Landscapes of RNA Architecture. Cell 2009, 136, 604–609. [Google Scholar] [CrossRef]
- Wilusz, J.E.; JnBaptiste, C.K.; Lu, L.Y.; Kuhn, C.-D.; Joshua-Tor, L.; Sharp, P.A. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012, 26, 2392–2407. [Google Scholar] [CrossRef]
- Jones, A.N.; Tikhaia, E.; Mourao, A.; Sattler, M. Structural effects of m6A modification of the Xist A-repeat AUCG tetraloop and its recognition by YTHDC1. Nucleic Acids Res 2022, 50, 2350–2362. [Google Scholar] [CrossRef]
- Lu, Z.; Guo, J.K.; Wei, Y.; Dou, D.R.; Zarnegar, B.; Ma, Q.; Li, R.; Zhao, Y.; Liu, F.; Choudhry, H. , et al. Structural modularity of the XIST ribonucleoprotein complex. Nat Commun 2020, 11, 6163. [Google Scholar] [CrossRef] [PubMed]
- Frank, F.; Kavousi, N.; Bountali, A.; Dammer, E.B.; Mourtada-Maarabouni, M.; Ortlund, E.A. The lncRNA Growth Arrest Specific 5 Regulates Cell Survival via Distinct Structural Modules with Independent Functions. Cell Reports 2020, 32, 107933. [Google Scholar] [CrossRef] [PubMed]
- Somarowthu, S.; Legiewicz, M.; Chillón, I.; Marcia, M.; Liu, F.; Pyle, A.M. HOTAIR Forms an Intricate and Modular Secondary Structure. Molecular Cell 2015, 58, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Mou, X.; Liew, S.W.; Kwok, C.K. Identification and targeting of G-quadruplex structures in MALAT1 long non-coding RNA. Nucleic Acids Research 2022, 50, 397–410. [Google Scholar] [CrossRef]
- Lin, Y.; Schmidt, B.F.; Bruchez, M.P.; McManus, C J. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Research 2018, 46, 3742–3752. [Google Scholar] [CrossRef]
- Simko, E.A.J.; Liu, H.; Zhang, T.; Velasquez, A.; Teli, S.; Haeusler, A.R.; Wang, J. G-quadruplexes offer a conserved structural motif for NONO recruitment to NEAT1 architectural lncRNA. Nucleic Acids Research, 2020. [Google Scholar] [CrossRef]
- Hirose, T.; Yamazaki, T.; Nakagawa, S. Molecular anatomy of the architectural NEAT1 noncoding RNA: The domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles. WIREs RNA 2019, 10, e1545. [Google Scholar] [CrossRef]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.M.; Schwarz, T.; Azzalin, C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun 2014, 5, 5220. [Google Scholar] [CrossRef]
- Hirashima, K.; Seimiya, H. Telomeric repeat-containing RNA/G-quadruplex-forming sequences cause genome-wide alteration of gene expression in human cancer cells in vivo. Nucleic Acids Research 2015, 43, 2022–2032. [Google Scholar] [CrossRef]
- Uroda, T.; Chillón, I.; Annibale, P.; Teulon, J.-M.; Pessey, O.; Karuppasamy, M.; Pellequer, J.-L.; Marcia, M. Visualizing the functional 3D shape and topography of long noncoding RNAs by single-particle atomic force microscopy and in-solution hydrodynamic techniques. Nat Protoc 2020, 15, 2107–2139. [Google Scholar] [CrossRef]
- Robinson, E.K.; Covarrubias, S.; Carpenter, S. The how and why of lncRNA function: An innate immune perspective. Biochim Biophys Acta Gene Regul Mech 2020, 1863, 194419. [Google Scholar] [CrossRef]
- Yu, J.; Han, Q.; Cui, Y. Decreased long non-coding RNA SPRY4-IT1 contributes to ovarian cancer cell metastasis partly via affecting epithelial–mesenchymal transition. Tumor Biology 2017, 39, 101042831770912. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol 2013, 10, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, Y.; Liu, H.; Su, D.; Luo, F.; Zhou, F. Long noncoding RNA CDKN2B-AS1 interacts with miR-411–3p to regulate ovarian cancer <i>in vitro</i> and in <i>vivo</i> through HIF-1a/VEGF/P38 pathway. Biochemical and Biophysical Research Communications 2019, 514, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhao, P.; Luo, Q.; Wu, X.; Wang, Y.; Nan, Y.; Liu, S.; Gao, W.; Li, B.; Liu, Z.; Cui, Z. RUNX1-IT1 acts as a scaffold of STAT1 and NuRD complex to promote ROS-mediated NF-κB activation and ovarian cancer progression. Oncogene 2024, 43, 420–433. [Google Scholar] [CrossRef]
- Aliperti, V.; Skonieczna, J.; Cerase, A. Long Non-Coding RNA (lncRNA) Roles in Cell Biology, Neurodevelopment and Neurological Disorders. Noncoding RNA 2021, 7, 36. [Google Scholar] [CrossRef]
- Avgeris, M.; Tsilimantou, A.; Levis, P.K.; Tokas, T.; Sideris, D.C.; Stravodimos, K.; Ardavanis, A.; Scorilas, A. Loss of GAS5 tumour suppressor lncRNA: an independent molecular cancer biomarker for short-term relapse and progression in bladder cancer patients. Br J Cancer 2018, 119, 1477–1486. [Google Scholar] [CrossRef]
- Lin, G.; Wu, T.; Gao, X.; He, Z.; Nong, W. Research Progress of Long Non-Coding RNA GAS5 in Malignant Tumors. Front Oncol 2022, 12. [Google Scholar] [CrossRef]
- Long, X.; Song, K.; Hu, H.; Tian, Q.; Wang, W.; Dong, Q.; Yin, X.; Di, W. Long non-coding RNA GAS5 inhibits DDP-resistance and tumor progression of epithelial ovarian cancer via GAS5-E2F4-PARP1-MAPK axis. J Exp Clin Cancer Res 2019, 38, 345. [Google Scholar] [CrossRef]
- Zhang, T.; Leng, Y.; Duan, M.; Li, Z.; Ma, Y.; Huang, C.; Shi, Q.; Wang, Y.; Wang, C.; Liu, D. , et al. LncRNA GAS5-hnRNPK axis inhibited ovarian cancer progression via inhibition of AKT signaling in ovarian cancer cells. Discov Oncol 2023, 14, 157. [Google Scholar] [CrossRef]
- Gao, J.; Liu, M.; Zou, Y.; Mao, M.; Shen, T.; Zhang, C.; Song, S.; Sun, M.; Zhang, S.; Wang, B. , et al. Long non-coding RNA growth arrest-specific transcript 5 is involved in ovarian cancer cell apoptosis through the mitochondria-mediated apoptosis pathway. Oncol Rep 2015, 34, 3212–3221. [Google Scholar] [CrossRef]
- Musahl, A.S.; Huang, X.; Rusakiewicz, S.; Ntini, E.; Marsico, A.; Kroemer, G.; Kepp, O.; Ørom, U.A. A long non-coding RNA links calreticulin-mediated immunogenic cell removal to RB1 transcription. Oncogene 2015, 34, 5046–5054. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-DiNardo, D.; Kanduri, C. Kcnq1ot1 Antisense Noncoding RNA Mediates Lineage-Specific Transcriptional Silencing through Chromatin-Level Regulation. Molecular Cell 2008, 32, 232–246. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A. , et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef]
- Blank-Giwojna, A.; Postepska-Igielska, A.; Grummt, I. lncRNA KHPS1 Activates a Poised Enhancer by Triplex-Dependent Recruitment of Epigenomic Regulators. Cell Reports 2019, 26, 2904–2915.e2904. [Google Scholar] [CrossRef]
- Niu, Z.-S.; Wang, W.-H.; Dong, X.-N.; Tian, L.-M.-L. Role of long noncoding RNA-mediated competing endogenous RNA regulatory network in hepatocellular carcinoma. World J Gastroenterol 2020, 26, 4240–4260. [Google Scholar] [CrossRef]
- Ha, J.H.; Radhakrishnan, R.; Nadhan, R.; Gomathinayagam, R.; Jayaraman, M.; Yan, M.; Kashyap, S.; Fung, K.-M.; Xu, C.; Bhattacharya, R. , et al. Deciphering a GPCR-lncrna-miRNA nexus: Identification of an aberrant therapeutic target in ovarian cancer. Cancer Letters 2024, 591, 216891. [Google Scholar] [CrossRef]
- Dong, B.; Li, C.; Xu, X.; Wang, Y.; Li, Y.; Li, X. LncRNA LINC01123 promotes malignancy of ovarian cancer by targeting hsa-miR-516b-5p/VEGFA. Genes & Genomics 2024, 46, 231–239. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef]
- Lin, X.; Spindler, T.J.; De Souza Fonseca, M.A.; Corona, R.I.; Seo, J.-H.; Dezem, F.S.; Li, L.; Lee, J.M.; Long, H.W.; Sellers, T.A. , et al. Super-Enhancer-Associated LncRNA UCA1 Interacts Directly with AMOT to Activate YAP Target Genes in Epithelial Ovarian Cancer. iScience 2019, 17, 242–255. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A. , et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nature Reviews Molecular Cell Biology 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Dai, L.; Niu, J.; Feng, Y. Knockdown of long non-coding RNA LINC00176 suppresses ovarian cancer progression by BCL3-mediated down-regulation of ceruloplasmin. Journal of Cellular and Molecular Medicine 2020, 24, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, J.; Yang, L.; Wu, M.; Ma, Q. The Role of Long Non-coding RNAs in Human Imprinting Disorders: Prospective Therapeutic Targets. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Monnier, P.; Martinet, C.; Pontis, J.; Stancheva, I.; Ait-Si-Ali, S.; Dandolo, L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci U S A 2013, 110, 20693–20698. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Zuo, X.; Hou, M.; Li, C.; Teng, Y. LncRNA-H19 regulates chemoresistance to carboplatin in epithelial ovarian cancer through microRNA-29b-3p and STAT3. J Cancer 2021, 12, 5712–5722. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular Cell Review Molecular Mechanisms of Long Noncoding RNAs. Molecular Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics, Proteomics & Bioinformatics 2016, 14, 42–54. [Google Scholar] [CrossRef]
- Gordon, M.A.; Babbs, B.; Cochrane, D.R.; Bitler, B.G.; Richer, J.K. The long non-coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2-mediated alternative splicing. Molecular Carcinogenesis 2019, 58, 196–205. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Wang, F.; Wang, L. Hypoxia-Related lncRNA Prognostic Model of Ovarian Cancer Based on Big Data Analysis. J Oncol 2023, 2023, 6037121. [Google Scholar] [CrossRef]
- Guzel, E.; Okyay, T.M.; Yalcinkaya, B.; Karacaoglu, S.; Gocmen, M.; Akcakuyu, M.H. Tumor suppressor and oncogenic role of long non-coding RNAs in cancer. North Clin Istanb 2019, 7, 81–86. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: what is functional and what is junk? Frontiers in Genetics 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Calanca, N.; Abildgaard, C.; Rainho, C.A.; Rogatto, S.R. The Interplay between Long Noncoding RNAs and Proteins of the Epigenetic Machinery in Ovarian Cancer. Cancers (Basel) 2020, 12, 2701. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Hu, X.; Ye, L.; Bai, P.; Jie, Y.; Shu, K. Long non-coding RNA ADAMTS9-AS1 attenuates ferroptosis by Targeting microRNA-587/solute carrier family 7 member 11 axis in epithelial ovarian cancer. Bioengineered 2022, 13, 8226–8239. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Hsieh, C.-H.; Alonso, L.C. ANRIL: A lncRNA at the CDKN2A/B Locus With Roles in Cancer and Metabolic Disease. Front Endocrinol (Lausanne) 2018, 9, 405. [Google Scholar] [CrossRef]
- Yuan, D.; Guo, T.; Zhu, D.; Ge, H.; Zhao, Y.; Huang, A.; Wang, X.; Cao, X.; He, C.; Qian, H.; Yu, H. Exosomal lncRNA ATB Derived from Ovarian Cancer Cells Promotes Angiogenesis via Regulating miR-204-3p/TGFβR2 Axis. Cancer Manag Res 2022, 14, 327–337. [Google Scholar] [CrossRef]
- Yu, G.; Wang, W.; Deng, J.; Dong, S. LncRNA AWPPH promotes the proliferation, migration and invasion of ovarian carcinoma cells via activation of the Wnt/β-catenin signaling pathway. Molecular Medicine Reports 2019, 49, 3615–3621. [Google Scholar] [CrossRef]
- Wang, D.-Y.; Li, N.; Cui, Y.-L. Long Non-coding RNA CCAT1 Sponges miR-454 to Promote Chemoresistance of Ovarian Cancer Cells to Cisplatin by Regulation of Surviving. Cancer Res Treat 2020, 52, 798–814. [Google Scholar] [CrossRef]
- Mu, Y.; Li, N.; Cui, Y.L. The lncRNA CCAT1 upregulates TGFβR1 via sponging miR-490-3p to promote TGFβ1-induced EMT of ovarian cancer cells. Cancer Cell International 2018, 18, 1–12. [Google Scholar] [CrossRef]
- Cao, Y.; Shi, H.; Ren, F.; Jia, Y.; Zhang, R. Long non-coding RNA CCAT1 promotes metastasis and poor prognosis in epithelial ovarian cancer. Experimental Cell Research 2017, 359, 185–194. [Google Scholar] [CrossRef]
- Lai, X.J.; Cheng, H.F. LncRNA colon cancer-associated transcript 1 (CCAT1) promotes proliferation and metastasis of ovarian cancer via miR-1290. European Review for Medical and Pharmacological Sciences 2018, 22, 322–328. [Google Scholar] [CrossRef]
- Hua, F.; Li, C.-H.; Chen, X.-G.; Liu, X.-P. Long Noncoding RNA CCAT2 Knockdown Suppresses Tumorous Progression by Sponging miR-424 in Epithelial Ovarian Cancer. Oncol Res 2018, 26, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhu, Y.; Fan, X.; Liu, Y.; Feng, Q. Upregulation of long non-coding RNA CCEPR is associated with poor prognosis and contributes to the progression of ovarian cancer through regulating the Wnt/β-catenin signaling pathway. Molecular Medicine Reports 2020, 21, 1950–1958. [Google Scholar] [CrossRef]
- Tan, W.-X.; Sun, G.; Shangguan, M.-Y.; Gui, Z.; Bao, Y.; Li, Y.-F.; Jia, Z.-H. Novel role of lncRNA CHRF in cisplatin resistance of ovarian cancer is mediated by miR-10b induced EMT and STAT3 signaling. Sci Rep 2020, 10, 14768. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, L.-X.; Zhang, C.-Y.; Bai, N.; Feng, C.; Zhang, Z.-M.; Wang, L.; Gao, Z.-Z. LncRNA CRNDE promotes cell proliferation, migration and invasion of ovarian cancer via miR-423-5p/FSCN1 axis. Mol Cell Biochem 2022, 477, 1477–1488. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Ni, X.; Yu, Z.; Wu, S.; Liu, Z. CRNDE inducing cisplatin resistance through SRSF1/TIA1 signaling pathway in ovarian cancer. Pathology - Research and Practice 2022, 235, 153957. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Qi, B.; Yao, H.; Zhang, L.; Li, Y.; Li, Q. Knockdown of DANCR Suppressed the Biological Behaviors of Ovarian Cancer Cells Treated with Transforming Growth Factor-β (TGF-β) by Sponging MiR-214. Medical Science Monitor : International Medical Journal of Experimental and Clinical Research 2020, 26, e922760. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, W.W.; Xu, T.H.; Xu, Z.F. Highly expressed long non-coding RNA DUXAP10 promotes proliferation of ovarian cancer. European Review for Medical and Pharmacological Sciences 2018, 22, 314–321. [Google Scholar] [CrossRef]
- Xu, Q.F.; Tang, Y.X.; Wang, X. LncRNA EBIC promoted proliferation, metastasis and cisplatin resistance of ovarian cancer cells and predicted poor survival in ovarian cancer patients. European Review for Medical and Pharmacological Sciences 2018, 22, 4440–4447. [Google Scholar] [CrossRef]
- Qiu, J.J.; Zhang, X.D.; Tang, X.Y.; Zheng, T.T.; Zhang, Y.; Hua, K.Q. ElncRNA1, a long non-coding RNA that is transcriptionally induced by oestrogen, promotes epithelial ovarian cancer cell proliferation. International Journal of Oncology 2017, 51, 507–514. [Google Scholar] [CrossRef]
- Yan, H.; Li, H.; Silva, M.A.; Guan, Y.; Yang, L.; Zhu, L.; Zhang, Z.; Li, G.; Ren, C. LncRNA FLVCR1-AS1 mediates miR-513/YAP1 signaling to promote cell progression, migration, invasion and EMT process in ovarian cancer. Journal of Experimental & Clinical Cancer Research 2019, 38, 356. [Google Scholar] [CrossRef]
- Zheng, Z.-G.; Xu, H.; Suo, S.-S.; Xu, X.-L.; Ni, M.-W.; Gu, L.-H.; Chen, W.; Wang, L.-Y.; Zhao, Y.; Tian, B.; Hua, Y.-J. The Essential Role of H19 Contributing to Cisplatin Resistance by Regulating Glutathione Metabolism in High-Grade Serous Ovarian Cancer. Sci Rep 2016, 6, 26093. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Wang, Y.; Ao, Y.; Sun, X. CREB1 induced lncRNA HAS2-AS1 promotes epithelial ovarian cancer proliferation and invasion via the miR-466/RUNX2 axis. 2019. [CrossRef]
- Zhang, F.; Luo, B.-H.; Wu, Q.-H.; Li, Q.-L.; Yang, K.-D. LncRNA HCG18 upregulates TRAF4/TRAF5 to facilitate proliferation, migration and EMT of epithelial ovarian cancer by targeting miR-29a/b. Mol Med 2022, 28, 2. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Wang, D.; Chen, X.; Peng, J. Propofol inhibits proliferation, migration, invasion and promotes apoptosis by regulating HOST2/JAK2/STAT3 signaling pathway in ovarian cancer cells. Cytotechnology 2021, 73, 243–252. [Google Scholar] [CrossRef]
- Zhang, Y.; Ai, H.; Fan, X.; Chen, S.; Wang, Y.; Liu, L. Knockdown of long non-coding RNA HOTAIR reverses cisplatin resistance of ovarian cancer cells through inhibiting miR-138-5p-regulated EZH2 and SIRT1. Biological Research 2020, 53, 18. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cui, Z.; Wu, Q.; Wang, H.; Xia, H.; Sun, Y. Long non-coding RNA HOXA11-AS knockout inhibits proliferation and overcomes drug resistance in ovarian cancer. Bioengineered 2022, 13, 13893–13905. [Google Scholar] [CrossRef]
- Dong, S.; Wang, R.; Wang, H.; Ding, Q.; Zhou, X.; Wang, J.; Zhang, K.; Long, Y.; Lu, S.; Hong, T. , et al. HOXD-AS1 promotes the epithelial to mesenchymal transition of ovarian cancer cells by regulating miR-186-5p and PIK3R3. J Exp Clin Cancer Res 2019, 38, 110. [Google Scholar] [CrossRef]
- Zhang, Y.; Dun, Y.; Zhou, S.; Huang, X.H. LncRNA HOXD-AS1 promotes epithelial ovarian cancer cells proliferation and invasion by targeting miR-133a-3p and activating Wnt/β-catenin signaling pathway. Biomedicine & Pharmacotherapy 2017, 96, 1216–1221. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.; Wang, Y.; Wang, S. HOXD-AS1 promotes cell proliferation, migration and invasion through miR-608/FZD4 axis in ovarian cancer. Am J Cancer Res 2018, 8, 170–182. [Google Scholar]
- Huang, B.; Wei, M.; Hong, L. Long noncoding RNA HULC contributes to paclitaxel resistance in ovarian cancer via miR-137/ITGB8 axis. Open Life Sciences 2021, 16, 667–681. [Google Scholar] [CrossRef]
- Lu, X.; Wang, F.; Fu, M.; Li, Y.; Wang, L. Long Noncoding RNA KCNQ1OT1 Accelerates the Progression of Ovarian Cancer via MicroRNA-212-3/LCN2 Axis. Oncol Res 2020, 28, 135. [Google Scholar] [CrossRef]
- Li, J.; Zhang, S.; Wu, L.; Pei, M. Interaction between LncRNA-ROR and miR-145 contributes to epithelial-mesenchymal transition of ovarian cancer cells. | General Physiology & Biophysics | EBSCOhost. 2019.
- Zou, H.; Li, H. Knockdown of long non-coding RNA LINC00152 increases cisplatin sensitivity in ovarian cancer cells. Experimental and Therapeutic Medicine, 3892. [Google Scholar] [CrossRef]
- Wang, S.; Weng, W.; Chen, T.; Xu, M.; Wei, P.; Li, J.; Lu, L.; Wang, Y. LINC00152 Promotes Tumor Progression and Predicts Poor Prognosis by Stabilizing BCL6 From Degradation in the Epithelial Ovarian Cancer. Front Oncol 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Fang, X.; Xia, B.; Zhao, Y.; Li, Q.; Wu, X. Long noncoding RNA LINC00152 promotes cell proliferation through competitively binding endogenous miR-125b with MCL-1 by regulating mitochondrial apoptosis pathways in ovarian cancer. Cancer Medicine 2018, 7, 4530–4541. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Niu, L.L.; Tian, S.C.; Jing, L.K.; Zhang, L.T.; Lin, Q.Q.; Cai, Y.H.; Liang, H.M.; Du, Q.; Li, H. Long non-coding RNA LINC00152 is up-regulated in ovarian cancer tissues and regulates proliferation and cell cycle of SKOV3 cells. European Review for Medical and Pharmacological Sciences 2019, 23, 9803–9813. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; You, J.; Han, Y.; Liu, Y.; Huang, M.; Lu, X.; Chen, J.; Zheng, Y. LINC00184 Promotes Ovarian Cancer Cells Proliferation and Cisplatin Resistance by Elevating CNTN1 Expression via Sponging miR-1305. OncoTargets and Therapy 2021, 14, 2711–2726. [Google Scholar] [CrossRef]
- Du, W.; Feng, Z.; Sun, Q. LncRNA LINC00319 accelerates ovarian cancer progression through miR-423-5p/NACC1 pathway. Biochemical and Biophysical Research Communications 2018, 507, 198–202. [Google Scholar] [CrossRef]
- Wang, K.; Zhu, G.; Bao, S.; Chen, S. Long Non-Coding RNA LINC00511 Mediates the Effects of ESR1 on Proliferation and Invasion of Ovarian Cancer Through miR-424-5p and miR-370-5p. Cancer Manag Res 2019, 11, 10807–10819. [Google Scholar] [CrossRef]
- Xu, D.; Song, Q.; Liu, Y.; Chen, W.; Xu, M.; Fang, X.; Zhao, W.; Zhou, H. LINC00665 promotes Ovarian Cancer progression through regulating the miRNA-34a-5p/E2F3 axis. J Cancer 2021, 12, 1755–1763. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Lu, J.; Wang, J. LncRNA LINC00665 Promotes Ovarian Cancer Cell Proliferation and Inhibits Apoptosis via Targeting MiR-181a-5p/FHDC. 2021. [CrossRef]
- Wang, S.; Liu, C.; Li, Y.; Qiao, J.; Chen, X.; Bao, J.; Li, R.; Xing, Y. LINC00665 affects the malignant biological behavior of ovarian cancer via the miR-148b-3p/KLF5. Systems Biology in Reproductive Medicine 2022, 68, 370–383. [Google Scholar] [CrossRef]
- Lin, X.; Li, P.; Feng, D.; Zheng, J.; Chen, G.; Wu, X.; Dong, Z.; Lv, Y. Regulation of Transcription Factor YAP-TEAD by Non-coding RNA LINC00857 and the Inhibitory Effects on Ovarian Cancer Cell Proliferation. Cellular and Molecular Biology 2022, 68, 162–170. [Google Scholar] [CrossRef]
- Lin, X.; Feng, D.; Li, P.; Lv, Y. LncRNA LINC00857 regulates the progression and glycolysis in ovarian cancer by modulating the Hippo signaling pathway. Cancer Medicine 2020, 9, 8122–8132. [Google Scholar] [CrossRef]
- Xue, H.; Wu, Z.; Rao, D.; Zhuo, B.; Chen, Q. Long non-coding RNA LINC00858 aggravates the oncogenic phenotypes of ovarian cancer cells through miR-134-5p/RAD18 signaling. Arch Gynecol Obstet 2020, 302, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Fu, Q.; Wang, P.P.; Cui, Y.L. STAT1-Induced Upregulation lncRNA LINC00958 Accelerates the Epithelial Ovarian Cancer Tumorigenesis by Regulating Wnt/β-Catenin Signaling. Disease Markers 2021, 2021. [Google Scholar] [CrossRef] [PubMed]
- Yao, N.; Sun, J.Q.; Yu, L.; Ma, L.; Guo, B.Q. LINC00968 accelerates the progression of epithelial ovarian cancer via mediating the cell cycle progression. European Review for Medical and Pharmacological Sciences 2019, 23, 4642–4649. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhai, Y.; Chen, Y. GATA1-induced upregulation of LINC01503 promotes carboplatin resistance in ovarian carcinoma by upregulating PD-L1 via sponging miR-766-5p. Journal of ovarian research 2021, 14. [Google Scholar] [CrossRef]
- Shu, C.; Yan, D.; Mo, Y.; Gu, J.; Shah, N.; He, J. Long noncoding RNA lncARSR promotes epithelial ovarian cancer cell proliferation and invasion by association with HuR and miR-200 family. Am J Cancer Res 2018, 8, 981–992. [Google Scholar]
- Xi, J.; Feng, J.; Zeng, S. Long noncoding RNA lncBRM facilitates the proliferation, migration and invasion of ovarian cancer cells via upregulation of Sox4. Am J Cancer Res 2017, 7, 2180–2189. [Google Scholar]
- Qiao, F.-H.; Tu, M.; Liu, H.-Y. Role of MALAT1 in gynecological cancers: Pathologic and therapeutic aspects. Oncol Lett 2021, 21, 333. [Google Scholar] [CrossRef]
- Zhou, S.; Xu, A.; Song, T.; Gao, F.; Sun, H.; Kong, X. lncRNA MIAT Regulates Cell Growth, Migration, and Invasion Through Sponging miR-150-5p in Ovarian Cancer. Cancer Biotherapy and Radiopharmaceuticals 2020, 35, 650–660. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, L.; Han, X.C.; Ma, H.Y.; Zhang, N.; Zhe, L. LncRNA MIF-AS1 aggravates the progression of ovarian cancer by sponging miRNA-31-5p. European Review for Medical and Pharmacological Sciences 2020, 24, 2248–2255. [Google Scholar] [CrossRef]
- Shen, Y.; Lv, M.; Fang, Y.; Lu, J.; Wu, Y. LncRNA MNX1-AS1 promotes ovarian cancer process via targeting the miR-744-5p/SOX12 axis. Journal of Ovarian Research 2021, 14, 161. [Google Scholar] [CrossRef]
- Yin, L.; Wang, Y. Long non-coding RNA NEAT1 facilitates the growth, migration, and invasion of ovarian cancer cells via the let-7 g/MEST/ATGL axis. Cancer Cell International 2021, 21, 437. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Yang, L.; Wang, X. NEAT1 Knockdown Suppresses the Cisplatin Resistance in Ovarian Cancer by Regulating miR-770-5p/PARP1 Axis. Cancer Manag Res 2020, 12, 7277–7289. [Google Scholar] [CrossRef] [PubMed]
- Tao, F.; Tian, X.; Lu, M.; Zhang, Z. A novel lncRNA, Lnc-OC1, promotes ovarian cancer cell proliferation and migration by sponging miR-34a and miR-34c. Journal of Genetics and Genomics 2018, 45, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Tabury, K.; Monavarian, M.; Listik, E.; Shelton, A.K.; Choi, A.S.; Quintens, R.; Arend, R.C.; Hempel, N.; Miller, C.R.; Györrfy, B.; Mythreye, K. PVT1 is a stress-responsive lncRNA that drives ovarian cancer metastasis and chemoresistance. Life Sci. Alliance 2022, 5, e202201370. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Jiang, J.; Wang, Z.; Xie, Y.; Wu, X. Long non-coding RNA SNHG1 is an unfavorable prognostic factor and promotes cell proliferation and migration by Wnt/β-catenin pathway in epithelial ovarian cancer. International Journal of Clinical and Experimental Pathology 2017, 10, 9284. [Google Scholar]
- Wu, Y.; Zhu, B.; Yan, Y.; Bai, S.; Kang, H.; Zhang, J.; Ma, W.; Gao, Y.; Hui, B.; Li, R. , et al. Long non-coding RNA SNHG1 stimulates ovarian cancer progression by modulating expression of miR-454 and ZEB1. Mol Oncol 2021, 15, 1584–1596. [Google Scholar] [CrossRef]
- Li Pei, M.; Xia Zhao, Z.; Shuang, T. Dysregulation of lnc-SNHG1 and miR-216b-5p correlate with chemoresistance and indicate poor prognosis of serous epithelial ovarian cancer. Journal of Ovarian Research 2020, 13, 144. [Google Scholar] [CrossRef]
- Wang, Y.; Ding, M.; Yuan, X.; Jiao, R.; Zhu, D.; Huang, W.; Deng, W.; Liu, Y. lncRNA SNHG15 Promotes Ovarian Cancer Progression through Regulated CDK6 via Sponging miR-370-3p. BioMed Research International 2021, 2021, e9394563. [Google Scholar] [CrossRef]
- Han, L.; Zhang, W.; Zhang, B.; Zhan, L. Long non-coding RNA SOX2OT promotes cell proliferation and motility in human ovarian cancer. Experimental and Therapeutic Medicine 2018, 15, 2182–2188. [Google Scholar] [CrossRef]
- Kim, L.K.; Park, S.A.; Yang, Y.; Kim, Y.T.; Heo, T.H.; Kim, H.J. LncRNA SRA mediates cell migration, invasion, and progression of ovarian cancer via NOTCH signaling and epithelial-mesenchymal transition. Bioscience Reports 2021, 41. [Google Scholar] [CrossRef]
- Li, Y.; Jiao, Y.; Hao, J.; Xing, H.; Li, C. Long noncoding RNA TP73-AS1 accelerates the epithelial ovarian cancer via epigenetically repressing p21. Am J Transl Res 2019, 11, 2447–2454. [Google Scholar] [PubMed]
- Wang, X.; Yang, B.; She, Y.; Ye, Y. The lncRNA TP73-AS1 promotes ovarian cancer cell proliferation and metastasis via modulation of MMP2 and MMP9. Journal of Cellular Biochemistry 2018, 119, 7790–7799. [Google Scholar] [CrossRef] [PubMed]
- Xiuyun, L.I.; Wang, X.; Li, M.A.O.; Zhao, S.; Haidong, W.E.I. LncRNA TP73-AS1 predicts poor prognosis and promotes cell proliferation in ovarian cancer via cell cycle and apoptosis regulation. Molecular Medicine Reports 2018, 18, 516–522. [Google Scholar] [CrossRef]
- Wu, W.; Gao, H.; Li, X.; Zhu, Y.; Peng, S.; Yu, J.; Zhan, G.; Wang, J.; Liu, N.; Guo, X. LncRNA TPT1-AS1 promotes tumorigenesis and metastasis in epithelial ovarian cancer by inducing TPT1 expression. Cancer Science 2019, 110, 1587–1598. [Google Scholar] [CrossRef]
- Fan, M.; Li, C.; He, P.; Fu, Y.; Li, M.; Zhao, X. Knockdown of long noncoding RNA-taurine-upregulated gene 1 inhibits tumor angiogenesis in ovarian cancer by regulating leucine-rich α-2-glycoprotein-1. Anti-Cancer Drugs 2019, 30, 562–570. [Google Scholar] [CrossRef]
- Pei, Y.; Li, K.; Lou, X.; Wu, Y.; Dong, X.; Wang, W.; Li, N.; Zhang, D.; Cui, W. MiR-1299/NOTCH3/TUG1 feedback loop contributes to the malignant proliferation of ovarian cancer. Oncol Rep 2020, 44, 438–448. [Google Scholar] [CrossRef]
- Dai, T.; Liang, J.; Liu, W.; Zou, Y.; Niu, F.; Li, M.; Zhang, H.; Li, C.; Fan, M.; Cui, G. The miRNA mir-582-3p suppresses ovarian cancer progression by targeting AKT/MTOR signaling via lncRNA TUG1. Bioengineered 2021, 12, 10771–10781. [Google Scholar] [CrossRef]
- Zhan, F.-L.; Chen, C.-F.; Yao, M.-Z. LncRNA TUG1 facilitates proliferation, invasion and stemness of ovarian cancer cell via miR-186-5p/ZEB1 axis. 2020. [CrossRef]
- Gu, L.; Li, Q.; Liu, H.; Lu, X.; Zhu, M. Long Noncoding RNA TUG1 Promotes Autophagy-Associated Paclitaxel Resistance by Sponging miR-29b-3p in Ovarian Cancer Cells. 2020. [CrossRef]
- Wambecke, A.; Ahmad, M.; Morice, P.-M.; Lambert, B.; Weiswald, L.-B.; Vernon, M.; Vigneron, N.; Abeilard, E.; Brotin, E.; Figeac, M. , et al. The lncRNA ‘UCA1’ modulates the response to chemotherapy of ovarian cancer through direct binding to miR-27a-5p and control of UBE2N levels. Mol Oncol 2021, 15, 3659–3678. [Google Scholar] [CrossRef]
- Li, Z.; Niu, H.; Qin, Q.; Yang, S.; Wang, Q.; Yu, C.; Wei, Z.; Jin, Z.; Wang, X.; Yang, A.; Chen, X. lncRNA UCA1 Mediates Resistance to Cisplatin by Regulating the miR-143/FOSL2-Signaling Pathway in Ovarian Cancer. Molecular Therapy - Nucleic Acids 2019, 17, 92–101. [Google Scholar] [CrossRef]
- Zhang, J.; Quan, L.-N.; Meng, Q.; Wang, H.-Y.; Wang, J.; Yu, P.; Fu, J.-T.; Li, Y.-J.; Chen, J.; Cheng, H. , et al. miR-548e Sponged by ZFAS1 Regulates Metastasis and Cisplatin Resistance of OC by Targeting CXCR4 and let-7a/BCL-XL/S Signaling Axis. Molecular Therapy - Nucleic Acids 2020, 20, 621–638. [Google Scholar] [CrossRef]
- Wang, A.; Jin, C.; Li, H.; Qin, Q.; Li, L. LncRNA ADAMTS9-AS2 regulates ovarian cancer progression by targeting miR-182-5p/FOXF2 signaling pathway. International Journal of Biological Macromolecules 2018, 120, 1705–1713. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Liu, Z.; Song, R. Antisense lncRNA As-SLC7A11 suppresses epithelial ovarian cancer progression mainly by targeting SLC7A11. Pharmazie 2017, 72, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Zhu, X.; Teng, Y. Long non-coding RNA CASC2 inhibits progression and predicts favorable prognosis in epithelial ovarian cancer. Molecular Medicine Reports 2018, 18, 5173–5181. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Li, Z.; Pan, J.; Sun, X. Long noncoding RNA DUXAP8 regulates proliferation and apoptosis of ovarian cancer cells via targeting miR-590-5p. Human Cell 2020, 33, 1240–1251. [Google Scholar] [CrossRef]
- Liu, S.; Zou, B.; Tian, T.; Luo, X.; Mao, B.; Zhang, X.; Lei, H. Overexpression of the lncRNA FER1L4 inhibits paclitaxel tolerance of ovarian cancer cells via the regulation of the MAPK signaling pathway. Journal of Cellular Biochemistry 2019, 120, 7581–7589. [Google Scholar] [CrossRef]
- Dong, Q.; Long, X.; Cheng, J.; Wang, W.; Tian, Q.; Di, W. LncRNA GAS5 suppresses ovarian cancer progression by targeting the miR-96-5p/PTEN axis. Ann Transl Med 2021, 9, 1770. [Google Scholar] [CrossRef]
- Liu, F.; Cao, L.; Zhang, Y.; Xia, X.; Ji, Y. LncRNA LIFR-AS1 overexpression suppressed the progression of serous ovarian carcinoma. Journal of Clinical Laboratory Analysis 2022, 36, e25470. [Google Scholar] [CrossRef]
- Hao, T.; Huang, S.; Han, F. LINC-PINT suppresses tumour cell proliferation, migration and invasion through targeting miR-374a-5p in ovarian cancer. Cell Biochemistry and Function 2020, 38, 1089–1099. [Google Scholar] [CrossRef]
- Li, Y.; Lv, M.; Wang, J.; Gao, C.; Wu, Y. LINC00641 inhibits the proliferation and invasion of ovarian cancer cells by targeting miR-320a. Transl Cancer Res 2021, 10, 4894–4904. [Google Scholar] [CrossRef]
- Gokulnath, P.; de Cristofaro, T.; Manipur, I.; Di Palma, T.; Soriano, A.A.; Guarracino, M.R.; Zannini, M. Long Non-Coding RNA MAGI2-AS3 is a New Player with a Tumor Suppressive Role in High Grade Serous Ovarian Carcinoma. Cancers (Basel) 2019, 11, 2008. [Google Scholar] [CrossRef]
- Li, Y.; Lou, S.; Zhang, J.; Zhao, S.; Lou, G. m6A methylation-mediated regulation of LncRNA MEG3 suppresses ovarian cancer progression through miR-885-5p and the VASH1 pathway. J Transl Med 2024, 22, 113. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Jiang, Y.; Wan, Y.; Zhang, L.; Liu, J.; Zhou, S.; Cheng, W. OncoTargets and Therapy Dovepress long noncoding rna nBaT-1 suppresses tumorigenesis and predicts favorable prognosis in ovarian cancer. OncoTargets and Therapy, 2147. [Google Scholar] [CrossRef]
- Tong, W.; Yang, L.; Yu, Q.; Yao, J.; He, A. A new tumor suppressor lncRNA RP11-190D6.2 inhibits the proliferation, migration, and invasion of epithelial ovarian cancer cells. OncoTargets and Therapy 2017, 10, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, C.; Zhang, A.; Wang, Q.; Ge, J.; Li, Q.; Xiao, J. Long non-coding RNA SDCBP2-AS1 delays the progression of ovarian cancer via microRNA-100-5p-targeted EPDR1. World J Surg Onc 2021, 19, 199. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.M.; Li, N. Downregulation of long noncoding RNA TUSC7 promoted cell growth, invasion and migration through sponging with miR-616-5p/GSK3β pathway in ovarian cancer. European Review for Medical and Pharmacological Sciences 2020, 24, 7253–7265. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Yuan, D.; Zhang, W.; Zhu, D.; Xiao, A.; Mao, G.; Jiang, W.; Lin, M.; Wang, J. Upregulation of long noncoding RNA XIST has anticancer effects on ovarian cancer through sponging miR-106a. Human Cell 2021, 34, 579–587. [Google Scholar] [CrossRef]
- Nam, E.J.; Kim, Y.T. Alteration of cell-cycle regulation in epithelial ovarian cancer. International Journal of Gynecologic Cancer 2008, 18. [Google Scholar] [CrossRef]
- Wang, B.-D.; Jiang, J.; Liu, M.-M.; Zhuang, R.-J.; Wang, H.; Li, P.-L. Silencing CCAT2 inhibited proliferation and invasion of epithelial ovarian carcinoma cells by regulating Wnt signaling pathway. International Journal of Clinical and Experimental Pathology 2017, 10, 11771–11778. [Google Scholar]
- Yong, W.; Yu, D.; Jun, Z.; Yachen, D.; Weiwei, W.; Midie, X.; Xingzhu, J.; Xiaohua, W. Long noncoding RNA NEAT1, regulated by LIN28B, promotes cell proliferation and migration through sponging miR-506 in high-grade serous ovarian cancer. Cell Death & Disease 2018, 9. [Google Scholar] [CrossRef]
- Yuan, J.; Yi, K.; Yang, L. LncRNA NEAT1 promotes proliferation of ovarian cancer cells and angiogenesis of co-incubated human umbilical vein endothelial cells by regulating FGF9 through sponging miR-365. Medicine 2021, 100, e23423. [Google Scholar] [CrossRef]
- Zhu, L.; Guo, Q.; Lu, X.; Zhao, J.; Shi, J.; Wang, Z.; Zhou, X. CTD-2020K17.1, a Novel Long Non-Coding RNA, Promotes Migration, Invasion, and Proliferation of Serous Ovarian Cancer Cells In Vitro. Medical Science Monitor 2018, 24, 1329–1339. [Google Scholar] [CrossRef]
- Qiu, J.-j.; Wang, Y.; Liu, Y.-l.; Zhang, Y.; Ding, J.-x.; Hua, K.-q. The long non-coding RNA ANRIL promotes proliferation and cell cycle progression and inhibits apoptosis and senescence in epithelial ovarian cancer. Oncotarget 2016, 7, 32478–32492. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhang, W.; Zhang, B.; Zhan, L. Long non-coding RNA SOX2OT promotes cell proliferation and motility in human ovarian cancer. Experimental and Therapeutic Medicine, 2017. [Google Scholar] [CrossRef]
- Ma, N.; Li, S.; Zhang, Q.; Wang, H.; Qin, H.; Wang, S. Long non-coding RNA GAS5 inhibits ovarian cancer cell proliferation via the control of microRNA-21 and SPRY2 expression. Experimental and Therapeutic Medicine 2018, 16, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Leng, Y.; Duan, M.; Li, Z.; Ma, Y.; Huang, C.; Shi, Q.; Wang, Y.; Wang, C.; Liu, D. , et al. LncRNA GAS5-hnRNPK axis inhibited ovarian cancer progression via inhibition of AKT signaling in ovarian cancer cells. Discov Onc 2023, 14, 157. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wei, Q.-M.; Zhang, X.-W.; Sheng, Q.; Yan, X.-T. MiR-376a promotion of proliferation and metastases in ovarian cancer: Potential role as a biomarker. Life Sciences 2017, 173, 62–67. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Zhao, Y.; Peng, H.; Bai, W.; Zhang, N. MEG3 sponges miRNA-376a and YBX1 to regulate angiogenesis in ovarian cancer endothelial cells. Heliyon 2023, 9. [Google Scholar] [CrossRef]
- Chavez-Dominguez, R.; Perez-Medina, M.; Lopez-Gonzalez, J.S.; Galicia-Velasco, M.; Aguilar-Cazares, D. The Double-Edge Sword of Autophagy in Cancer: From Tumor Suppression to Pro-tumor Activity. Front Oncol 2020, 10. [Google Scholar] [CrossRef]
- Boroughs, L.K.; DeBerardinis, R.J. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 2015, 17, 351–359. [Google Scholar] [CrossRef]
- Chen, S.; Wu, D.-D.; Sang, X.-B.; Wang, L.-L.; Zong, Z.-H.; Sun, K.-X.; Liu, B.-L.; Zhao, Y. The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death & Disease 2017, 8, e3118–e3118. [Google Scholar] [CrossRef]
- Tao, P.; Yang, B.; Zhang, H.; Sun, L.; Wang, Y.; Zheng, W. The overexpression of lncRNA MEG3 inhibits cell viability and invasion and promotes apoptosis in ovarian cancer by sponging miR-205-5p. International Journal of Clinical and Experimental Pathology 2020, 13, 869–879. [Google Scholar]
- Jin, Y.; Qiu, J.; Lu, X.; Ma, Y.A.N.; Li, G. LncRNA CACNA1G-AS1 up-regulates FTH1 to inhibit ferroptosis and promote malignant phenotypes in ovarian cancer cells. Oncol Res 2023, 31, 169–179. [Google Scholar] [CrossRef]
- Nazio, F.; Bordi, M.; Cianfanelli, V.; Locatelli, F.; Cecconi, F. Autophagy and cancer stem cells: molecular mechanisms and therapeutic applications. Cell Death & Differentiation 2019, 26, 690–702. [Google Scholar] [CrossRef]
- Xu, P.; Xu, S.; Pan, H.; Dai, C.; Xu, Y.; Wang, L.; Cong, Y.; Zhang, H.; Cao, J.; Ge, L.; Jia, X. Differential effects of the LncRNA RNF157-AS1 on epithelial ovarian cancer cells through suppression of DIRAS3- and ULK1-mediated autophagy. Cell Death Dis 2023, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.-J.; Jiang, P.; Zhai, H.; Dong, J.-S. <p>LncRNA GAS8-AS1 Inhibits Ovarian Cancer Progression Through Activating Beclin1-Mediated Autophagy</p>. OncoTargets and Therapy 2020, Volume 13, 10431–10440. [Google Scholar] [CrossRef]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef] [PubMed]
- Nadhan, R.; Dhanasekaran, D.N. Regulation of Tumor Metabolome by Long Non-Coding RNAs. Journal of Molecular Signalling, 2022. [Google Scholar] [CrossRef]
- Nadhan, R.; Kashyap, S.; Ha, J.H.; Jayaraman, M.; Song, Y.S.; Isidoro, C.; Dhanasekaran, D.N. Targeting Oncometabolites in Peritoneal Cancers: Preclinical Insights and Therapeutic Strategies. Metabolites 2023, 13. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, N. Noncoding RNAs in the Glycolysis of Ovarian Cancer. Frontiers in Pharmacology 2022, 13. [Google Scholar] [CrossRef]
- Li, N.; Zhan, X.; Zhan, X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol 2018, 150, 343–354. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Lee, J.; Haemmerle, M.; Ling, H.; Previs, Rebecca A. ; Pradeep, S.; Wu, Sherry Y.; Ivan, C.; Ferracin, M.; Dennison, Jennifer B., et al. Long Noncoding RNA Ceruloplasmin Promotes Cancer Growth by Altering Glycolysis. Cell Reports 2015, 13, 2395–2402. [Google Scholar] [CrossRef]
- Nieman, K.M.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.L.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S. , et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med 2011, 17, 1498–1503. [Google Scholar] [CrossRef]
- Li, M.; Yan, Y.; Liu, Y.; Zhao, J.; Guo, F.; Chen, J.; Nie, L.; Zhang, Y.; Wang, Y. Comprehensive analyses of fatty acid metabolism-related lncRNA for ovarian cancer patients. Scientific Reports 2023, 13, 14675. [Google Scholar] [CrossRef]
- Ye, L.; Jiang, Z.; Zheng, M.; Pan, K.; Lian, J.; Ju, B.; Liu, X.; Tang, S.; Guo, G.; Zhang, S. , et al. Fatty Acid Metabolism-Related lncRNA Prognostic Signature for Serous Ovarian Carcinoma. Epigenomics 2024, 16, 309–329. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Qu, S.; Zhai, Y.; Yang, X. circ_0025033 promotes ovarian cancer development via regulating the hsa_miR-370-3p/SLC1A5 axis. Cellular & molecular biology letters 2022, 27, 94. [Google Scholar] [CrossRef]
- Yuan, J.; Liu, Z.; Song, R. Antisense lncRNA As-SLC7A11 suppresses epithelial ovarian cancer progression mainly by targeting SLC7A11. Pharmazie 2017, 72, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Ji, G.; Le, X.; Wang, C.; Xu, L.; Feng, M.; Zhang, Y.; Yang, H.; Xuan, Y.; Yang, Y. , et al. Long Noncoding RNA LINC00092 Acts in Cancer-Associated Fibroblasts to Drive Glycolysis and Progression of Ovarian Cancer. Cancer Research 2017, 77, 1369–1382. [Google Scholar] [CrossRef]
- Nadhan, R.; Ha, J.H.; Jayaraman, M.; Kashyap, S.; Dhanasekaran, D.N. Abstract LB039: Ovarian cancer cell-derived exosomal UCA1 reprograms glucose metabolism in stromal fibroblasts. Cancer Research 2023, 83, LB039–LB039. [Google Scholar] [CrossRef]
- Friedl, P.; Wolf, K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer 2003, 3, 362–374. [Google Scholar] [CrossRef]
- Lampropoulou, D.I.; Papadimitriou, M.; Papadimitriou, C.; Filippou, D.; Kourlaba, G.; Aravantinos, G.; Gazouli, M. The Role of EMT-Related lncRNAs in Ovarian Cancer. International Journal of Molecular Sciences 2023, 24, 10079. [Google Scholar] [CrossRef]
- Xiong, T.; Wang, Y.; Zhang, Y.; Yuan, J.; Zhu, C.; Jiang, W. lncRNA AC005224.4/miR-140-3p/SNAI2 regulating axis facilitates the invasion and metastasis of ovarian cancer through epithelial-mesenchymal transition. Chinese Medical Journal 2023, 136, 1098–1110. [Google Scholar] [CrossRef]
- Li, J.; Wang, J.; Zhong, Y.; Guo, R.; Chu, D.; Qiu, H.; Yuan, Z. HOTAIR: a key regulator in gynecologic cancers. Cancer Cell International 2017, 17, 65. [Google Scholar] [CrossRef]
- Qiu, J.J.; Lin, Y.Y.; Ye, L.C.; Ding, J.X.; Feng, W.W.; Jin, H.Y.; Zhang, Y.; Li, Q.; Hua, K.Q. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol 2014, 134, 121–128. [Google Scholar] [CrossRef]
- Gao, Z.; Liu, R.; Ye, N.; Liu, C.; Li, X.; Guo, X.; Zhang, Z.; Li, X.; Yao, Y.; Jiang, X. FOXO1 Inhibits Tumor Cell Migration via Regulating Cell Surface Morphology in Non-Small Cell Lung Cancer Cells. Cellular Physiology and Biochemistry 2018, 48, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Zamaraev, A.V.; Volik, P.I.; Sukhikh, G.T.; Kopeina, G.S.; Zhivotovsky, B. Long non-coding RNAs: A view to kill ovarian cancer. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer 2021, 1876, 188584. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Guo, R.; Yuan, Z.; Shi, H.; Zhang, D. LncRNA HOTAIR Regulates CCND1 and CCND2 Expression by Sponging miR-206 in Ovarian Cancer. Cellular Physiology and Biochemistry 2018, 49, 1289–1303. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, W.; He, Y.; Xia, Q.; Liu, S. LncRNA MEG3 impacts proliferation, invasion, and migration of ovarian cancer cells through regulating PTEN. Inflammation Research 2018, 67, 927–936. [Google Scholar] [CrossRef]
- Saman, H.; Raza, S.S.; Uddin, S.; Rasul, K. Inducing Angiogenesis, a Key Step in Cancer Vascularization, and Treatment Approaches. Cancers (Basel) 2020, 12, 1172. [Google Scholar] [CrossRef]
- Teppan, J.; Barth, D.A.; Prinz, F.; Jonas, K.; Pichler, M.; Klec, C. Involvement of Long Non-Coding RNAs (lncRNAs) in Tumor Angiogenesis. Noncoding RNA 2020, 6, 42. [Google Scholar] [CrossRef]
- Lin, X.; Yang, F.; Qi, X.; Li, Q.; Wang, D.; Yi, T.; Yin, R.; Zhao, X.; Zhong, X.; Bian, C. LncRNA DANCR promotes tumor growth and angiogenesis in ovarian cancer through direct targeting of miR-145. Molecular Carcinogenesis 2019, 58, 2286–2296. [Google Scholar] [CrossRef]
- Yuan, D.; Guo, T.; Zhu, D.; Ge, H.; Zhao, Y.; Huang, A.; Wang, X.; Cao, X.; He, C.; Qian, H.; Yu, H. Exosomal lncRNA ATB Derived from Ovarian Cancer Cells Promotes Angiogenesis via Regulating miR-204-3p/TGFβR2 Axis. Cancer Management and Research 2022, Volume 14, 327–337. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Wu, Y.; Zhao, Y.; Hu, X.; Sun, C. The long non-coding RNA NEAT1 promotes the progression of human ovarian cancer through targeting miR-214-3p and regulating angiogenesis. Journal of Ovarian Research 2023, 16. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Zhao, Y.; Peng, H.; Bai, W.; Zhang, N. MEG3 sponges miRNA-376a and YBX1 to regulate angiogenesis in ovarian cancer endothelial cells. Heliyon 2023, 9, e13204. [Google Scholar] [CrossRef]
- Kenda Suster, N.; Virant-Klun, I. Presence and role of stem cells in ovarian cancer. World J Stem Cells 2019, 11, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Królewska-Daszczyńska, P.; Wendlocha, D.; Smycz-Kubańska, M.; Stępień, S.; Mielczarek-Palacz, A. Cancer stem cells markers in ovarian cancer: Clinical and therapeutic significance (Review). Oncol Lett 2022, 24, 465. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Wu, Y.; Qiu, L.; Zhao, R.; Jiang, M.; Zhang, H. LncRNAs link cancer stemness to therapy resistance. American Journal of Cancer Research 2021, 11, 1051–1068. [Google Scholar] [PubMed]
- Zhang, Y.; Guo, J.; Cai, E.; Cai, J.; Wen, Y.; Lu, S.; Li, X.; Han, Q.; Jiang, J.; Li, T.; Wang, Z. HOTAIR maintains the stemness of ovarian cancer stem cells via the miR-206/TBX3 axis. Experimental Cell Research 2020, 395, 112218. [Google Scholar] [CrossRef]
- Mamun, M.A.; Mannoor, K.; Cao, J.; Qadri, F.; Song, X. SOX2 in cancer stemness: tumor malignancy and therapeutic potentials. J Mol Cell Biol 2018, 12, 85–98. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Y.; Zhong, W.; Cheng, H.; Tian, Z. The Long Non-Coding RNA MALAT1 Enhances Ovarian Cancer Cell Stemness by Inhibiting YAP Translocation from Nucleus to Cytoplasm. Med Sci Monit 2020, 26, 0–0. [Google Scholar] [CrossRef]
- Hou, R.; Jiang, L. LINC00115 promotes stemness and inhibits apoptosis of ovarian cancer stem cells by upregulating SOX9 and inhibiting the Wnt/β-catenin pathway through competitively binding to microRNA-30a. Cancer Cell International 2021, 21, 360. [Google Scholar] [CrossRef]
- Varier, K.M.; Dhandapani, H.; Liu, W.; Song, J.; Wang, C.; Hu, A.; Ben-David, Y.; Shen, X.; Li, Y.; Gajendran, B. An immunotherapeutic approach to decipher the role of long non-coding RNAs in cancer progression, resistance and epigenetic regulation of immune cells. Journal of Experimental & Clinical Cancer Research 2021, 40. [Google Scholar] [CrossRef]
- Liu, J.; Yan, C.; Xu, S. LncRNA IL21-AS1 facilitates tumour progression by enhancing CD24-induced phagocytosis inhibition and tumorigenesis in ovarian cancer. Cell Death & Disease 2024, 15. [Google Scholar] [CrossRef]
- Lei, J.; He, Z.Y.; Wang, J.; Hu, M.; Zhou, P.; Lian, C.L.; Hua, L.; Wu, S.G.; Zhou, J. Identification of MEG8/miR-378d/SOBP axis as a novel regulatory network and associated with immune infiltrates in ovarian carcinoma by integrated bioinformatics analysis. Cancer Medicine 2021, 10, 2924–2939. [Google Scholar] [CrossRef]
- Qian, M.; Ling, W.; Ruan, Z. Long non-coding RNA SNHG12 promotes immune escape of ovarian cancer cells through their crosstalk with M2 macrophages. Aging 2020, 12, 17122–17136. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xie, H.-J.; Li, Y.-Y.; Wang, X.; Liu, X.-X.; Mai, J. Molecular mechanisms of platinum-based chemotherapy resistance in ovarian cancer (Review). Oncol Rep 2022, 47, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Wang, M. LINC01118 Modulates Paclitaxel Resistance of Epithelial Ovarian Cancer by Regulating miR-134/ABCC1. Med Sci Monit 2018, 24, 8831–8839. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, T.; Xia, L.; Zhang, M. LncRNA WDFY3-AS2 promotes cisplatin resistance and the cancer stem cell in ovarian cancer by regulating hsa-miR-139-5p/SDC4 axis. Cancer Cell International 2021, 21, 284. [Google Scholar] [CrossRef] [PubMed]
- Penning, T.M.; Jonnalagadda, S.; Trippier, P.C.; Rižner, T.L. Aldo-Keto Reductases and Cancer Drug Resistance. Pharmacological Reviews 2021, 73, 1150–1171. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, J.; Xu, X.; Li, L. Curcumin suppresses cisplatin resistance development partly via modulating extracellular vesicle-mediated transfer of MEG3 and miR-214 in ovarian cancer. Cancer Chemother Pharmacol 2017, 79, 479–487. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, M.; Shi, C.; Shi, F.; Pei, C. Long non-coding RNA Linc00312 modulates the sensitivity of ovarian cancer to cisplatin via the Bcl-2/Caspase-3 signaling pathway. BioScience Trends 2018, 12, 309–316. [Google Scholar] [CrossRef]
- Xiao, L.; Shi, X.-Y.; Li, Z.-L.; Li, M.; Zhang, M.-M.; Yan, S.-J.; Wei, Z.-L. Downregulation of LINC01508 contributes to cisplatin resistance in ovarian cancer via the regulation of the Hippo-YAP pathway. Journal of Gynecologic Oncology 2021, 32. [Google Scholar] [CrossRef]
- Guo, J.; Pan, H. Long Noncoding RNA LINC01125 Enhances Cisplatin Sensitivity of Ovarian Cancer via miR-1972. Med Sci Monit 2019, 25, 9844–9854. [Google Scholar] [CrossRef]
- Jurkovicova, D.; Neophytou, C.M.; Gašparović, A.Č.; Gonçalves, A.C. DNA Damage Response in Cancer Therapy and Resistance: Challenges and Opportunities. International Journal of Molecular Sciences 2022, 23, 14672. [Google Scholar] [CrossRef]
- Hara, T.; Omura-Minamisawa, M.; Chao, C.; Nakagami, Y.; Ito, M.; Inoue, T. Bcl-2 inhibitors potentiate the cytotoxic effects of radiation in Bcl-2 overexpressing radioresistant tumor cells. International Journal of Radiation Oncology*Biology*Physics 2005, 61, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Dou, Q.; Xu, Y.; Zhu, Y.; Hu, Y.; Yan, Y.; Yan, H. LncRNA FAM83H-AS1 contributes to the radioresistance, proliferation, and metastasis in ovarian cancer through stabilizing HuR protein. European Journal of Pharmacology 2019, 852, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Salamini-Montemurri, M.; Lamas-Maceiras, M.; Barreiro-Alonso, A.; Vizoso-Vázquez, Á.; Rodríguez-Belmonte, E.; Quindós-Varela, M.; Cerdán, M.E. The Challenges and Opportunities of LncRNAs in Ovarian Cancer Research and Clinical Use. Cancers (Basel) 2020, 12, 1020. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Lei, N.; Tian, W.; Li, Y.; Chang, L. Recent advances of non-coding RNAs in ovarian cancer prognosis and therapeutics. Ther Adv Med Oncol 2022, 14, 17588359221118010. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-N.; Zhang, H.-Y. Serum lncRNA LOXL1-AS1 is a diagnostic and prognostic marker for epithelial ovarian cancer. The Journal of Gene Medicine 2020, 22, e3233. [Google Scholar] [CrossRef]
- Zuo, K.; Zhao, Y.; Zheng, Y.; Chen, D.; Liu, X.; Du, S.; Liu, Q. Long non-coding RNA XIST promotes malignant behavior of epithelial ovarian cancer. OncoTargets and Therapy 2019, 12, 7261–7267. [Google Scholar] [CrossRef]
- Wang, J.; Tian, Y.; Zheng, H.; Ding, Y.; Wang, X. An integrated analysis reveals the oncogenic function of lncRNA LINC00511 in human ovarian cancer. Cancer Medicine 2019, 8, 3026–3035. [Google Scholar] [CrossRef]
- Chen, Y.; Bi, F.; An, Y.; Yang, Q. Identification of pathological grade and prognosis-associated lncRNA for ovarian cancer. Journal of Cellular Biochemistry 2019, 120, 14444–14454. [Google Scholar] [CrossRef]
- Zhou, M.; Sun, Y.; Sun, Y.; Xu, W.; Zhang, Z.; Zhao, H.; Zhong, Z.; Sun, J. Comprehensive analysis of lncRNA expression profiles reveals a novel lncRNA signature to discriminate nonequivalent outcomes in patients with ovarian cancer. Oncotarget 2016, 7, 32433–32448. [Google Scholar] [CrossRef]
- Gong, M.; Luo, C.; Meng, H.; Li, S.; Nie, S.; Jiang, Y.; Wan, Y.; Li, H.; Cheng, W. Upregulated LINC00565 Accelerates Ovarian Cancer Progression By Targeting GAS6. OncoTargets and Therapy 2019, 12, 10011–10022. [Google Scholar] [CrossRef]
- Yang, K.; Hou, Y.; Li, A.; Li, Z.; Wang, W.; Xie, H.; Rong, Z.; Lou, G.; Li, K. Identification of a six-lncRNA signature associated with recurrence of ovarian cancer. Sci Rep 2017, 7, 752. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Biglia, N.; Wang, Z.; Shen, Y.; Risch, H.A.; Lu, L.; Canuto, E.M.; Jia, W.; Katsaros, D.; Yu, H. Long non-coding RNAs, <i>ASAP1-IT1</i>, <i>FAM215A</i>, and <i>LINC00472</i>, in epithelial ovarian cancer. Gynecologic Oncology 2016, 143, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Salamini-Montemurri, M.; Lamas-Maceiras, M.; Lorenzo-Catoira, L.; Vizoso-Vázquez, Á.; Barreiro-Alonso, A.; Rodríguez-Belmonte, E.; Quindós-Varela, M.; Cerdán, M.E. Identification of lncRNAs Deregulated in Epithelial Ovarian Cancer Based on a Gene Expression Profiling Meta-Analysis. International Journal of Molecular Sciences 2023, 24, 10798. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Girisa, S.; Alqahtani, M.S.; Abbas, M.; Hegde, M.; Sethi, G.; Kunnumakkara, A.B. Targeting Autophagy Using Long Non-Coding RNAs (LncRNAs): New Landscapes in the Arena of Cancer Therapeutics. Cells 2023, 12, 810. [Google Scholar] [CrossRef]
- Zhou, L.; Zhu, Y.; Sun, D.; Zhang, Q. Emerging Roles of Long non-coding RNAs in The Tumor Microenvironment. Int J Biol Sci 2020, 16, 2094–2103. [Google Scholar] [CrossRef]
- Fathi Dizaji, B. Strategies to target long non-coding RNAs in cancer treatment: progress and challenges. Egypt J Med Hum Genet 2020, 21, 41. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhu, L.; Wang, X.; Jin, H. RNA-based therapeutics: an overview and prospectus. Cell Death & Disease 2022, 13. [Google Scholar] [CrossRef]
- Tanioka, M.; Nokihara, H.; Yamamoto, N.; Yamada, Y.; Yamada, K.; Goto, Y.; Fujimoto, T.; Sekiguchi, R.; Uenaka, K.; Callies, S.; Tamura, T. Phase I study of LY2181308, an antisense oligonucleotide against survivin, in patients with advanced solid tumors. Cancer Chemother Pharmacol 2011, 68, 505–511. [Google Scholar] [CrossRef]
- Oza, A.M.; Elit, L.; Swenerton, K.; Faught, W.; Ghatage, P.; Carey, M.; McIntosh, L.; Dorr, A.; Holmlund, J.T.; Eisenhauer, E. Phase II study of CGP 69846A (ISIS 5132) in recurrent epithelial ovarian cancer: an NCIC clinical trials group study (NCIC IND.116)☆. Gynecologic Oncology 2003, 89, 129–133. [Google Scholar] [CrossRef]
| LncRNA | Function | Mechanism | |
|---|---|---|---|
| 1. | ADAMTS9-AS1 | Inhibits ferroptosis resulting in increased cell proliferation and migration | Acts as a ceRNA to miR-587 downregulating the expression of SLC7A11 [76] |
| 2. | ANRIL | Promotes cell proliferation and invasion. | Activation of Wnt/ β-catenin pathway [77] |
| 3. | ATB | Promotes tumorigenesis | Acts as a ceRNA towards miR-204-3p upregulating TGFβR2 pathway [78] |
| 4. | AWPPH | Promotes overall OC development | Up-regulates β-catenin expression by activating the Wnt/β-catenin pathway [79] |
| 5. | CCAT1 | Confers cisplatin resistance | Acts as a ceRNA against miR-454, inducing the expression of survivin [80] |
| Induces EMT of OC cells | Upregulates TGFβR1 through sponging of miR-490-3p [81] | ||
| Contributes to metastasis and progression in epithelial ovarian cancer (EOC) | Modulates the ADAM17/Wnt1/STAT3/ZEB1 regulatory network via miR-152 and miR-130b [82] | ||
| Promotes proliferation of OC | Sequesters miR-1290 and suppresses its tumorigenic role [83] | ||
| 6. | CCAT2 | Acts as an oncogene | Sequesters to miR-424 resulting in its downregulation [84] |
| 7. | CCEPR | Promotes cellular invasion and poor prognosis | Activation of Wnt/ β-catenin pathway [85] |
| 8. | CHRF | Confers cisplatin resistance | Acts as ceRNA to miR-10b activating STAT3 pathway [86] |
| 9. | CRNDE | Promotes cell migration, invasion, and proliferation | Acts as a ceRNA against miR-423-5p resulting in its downregulation [87] |
| Confers cisplatin resistance | Activation of SRSF1/TIA1 signaling pathway [88] | ||
| 10. | DANCR | Promotes cell proliferation and migration | Negative regulation of TGF- β by acting a ceRNA for miR-214 [89] |
| 11. | DUXAP10 | Promotes cell proliferation | Increased expression of DUXAP10 positively regulates the proliferation of OC cells [90] |
| 12. | EBIC | Promotes cell proliferation, invasion and migration. Confers cisplatin resistance | Activation of Wnt/ β-catenin pathway [91] |
| 13. | ElncRNA1 | Oncogenic role in overall EOC progression | E2 (estrogen) transcriptionally induces ElncRNA1, which modulates cyclin D1-CDK4/6 [92] |
| 14. | FLVCR1-AS1 | Promotes EMT | Acts as ceRNA to miR-513 upregulating YAP1 expression [93] |
| 15. | H19 | Promotes cisplatin-resistance | Glutathione metabolism [94] |
| 16. | HAS2-AS1 | Accelerates EOC tumorigenesis, facilitates invasion and proliferation | HAS2-AS1, induced by CREB1, sequesters miR-466, thus positively regulates RUNX2 gene [95] |
| 17. | HCG18 | Cell proliferation and migration | Acts as a ceRNA for miR-29a/b downregulating TRAF4/5, activating NF-κB pathway [96] |
| 18. | HOST2 | Promotes cell proliferation, migration and invasion | Activation of JAK2/STAT3 pathway [97] |
| 19. | HOTAIR | Confers cisplatin resistance | Regulates Her2 expression by acting as a ceRNA against miR-138-5p [98] |
| 20. | HOXA11-AS | Confers cisplatin resistance | Inhibits intracellular autophagy and cell cycle arrest [99] |
| 21. | HOXD-AS1 | Regulates cell migration, invasion, and EMT in EOC | Elevated HOXD-AS1 leads to increased levels of PIK3R3 by sequestering miR-186-5p (acts as a ceRNA) [100] |
| Promotes cell proliferation, migration, and invasion and EMT in EOC cells | Activates the Wnt/ β-catenin pathway by sequestering miR-133a-3p [101] | ||
| Positively regulates proliferation, migration, and invasion in OC cells | HOXD-AS1 mediates this effect partially through the miR-608/FZD4 axis [102] | ||
| 22. | HULC | Confers paclitaxel resistance | Acts as a ceRNA against miR-199a-3p upregulating the expression of ITGB8 [103] |
| 23. | KCNQ1OT1 | Enhances cell growth, migration, and invasion and inhibits cell apoptosis | Positively regulates LCN2 expression by repressing miR-212-3p [104] |
| 24. | LINC-ROR | Promotes EMT | Suppresses miR-145, promoting the expression of FLNB [105] |
| 25. | LINC00152 | Confers cisplatin resistance in COC1/DDP cells | Modulates apoptosis and expression of MDR1, GSTn, and MRP1 [106] |
| Increased levels facilitate invasion and tumor proliferation in EOC | Prevents ubiquitination of Bcl6 by binding to its Ser 333/Ser 343 site [107] | ||
| Mediates cell proliferation and survival in OC | Affects MCL1- dependent mitochondrial apoptosis and acts as a ceRNA of miR-125b [108] | ||
| Regulates cell cycle and cell proliferation in EOC cells | Modulates the Tumor Necrosis Factor (TNF) signaling pathway [109] | ||
| 26. | LINC00184 | Promotes cellular proliferation and confers cisplatin resistance | Promotes CNTN1 expression by acting as a ceRNA towards miR-1305 [110] |
| 27. | LINC00319 | Facilitates proliferation, migration, invasion, and tumor growth | Upregulates NACC1 by sequestering miR-423-5p [111] |
| 28. | LINC00511 | Promotes cell proliferation and invasion | Acts as a ceRNA against miR-424-5p and miR-370-5p which are responsible for anti-tumor effects [112] |
| 29. | LINC00665 | Promotes tumor progression | Regulates the miRNA-34a-5p/E2F3 axis [113] |
| Facilitates cancer cell proliferation and inhibits apoptosis | Upregulates FHDC1 by sequestering miR-181a-5p [114] |
||
| Promotes cancer cell proliferation and migration | Positively regulates KLF5 via sponging miR-148b-3p [115] | ||
| 30. | LINC00857 | Modulates OC progression and glycolysis | Regulates Hippo signaling Pathway through the miR-486-5p/YAP1 axis [116] |
| Reduces the proliferative, invasive, and migratory capacity of OC cells and facilitates cell apoptosis | Reduces YAP-TEAD expression via Hippo signaling pathway [117] | ||
| 31. | LINC00858 | Contributes to the metastatic nature of OC | Acts as a ceRNA towards miR-134-5p, upregulating RAD18 expression [118] |
| 32. | LINC00958 | STAT1-induced over-expression promotes overall EOC progression (proliferation, invasion, and migration) | Epigenetic modulation of the Wnt/ β-catenin pathway [119] |
| 33. | LINC00968 | Accelerates EOC progression | Arrests cell cycle in the G1 phase by inhibiting the MAPK and PI3K/Akt/mTOR pathways [120] |
| 34. | LINC01503 | Contributes to carboplatin resistance in OC | Up-regulates PD-L1 levels by sequestering miR-766-5p [121] |
| 35. | lncARSR | Enhances EOC cells' proliferative and invasive property | Upregulates β-catenin and ZEB1/2 via association with HuR and miR-200 family respectively [122] |
| 36. | lncBRM | Facilitates migration, invasion, and proliferation in OC cells | Up-regulates SOX4 via sequestering miR-204 [123] |
| 37. | MALAT1 | Induces cell proliferation, migration and EMT- transition. | Activation of PI3K/AKT pathway [124] |
| 38. | MIAT | Promotes EMT, migration, invasion and proliferation | Acts as a ceRNA towards resulting in its suppression [125] |
| 39. | MIF-AS1 | Promotes cell proliferation, migration and invasion | Acts as a ceRNA to miR-NA-31-5p downregulating PLCB1 expression [126] |
| 40. | MNX1-AS1 | Promotes overall OC carcinogenesis | Upregulates SOX12 by repressing miR-744-5p [127] |
| 41. | NEAT1 | Promotes cell proliferation and migration. | Acts as ceRNA binding to let-7g promoting MEST and inhibiting ATGL expression [128] |
| Confers cisplatin resistance | Regulates the expression of PARP1 and acts as a ceRNA against miR-770-5p [129] | ||
| 42. | OC1 | Promotes cell proliferation and migration | Acts as a ceRNA to miR-34a and miR-34c which regulates tumorigenesis [130] |
| 43. | PVT1 | Promotes cell migration and survival | Activation of YAP1-mediated tumorigenesis [131] |
| 44. | SNHG1 | Promotes proliferation and migration in EOC | Activates downstream effectors of the Wnt/β-catenin pathway[132] |
| Facilitates migration and invasion of OC cells | Modulates via SNHG1/miR-454/ZEB1 axis[133] | ||
| Modulates chemoresistance in SOC cells and patients (paclitaxel) | Functions as a ceRNA for miR-216b-5p in conferring paclitaxel resistance in OC [134] | ||
| 45. | SNHG25 | Promotes overall EOC progression | Positively regulates COMP (cartilage oligomeric matrix protein) contributing to the more invasiveness nature of the tumor [135] |
| 46. | SOX2OT | Facilitates OC progression | SOX2-OT contributed to OC malignancy through miR-181b-5p/SCD1 axis [136] |
| 47. | SRA | Facilitates cell proliferation, migration, and tumor invasion | Via EMT and NOTCH signaling pathway [137] |
| 48. | TP73-AS1 | Contributes to EOC carcinogenesis | Epigenetically suppresses p21 via trimethylation of H3K27 by recruiting EZH2 [138] |
| Positively regulates tumor growth and metastasis, and facilitates overall OC progression | Increased expression of TP73-AS1 enhances levels of MMP2 and MMP9 [139] | ||
| Promotes proliferation and overall OC progression | Negatively regulates cellular apoptosis and cell cycle [140] | ||
| 49. | TPT1-AS1 | Contributed to EOC tumor development and metastasis, and inhibited cellular adhesion | Induces TPT1 expression and activates the PI3K/AKT pathway [141] |
| 50. | TUG1 | Facilitates angiogenesis of endothelial cells in OC cells | Regulates LRG1 secretion levels partially via the TGF-β pathway [142] |
| Promotes OC cell proliferation and malignancy | Acts as a ceRNA for miR-1299, thus positively regulating NOTCH3 expression levels [143] | ||
| Affects OC progression and carcinogenesis | Works as an interacting component of the miR-582-3p/AKT/mTOR axis [144] | ||
| Contributes to stemness, proliferation, and invasion of OC cells | TUG1 sequesters miR-186-5p to release ZEB1 [145] | ||
| Confers autophagy-associated paclitaxel resistance in OC cells | Sequesters miR-29b-3p and consequently mediates paclitaxel resistance via autophagy induction [146] | ||
| 51. | UCA1 | Confers cisplatin resistance | Acts as ceRNA to miR-27a-5p regulating the expression of UBE2N [147] Acts as a ceRNA for miR-143 upregulating FOSL2 expression [148] |
| Promotes proliferation, invasive migration, and therapy resistance | Sequesters a panel of the let-7 family of miRNAs negatively regulating their tumor suppressive roles [60] | ||
| 52. | ZFAS1 | Promotes cell proliferation and metastasis | Sequesters tumor suppressive roles of miR-548e [149] |
| Confers cisplatin resistance | Suppresses the expression of let-7a further elevating BCL-XL/S levels [149] |
| LncRNA | Function | Mechanism | |
|---|---|---|---|
| 1. | ADAMTS9-AS2 | Inhibits cell proliferation and invasion | Acts as a ceRNA against miR-182-5p modulating FOXF2 pathway [150] |
| 2. | AS-SLC7A11 | Reduced AS-SLC7A11 promotes EOC progression | AS-SLC7A11 mainly deregulates SLC7A11 to suppress EOC progression [151] |
| 3. | CASC2 | Inhibits migration, invasion and proliferation | Reduced expression can be linked with poor prognosis in patient samples [152] |
| 4. | DUXAP8 | Regulate the proliferation and apoptosis of OC cells | Mediates YAP1 regulation via the suppression of miR-590-5p[153] |
| 5. | FER1L4 | Higher levels of FER1L4 facilitate paclitaxel sensitivity of OC cells | Suppresses paclitaxel resistance via inhibition of the MAPK pathway [154] |
| 6. | GAS5 | Inhibition of cell proliferation, migration, and invasion. | Activation of AKT/PTEN pathway by sequestering miR-96-5p [155] |
| 7. | LIFR-AS1 | Deregulation in OC cells and subsequent patients correlates to poor prognosis and increased carcinogenesis | Overexpression of LIFR-AS1 is associated with decreased invasion, migration, proliferation and viability in SOC cells [156] |
| 8. | LINC-PINT | Inhibits cell migration, invasion, EMT, and proliferation, and promotes cellular apoptosis (acts as a tumor suppressor) | Increased levels of LINC-PINT sequester miR-374a-5p (acts as an oncogene) [157] |
| 9. | LINC00641 | Suppresses the oncogenic role of miR-320a | Acts as a ceRNA for miR-320a which promotes cell migration and invasion [158] |
| 10. | MAGI2-AS3 | Suppresses the oncogenic role of miRNAs | Sequesters towards miR-15-5p, miR-374a-5p and miR-374b-5p [159] |
| 11. | MEG3 | Inhibits cellular proliferation and metastasis | Acts as a ceRNA against miR-885-5p increasing VASH1 expression [160] |
| 12. | NBAT-1 | Suppresses tumorigenesis | Mediates its effect by targeting the AKT and ERK pathway [161] |
| 13. | RP11-190D6.2 | Low levels of RP11-190D6.2 associates with increased proliferative, invasive, and migratory properties in EOC | RP11-190D6.2 acts like a tumor suppressor where it confers its effects partly by regulating the expression of the gene WWOX [162] |
| 14. | SDCBP2-AS1 | Inhibits cell migration, invasion and increased apoptotic rate | Sequesters miR-100-5p, upregulating its expression and downregulating EPDR1 expression [163] |
| 15. | TUSC7 | Low levels of TUSC7 mediate proliferation, migration, and invasion of OC cells | Regulates GSK3β/β-catenin pathway through sponging of miR-616-5p [164] |
| 16. | XIST | Reduces tumor growth by inducing apoptosis | Sequesters against miR-106a [165] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





