1. Introduction
Rheumatoid arthritis is a chronic inflammatory autoimmune disease that affects approximately 1% of the population worldwide and is characterized by the destruction of articular cartilage and bone leading to eventual disability [
1]. RA is a sophisticated condition that has clear evidence for its etiology until now [
2]. Females are more frequently susceptible than males, and it strikes often at 40 to 60 years of age [
3].
One of the important RA clinically relevant markers is the peptidyl arginine deiminase (PAD) gene which encodes enzymes inducing catalysis of arginine into citrulline [
4]. Thus, the family of PAD proteins is responsible for autoantibody production against these citrullinated proteins (ACPA). Measuring ACPA in patients’ sera is considered an important immunological marker for the diagnosis of RA [
5].
The PAD family contains five isoenzymes identified as PAD1, PAD2, PAD3, PAD4, and PAD6 [
6].
PADI2 and
PADI4 are the two types of interest due to their expression on immune cells including monocytes and neutrophils as well as macrophages; therefore, these two
PADIs involve in pathogenic mechanisms of several immune disorders such as systematic lupus erythematosus, rheumatoid arthritis and multiple sclerosis [
7,
8].
According to the Saudi General Authority for Statistics in 2017, a statistical survey performed on elder Saudis (65 years and above), a total of 854281 included, 160979 Saudis were diagnosed with arthritis which represents 5.3% of Saudi elders. The prevalence of disease in elders is only considered as an alarm regarding unenclosed age categories and undiagnosed patients. Studies on disease etiology in Saudi Arabia are insufficient, with the increase of diagnosed RA Saudis, further research investigation is a necessity. Specifically, DNA studies include extraction and isolation of desired genes from Saudi RA patients to determine whether specific gene alleles are involved in the disease and its severity.
Previous research confirmed the relationship between
PADI4 single nucleotide polymorphisms (SNPs) and RA risk in Japanese patients [
9]. After that, various studies verified this relationship among RA patients in East Asia, North America, and Germany. In 2023, Bagheri-Hosseinabadi et al. assessed the relationship between
PADI4 gene rs1748033 and rs11203367 SNPs and RA in the Iranian population [
10]. They found out that
PADI4 rs1748033 SNP was linked with an enlarged risk of RA in the Iranian population.
Chronic inflammation in RA is a complex process regulated by a balance of numerous pro- and anti-inflammatory cytokines that play fundamental roles in the processes that cause inflammation, articular destruction, and the co-morbidities related to RA. One of the major consequences of this imbalance is the production of TNF-α, which leads to chronic inflammation and destruction of bone and cartilage. Th17 cytokines play a key role in rheumatoid arthritis [
11,
12,
13]. Th17 cytokines profile in the rheumatoid arthritis of the Saudi population is still unknown.
This research aims to investigate the association of PADI4 gene rs11203366 and rs11203367 SNPs and RA in the Saudi population. It also identified the levels of Th17 cytokines profile (IL-6, IL-17, IL-22, IL-23, TGF-β, TNF-α, and TNF-RN) in Saudi rheumatoid arthritis for the first time.
2. Materials and Methods
2.1. Patients
A total of 45 participants from King Abdulaziz University Hospital in Jeddah were included in this study, 37 of them with established RA, and 8 were healthy controls. Blood samples were taken in 4 ml collection tubes containing EDTA. The RA participants have ages of 29-72 years and a disease duration of 5-34 years. Females were a higher percentage than males with 67% of the total samples were females and 33% were males. Only ten (8 females and 2 males) of the patients had a family genetic history. All subjects gave their informed consent for inclusion before they participated in the study. Patients who had liver and kidney complications were excluded as well as other patients who had other autoimmune diseases. The Ethics Committee approved the study. Also, all subjects were tested for erythrocyte sedimentation rate (ESR), anti-CCP, C-reactive protein (CRP), and rheumatoid factor (RF) as diagnostic and monitoring tools for RA. The results of rheumatic factor ranged between 9.2 and 1220 IU/ml.
2.2. Kits for PADI4 rs11203366 and rs11203367 SNPs and Th17 Cytokines Detection
QIAamp DNA mini extraction kit was purchased from Qiagen (Hilden, Germany). TaqMan Single nucleotide polymorphisms genotyping Assay was pre-designated by Thermo Fisher Scientific (Bedford, MA, USA) for the SNPs of interest; rs11203366 and rs11220367. TaqMan genotyping master mix (2X) was also obtained from Thermo Fisher Scientific. Kits for measurement of IL-6, IL-17, IL-22, IL-23, TGF-β, TNF-α, and TNF-RN in peripheral blood were purchased from Thermo Fisher Scientific.
2.3. DNA Extraction
QIAamp DNA mini extraction kit was used for the extraction of DNA from EDTA-blood samples, spin protocol was followed as recommended by the manufacturer. Washing was done by adding AW1 (25 ml of Ethanol) and AW2 buffer (30 ml of Ethanol). All extracted DNA samples were stored in 1.5 ml Microtubes at -20 °C. Extracted DNA samples were run on agarose gel electrophoresis. The used DNA marker was GENESTA 1 Kbp Ladder (Surabaya, Indonesia), which consists of 14 fragments (250, 500, 750, 1,000, 1,500, 2,000, 2,500, 3,000, 4,000, 5,000, 6,000, 8,000, 10,000, and 25,000 bp).
2.4. Real-Time PCR Genotyping and SNP Detection
TaqMan probes were used to discriminate between the allelic variants. The PCR reaction was prepared as follows: 5 μl TaqMan genotyping master mix (2X), 1 μl DNA sample (10-20 ng\μl), 0.25 μl SNP genotyping assay (40 X), and 3.75 μl nuclease-free H2O. The Real-Time PCR was performed using 7500 fast real-time PCR. The following program was run: 10 min at 95 ºC, 40 cycles of 15 sec denaturation at 92 ºC, and 1 min of annealing/extension at 60ºC.
2.5. Data Analysis for SNP Detection
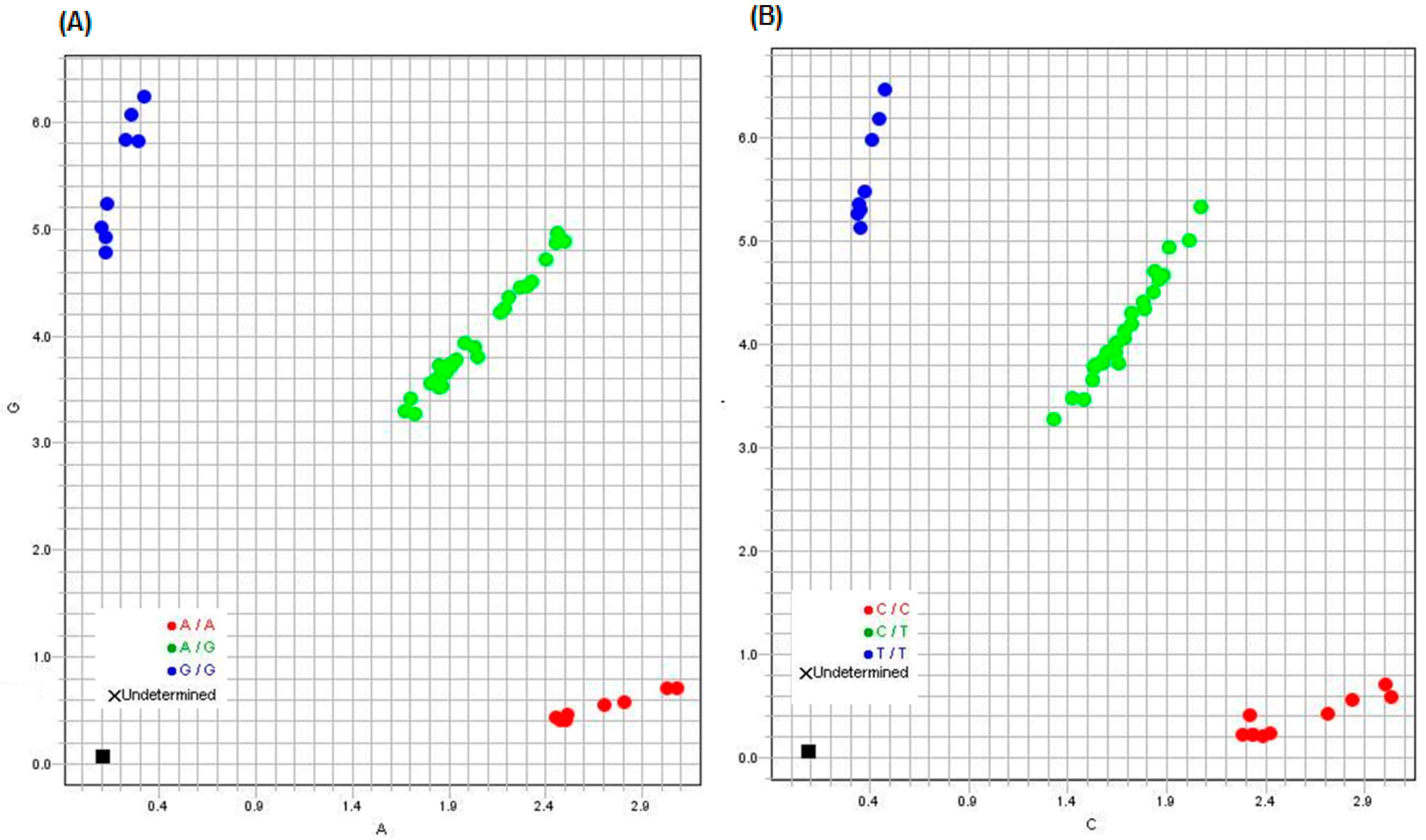
TaqMan Genotyper Software (v1.6) was used to analyze the results. The software analyzes the data automatically and displays the results for each assay in a scatter plot color-coded by genotype calls. Classification schemes enable defining regions associated with each genotype call category by defining linear boundaries (i.e., homozygote regions, heterozygote regions, and undetermined regions).
2.6. Th17 Cytokines Determination
IL-6, IL-17, IL-22, IL-23, TGF-β, TNF-α, and TNF-RN were measured in peripheral blood samples of Saudi RA patients and healthy controls using corresponding ELISA kits according to manufacturer instructions.
2.7. Statistical Analysis for Th17 Cytokines Determination
The data of Th17 cytokines were statistically analysed using SPSS, version 23 statistical program, the significance will be considered when the p-value is less than 0.01 (p < 0.01).
3. Results
Samples involved in this work were collected in the period from 2019 until 2020 at King Abdulaziz University Hospital. The final samples were selected based on different aspects. Firstly, age categories were divided into three subgroups 20s-30s years, 40s – 50s years, and above 60s group. Secondly, hemolyzed and insufficient samples were excluded. Thirdly, samples with no sufficient DNA were excluded as well. All samples are taken from patients with confirmed RA condition. Diagnostic markers were performed, and patients’ sera were tested for RF and anti-CCP autoantibodies.
3.1. DNA Extraction
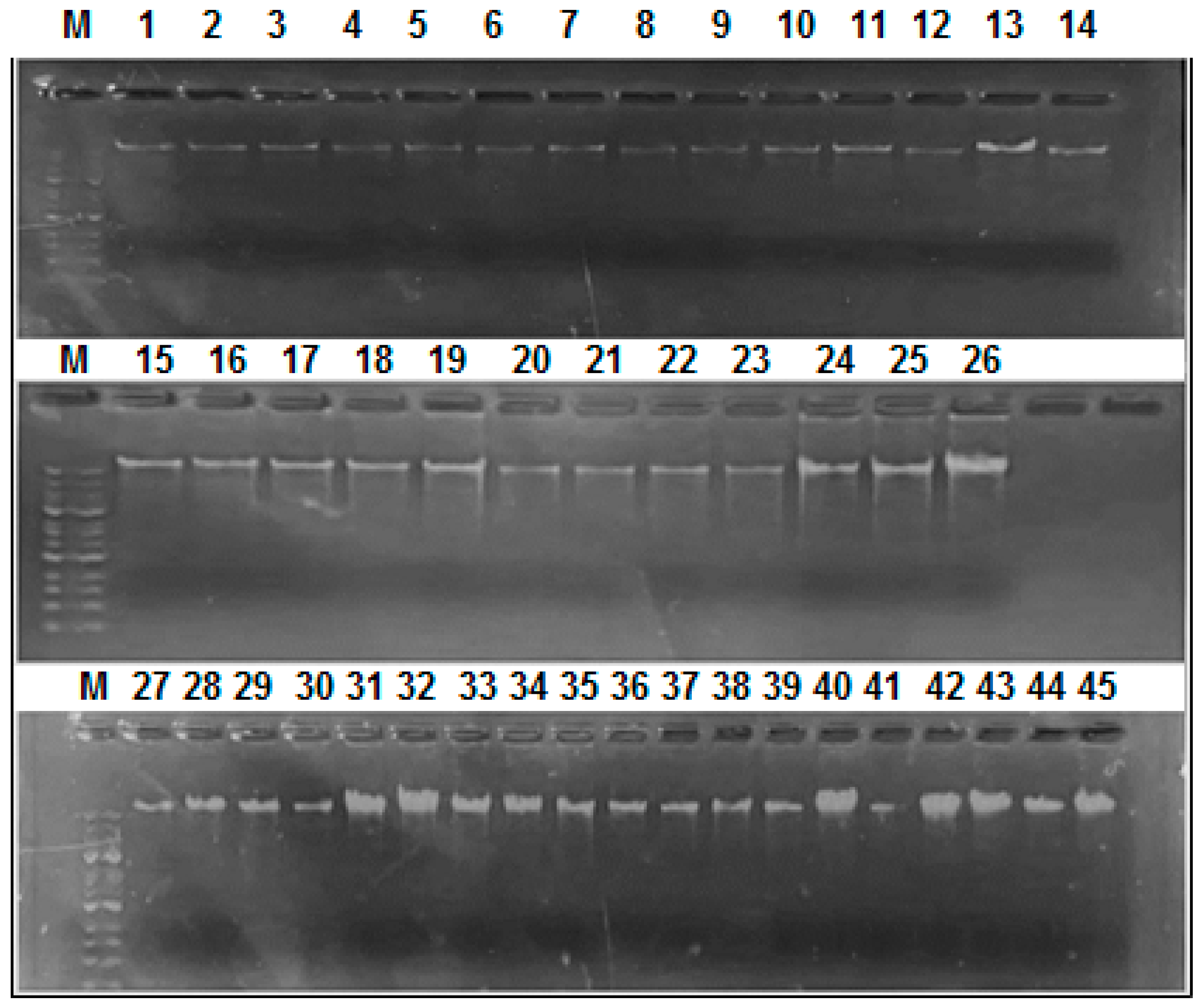
About 200 µl of EDTA blood of patients gave a total of 20 µl (with a concentration of 16-28 ng\μl) of wet DNA. Around 2 µl of extracted DNA were loaded on agarose gel from each sample and gave the results shown in
Figure 1. The extracted DNA ranged in size between 10 and 25 Kbp.
3.2. Real-Time PCR Genotyping and SNP Detection
Results of genotyping for rs11203366 and rs11203367 in all 45 participants (37 RA patients and 8 healthy subjects) are represented in
Table 1. The two SNPs were significantly associated with RA in the Saudi population.
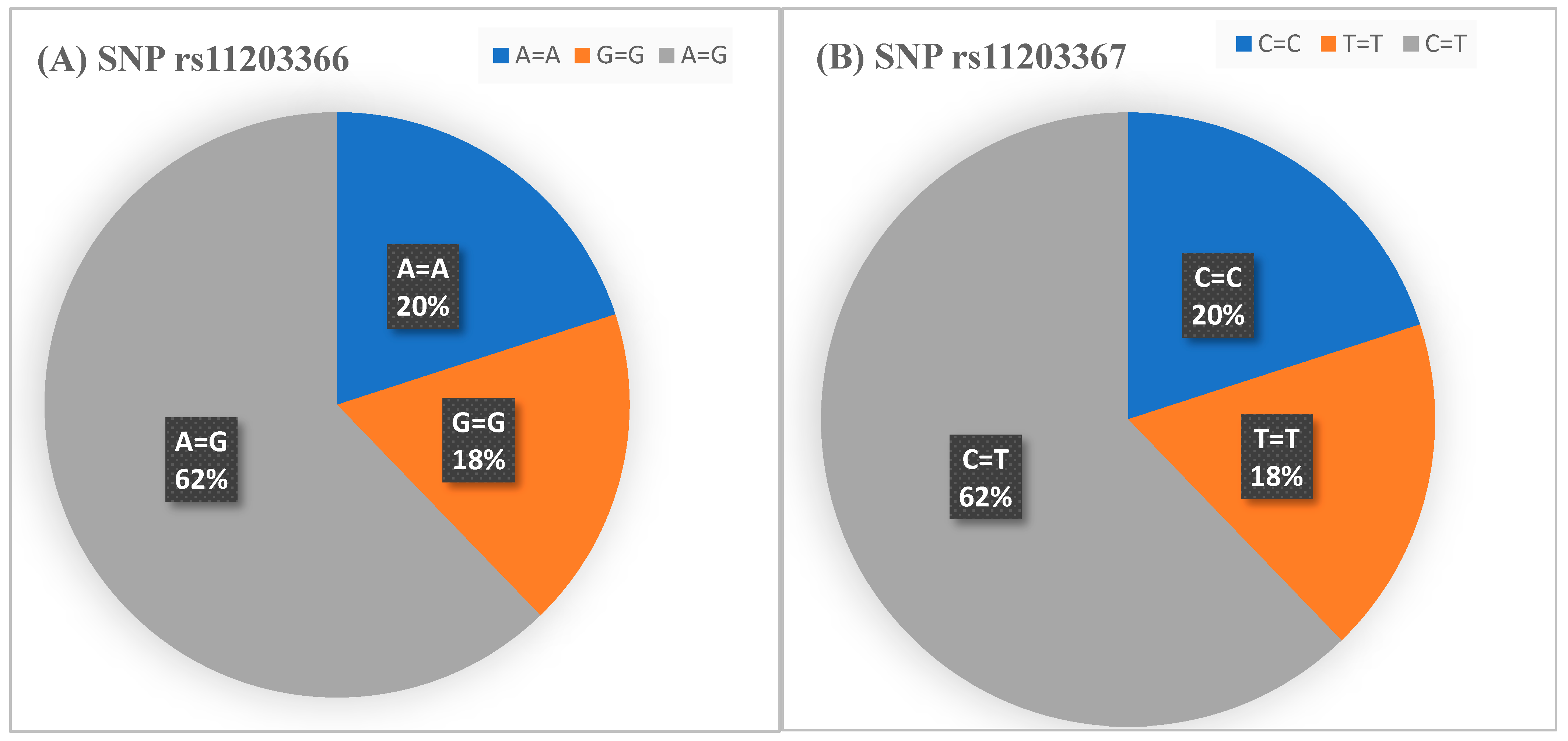
Table 1 revealed that around 46% of Saudi RA subjects were homo allelic, 64% were hetero allelic while all the healthy control group was found to be hetero allelic. Based on the genotyping results in
Table 1, the two SNPs results are related to each other for each subject. For example, the patients who have the homo allelic frequency on the first SNP have also homo allelic frequency for the second SNP.
SNP 112203366 and rs11203367 allelic distribution in all participants are shown in
Figure 2.
Figure 3 demonstrates allelic discrimination plots for the SNP rs11203366 and rs11203367.
All subjects were tested for CRP, ESR, anti-CCP, and RF as diagnostic and monitoring tools for RA. Healthy control subjects were negative for all the tested parameters. Noticeably, about 24% of Saudi RA patients were positive for CRP and ESR, 21% were positive for all tests, 21% were positive for CRP only, while only 13% of them were negative for RF and positive for the three other tests (
Table 2).
The association between
PADI4 genotypes (rs11203366 and rs11203367) and Saudi RA patients’ serological results and disease duration was studied. Our finding on Saudi RA patients was that homogenous RA individuals had elevated mean RF in comparison to heterogenous individuals (
Table 3).
3.3. Th17 Cytokines Determination
Table 4 shows the comparison in means of cytokines proteins between healthy control and rheumatoid arthritis patients, means of IL-6, IL-17, IL-22, and TNF-α were significantly greater (
p < 0.01) in Saudi rheumatoid arthritis patients than those of healthy control, while the mean of TGF-β for rheumatoid arthritis patients was lower than that of healthy control. However, the means of IL-23 and TNF-RN did not significantly change compared to healthy subjects.
4. Discussion
Many studies investigated the association between PADI4 polymorphism and RA susceptibility especially in Asian populations [
14,
15,
16]. By employing electronic searching approach on the relationship between PADI4 polymorphism and RA susceptibility, Hou et al. (2013) found 6 SNPs to considerably associate with RA in Asian populations [
14]. Three of them (
PADI4 104, 90, 89) also exhibited noteworthy associations in the European population, while the other three (
PADI4 94, 92, 100) had none of these associations [
14]. Vetchinkina et al. (2021) revealed that alleles of numerous
PADI4 genotypes have a moderately small impact on RA risk when they appear distinctly, but in combination (as in the case of rs2240340, rs11203367, rs874881, and rs11203366), they prompt an individual to develop RA [
16].
In a previous study performed by Ehnert et al., they carried out a comparison of four different SNPs genotyping methods [
17]. Firstly, RT-PCR using SYBR-green in addition to high-resolution melting curve analysis HRM-PCR, a second method involved PCR followed by detection of restriction fragment length polymorphisms PCR-RFLP, third method included conventional tetra-primer amplification refractory mutation system PCR, and lastly one-step real-time ARMS-PCR [
17]. Considering the cost and preparation/handling simplicity, ARMS-PCR was reported to be the most efficient method [
17]. While in the present work, the extracted DNA from Saudi RA patients was tested for rs11203366 and rs11203367 SNPs using the TaqMan SNPs genotyping assay by RT-PCR.
According to previous literature, different SNPs were related to RA that differ regarding ethnic groups. In the Japanese population, a study on 830 RA individuals found that rs11203366, rs11203367, rs874881, and rs1748033 SNPs were associated with RA [
9]. In 2023, Bagheri-Hosseinabadi et al. assessed the relationship between
PADI4 gene rs1748033 and rs11203367 SNPs and RA in the Iranian population [
10]. They found out that
PADI4 rs1748033 SNP was linked with an enlarged risk of RA in the Iranian population. In southern Mexico, a total of 200 RA subjects were included and three SNPs associations were confirmed (rs11203366, rs11203367, and rs874881) [
15]. In a Russian study, 125 RA patients were genotyped for the following SNPs: rs11203367, rs2240340, rs11203366, and rs874881 [
16]. All of them were found to be associated with RA. In the case of Chinese, Indian, and Malaysian populations, rs2240340 and rs1748021 SNPs were related to RA [
18]. The interest in this research was on rs11203366 and rs11203367. These two SNPs were selected due to their common mention in all of these above-mentioned research studies, they are thought to be a stabilizing factor for mRNA in PAD4 and can influence their level and activity [
17]. The present work found that SNPs rs11203366 and rs11203367 are associated with rheumatoid arthritis in Saudi patients.
Based on the genotyping results we obtained here that the two SNPs results related to each other for each subject. For instance, the patients that have the homo allelic frequency on the first SNP has also homo allelic frequency for the second one. Of all RA subjects, 17 were homo allelic on the two tested SNPs and 20 were hetero allelic. While all the healthy controls showed heteroallelic SNPs. The present study showed that homogeneous haplotypes had susceptibility to developing RA in the Saudi population. These findings agree with studies that had been reported on Japanese [
9], western Mexican [
19], and Russian [
16] populations. In Russians, Vetchinkina et al. suggested that rs1123366 and rs1123367 are associated with disease severity but not serological tests [
16]. A similar finding was proposed by Baños-Hernández et al. who suggested that homogeneity had no significant effect on Mexican RA patients’ serological results (clinical parameters and anti-CCP antibody levels). Additionally, Abd-Allah et al. reported that PADI4 SNPs and haplotypes were linked to susceptibility to RA in Egyptians, though no association was detected between these haplotypes and levels of anti-mutated citrullinated vimentin antibodies (anti-MCV) in serum [
20]. Our present findings on Saudis are consistent with Russians, Mexicans, and Egyptians except for RF. We found that homogenous RA individuals had elevated mean RF in comparison to heterogenous individuals.
Enormous analysis of cytokines proteins in rheumatoid arthritis has the potential to not only provide novelty insights into the immunopathogenesis of the disease but also to identify subgroups of patients on the verge of developing more severe conditions and suggest possible targets for therapy [
21]. As illustrated in this work, Saudi RA patients have a greater mean of IL-6 than that healthy controls. This is a positive sign because greater interleukin-6 levels in RA patients are linked with better advances in health-related quality of life [
22]. Moreover, high IL-6 among RA patients indicates augmented cytokines levels, which can aid in imitating inflammation besides being predictive in elucidating how bodies of patients can accomplish coping with depression, therapeutic responses, fatigue, or pain [
23]. The levels of IL-6 in serum were previously correlated with RA activity (e.g. Bristol RA Fatigue Multi-Dimensional Questionnaire (BRAF-MDQ) and disease activity score-28 for RA with ESR (DAS28-ESR)) and this interleukin remains a practicable target for therapy [
24].
This study also demonstrated a greater mean of IL-17 in Saudi RA patients compared with that of healthy control which agrees with previous reports on the importance of serum IL-17 and Th-17 cells and their contribution to the destructive pattern and inflammatory pathogenesis of RA [
25,
26]. Research on animal/human models proposed that the net influence of IL-17 signaling stimulates RA development [
27].
IL-22 is raised in the serum of Saudi patients with rheumatoid arthritis. In 2005, Ikeuchi et al. stated the expression of high levels of IL-22 in the lining/subliming layers of synovial tissues of RA [
28]. They proposed that the expressed IL-22 can endorse inflammatory responses in synovial tissues of RA by prompting the production of chemokine and the proliferation of these tissues' fibroblasts [
28].
As per our research findings regarding TNF-α levels in control and RA patients, RA patients had higher levels of TNF. TNF-α was found to be a crucial controller of the damage of homeostatic immune-inflammatory reactions in RA and thus a worthy target for therapy [
29]. Consequently, TNF blockade is a potential competitor to typical anti-rheumatoid drugs.
The means of IL-23 and TNF-RN did not significantly change compared to healthy subjects, while the mean of TGF-β for Saudi RA patients was lower than that of healthy control. TGF-β 1 is highly expressed in joints in RA and thus it is an anti-inflammation regulator in RA [
30]. TGF-β 1 production was linked with an increased risk of RA.
5. Conclusions
PADI4 rs1123366 and rs1123367 are associated with RA susceptibility in the Saudi population. Additionally, we observed that disease duration and serological tests have no significant correlation with homo allelic and hetero allelic subgroups except for RF-positive homo allelic participants who have noticeably higher RF than hetero allelic ones. Further investigation with a larger sample size from different regions in Saudi Arabia is recommended. Also including all SNPs in the PADI4 gene loci is advised as it may show a better approach to the pathogenesis of this disease in Saudi Arabia.
As cytokines prove to be crucial in regulating inflammation, and autoimmunity in patients with rheumatoid arthritis, they should be measured and exhaustively analyzed. In addition, differentiation in the levels between the cytokines (especially IL-6, IL-17, IL-22, and TNF-α) of rheumatoid arthritis in Saudi patients may predict the quality of life expected among them. Enormous analysis of cytokines proteins in Saudi RA patients has the potential to not only provide novelty insights into the immunopathogenesis of the disease but also to suggest possible targets for therapy.
Author Contributions
Conceptualization, E.M.R.; literature search, E.H.M, D.A., A.A.; validation, E.H.M, D.A., A.A, N.A.E.; formal analysis, EH.M. and NA.E.; writing—original draft preparation, E.M.R., E.H.M., D.A., A.A., N.A.E.; writing—review and editing, E.M.R., E.H.M., D.A., A.A., N.A.E.; visualization, E.M.R.; supervision, E.M.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the research ethics committee of the Faculty of Medicine at King Abdulaziz University (Approval No 468-19).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets used and analyzed in this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Coenen, M.J.; Trynka, G.; Heskamp, S.; Franke, B.; van Diemen, C.C.; Smolonska, J.; van Leeuwen, M.; Brouwer, E.; Boezen, M.H.; Postma, D.S.; Platteel, M.; Zanen, P.; Lammers, J.W.; Groen, H.J.; Mali, W.P.; Mulder, C.J.; Tack, G.J.; Verbeek, W.H.; Wolters, V.M.; Houwen, R.H.; Mearin, M.L.; van Heel, D.A.; Radstake, T.R.; van Riel, P.L.; Wijmenga, C.; Barrera, P.; Zhernakova, A. Common and different genetic background for rheumatoid arthritis and coeliac disease. Hum. Mol. Genet. 2009, 18, 4195–4203. [Google Scholar] [CrossRef] [PubMed]
- Orozco, G.; Rueda, B.; Martin, J. Genetic basis of rheumatoid arthritis. Biomed. Pharmacother. 2006, 60, 656–662. [Google Scholar] [CrossRef]
- Castro-Santos, P.; Díaz-Peña, R. Genetics of rheumatoid arthritis: a new boost is needed in Latin American populations. Rev. Bras. Reumatol. Engl. Ed. 2016, 56, 171–177. [Google Scholar] [CrossRef]
- Hua, J.; Huang, W. Peptidylarginine deiminase 4 -104C/T polymorphism and risk of rheumatoid arthritis: A pooled analysis based on different populations. PLoS One. 2018, 13, e0193674. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Castillo, Z.; Palafox-Sánchez, C.A.; Parra-Rojas, I.; Martínez-Bonilla, G.E.; del Toro-Arreola, S.; Ramírez-Dueñas, M.G.; Ocampo-Bermudes, G.; Muñoz-Valle, J.F. Comparative analysis of autoantibodies targeting peptidylarginine deiminase type 4, mutated citrullinated vimentin and cyclic citrullinated peptides in rheumatoid arthritis: associations with cytokine profiles, clinical and genetic features. Clin. Exp. Immunol. 2015, 182, 119–131. [Google Scholar] [CrossRef]
- Vossenaar, E.R.; Zendman, A.J.; van Venrooij, W.J.; Pruijn, G.J. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays. 2003, 25, 1106–1118. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lightfoot, Y. L.; Seto, N.; Carmona-Rivera, C.; Moore, E.; Goel, R.; O'Neil, L.; Mistry, P.; Hoffmann, V.; Mondal, S.; Premnath, P. N.; Gribbons, K.; Dell'Orso, S.; Jiang, K.; Thompson, P. R.; Sun, H.W.; Coonrod, S. A.; Kaplan, M. J. Peptidylarginine deiminases 2 and 4 modulate innate and adaptive immune responses in TLR-7-dependent lupus. JCI Insight. 2018, 3, e124729. [Google Scholar] [CrossRef]
- Nagar, M.; Tilvawala, R.; Thompson, P.R. Thioredoxin Modulates Protein Arginine Deiminase 4 (PAD4)-Catalyzed Citrullination. Front. Immunol. 2019, 10, 244. [Google Scholar] [CrossRef]
- Suzuki, A.; Yamada, R.; Chang, X.; Tokuhiro, S.; Sawada, T.; Suzuki, M.; et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidyl arginine deiminase 4, are associated with rheumatoid arthritis. Nat. Genet. 2003, 34, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Bagheri-Hosseinabadi, Z.; Mirzaei, M.R.; Esmaeili, O.; Asadi, F.; Ahmadinia, H.; Shamsoddini, B.; Abbasifard, M. Implications of Peptidyl Arginine Deiminase 4 gene transcription and polymorphisms in susceptibility to rheumatoid arthritis in an Iranian population. BMC Med. Genomics. 2023, 16, 104. [Google Scholar] [CrossRef]
- Singh, R.; Aggarwal, A.; Misra, R. Th1/Th17 cytokine profiles in patients with reactive arthritis/undifferentiated spondyloarthropathy. J. Rheumatol. 2007, 34, 2285–2290. [Google Scholar] [PubMed]
- Sarkar, S.; Cooney, L. A.; Fox, D. A. The role of T helper type 17 cells in inflammatory arthritis. Clin. Exp. Immunol. 2010, 159, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Lubberts, E. Th17 cytokines and arthritis. Semin. Immunopathol. 2010, 32, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Gao, G.P.; Zhang, X.J.; Sun, L.; Peng, W.J.; Wang, H.F.; Ge, X.J.; Huang, W.; Sun, Y.H. PADI4 polymorphisms and susceptibility to rheumatoid arthritis: a meta-analysis. Mod. Rheumatol. 2013, 23, 50–60. [Google Scholar] [CrossRef]
- Baños-Hernández, C.J.; Navarro-Zarza, J.E.; Parra-Rojas, I.; Vázquez-Villamar, M.; Ramón Padilla-Gutiérrez, J.; Valle, Y.; Reyes-Castillo, Z.; Magdalena Torres-Carrillo, N.; García-Arellano, S.; Brennan-Bourdon, L.M.; Muñoz-Valle, J.F. PADI4 polymorphisms and the functional haplotype are associated with increased rheumatoid arthritis susceptibility: A replication study in a Southern Mexican population. Hum Immunol. 2017, 78, 553–558. [Google Scholar] [CrossRef]
- Vetchinkina, E.A.; Mikhaylenko, D.S.; Kuznetsova, E.B.; Deryagina, T.A.; Alekseeva, E.A.; Bure, I.V.; Zamyatnin, A.A. Jr; Nemtsova, M.V. Genetic Factors of Predisposition and Clinical Characteristics of Rheumatoid Arthritis in Russian Patients. J. Pers Med. 2021, 11, 469. [Google Scholar] [CrossRef]
- Ehnert, S.; Linnemann, C.; Braun, B.; Botsch, J.; Leibiger, K.; Hemmann, P.; Nussler, A.A.K. One-Step ARMS-PCR for the Detection of SNPs-Using the Example of the PADI4 Gene. Methods Protoc. 2019, 2, 63. [Google Scholar] [CrossRef] [PubMed]
- Koushik, S.; Joshi, N.; Nagaraju, S.; Mahmood, S.; Mudeenahally, K.; Padmavathy, R.; Jegatheesan, S.K.; Mullangi, R.; Rajagopal, S. PAD4: pathophysiology, current therapeutics and future perspective in rheumatoid arthritis. Expert Opin. Ther. Targets. 2017, 21, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Guzmán, I.P.; Reyes-Castillo, Z.; Muñoz-Barrios, S.; Ruiz-Noa, Y.; Martínez-Bonilla, G.E.; Parra-Rojas, I.; Palafox-Sánchez, C.A.; Muñoz-Valle, J.F. Polymorphisms and functional haplotype in PADI4: further evidence for contribution on rheumatoid arthritis susceptibility and anti-cyclic citrullinated peptide antibodies in a western Mexican population. Immunol Lett. 2015, 163, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Abd-Allah, S.H.; el-Shal, A.S.; Shalaby, S.M.; Pasha, H.F.; Abou el-Saoud, A.M.; el-Najjar, A.R.; el-Shahawy, E.E. PADI4 polymorphisms and related haplotype in rheumatoid arthritis patients. Joint Bone Spine. 2012, 79, 124–128. [Google Scholar] [CrossRef]
- Feldmann, M.; Brennan, F.M.; Maini, R.N. Role of cytokines in rheumatoid arthritis. Annu Rev Immunol. 1996, 14, 397–440. [Google Scholar] [CrossRef] [PubMed]
- Strand, V.; Boklage, S.H.; Kimura, T.; Joly, F.; Boyapati, A.; Msihid, J. High levels of interleukin-6 in patients with rheumatoid arthritis are associated with greater improvements in health-related quality of life for sarilumab compared with adalimumab. Arthritis Res Ther. 2020, 22, 250. [Google Scholar] [CrossRef]
- Hashizume, M.; Mihara, M. The roles of interleukin-6 in the pathogenesis of rheumatoid arthritis. Arthritis. 2011, 2011, 765624. [Google Scholar] [CrossRef] [PubMed]
- Sharma, Y.; Kumar, N.; Thakur, D. Interleukin 6 in Patients with Rheumatoid Arthritis. In Interleukins - The Immune and Non-Immune Systems’ Related Cytokines; IntechOpen: London, UNITED KINGDOM, 2021. [Google Scholar] [CrossRef]
- Al-Saadany, H. M.; Hussein, M. S.; Gaber, R. A.; Zaytoun, H. A. Th-17 cells and serum IL-17 in rheumatoid arthritis patients: Correlation with disease activity and severity. The Egyptian Rheumatologist 2016, 38, 1–7. [Google Scholar] [CrossRef]
- Tian, Y.; Shen, H.; Xia, L.; Lu, J. Elevated serum and synovial fluid levels of interleukin-34 in rheumatoid arthritis: possible association with disease progression via interleukin-17 production. J. Interferon Cytokine Res. 2013, 33, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Zenobia, C.; Hajishengallis, G. Basic biology and role of interleukin-17 in immunity and inflammation. Periodontol 2000. 2015, 69, 142–159. [Google Scholar] [CrossRef]
- Ikeuchi, H.; Kuroiwa, T.; Hiramatsu, N.; Kaneko, Y.; Hiromura, K.; Ueki, K.; Nojima, Y. Expression of interleukin-22 in rheumatoid arthritis: potential role as a proinflammatory cytokine. Arthritis Rheum. 2005, 52, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, M.; Maini, S.R. Role of cytokines in rheumatoid arthritis: an education in pathophysiology and therapeutics. Immunol Rev. 2008, 223, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, Y.; Niimi, T.; Sato, S.; Yoshinouchi, T.; Banno, S.; Naniwa, T.; Maeda, H.; Shimizu, S.; Ueda, R. Transforming growth factor beta1 gene polymorphism in rheumatoid arthritis. Ann. Rheum. Dis. 2002, 61, 826–828. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).