1. Introduction
Viral vector delivery of gene therapy reagents holds great promise as an effective approach for treating a number of genetic diseases and has now been approved for several clinical therapies [
1]. Primarily, the delivery vehicles have been adeno-associated viral vectors (AAV) owing to their desirable properties including low chromosomal integration and moderate stimulation of immune responses in the eye. In the context of retinal disease, AAV has been successfully utilised to deliver a
RPE65 transgene in the first clinically available retinal gene therapy, and a number of studies have shown AAV to be an effective vehicle for supplementation of retinal genes in several disease models [
2,
3]. Despite the popularity of AAV as a delivery vector, a substantial limitation consists of the relatively small packaging capacity of ~4.7 kb. This precludes the delivery of a number of large, yet clinically relevant retinal genes, such as
USH2A (15.6 kb),
CDH23 (10.1 kb), EYS (9.4 kb),
CEP290 (7.4 kb),
GPR179 (7.1 kb),
ABCA4 (6.8 kb),
RP1 (6.5 kb),
MYO7A (6.6 kb), or
CRB1 (4.2 kb) or gene editing components such as full-length prime editing (6.2 kb) and base editing (4.2-5.2 kb) constructs [
4,
5].
Alternative viral vectors for gene delivery consist of high-capacity human adenoviral vector (HC-AdV) particles, which are capable of carrying cargo up to 36 kb [
6]. Indeed, the HC-AdV platform is able to deliver expression units encoding the human full-length dystrophin coding sequence (11.1 kb) to muscle cells [
7]. These 3rd generation adenovectors are devoid of all viral genes and have been shown to have a reduced cytotoxicity profile in vitro [
8] and inflammatory response in vivo when compared to earlier generation adenovectors deleted exclusively in a subset of viral regulatory functions [
6,
9]. The capsid shell of an AdV comprises multiple copies of three major capsid proteins (i.e., hexon, penton base, and fiber) and four minor/cement proteins (i.e., IX, VIII, VI, and IIIa). In addition, six other proteins (i.e., μ, IVa2, V, VII, terminal protein, and adenovirus protease) are encapsidated along with the 36-kb dsDNA genome [
10]. As there is an unmet need for vectors capable of delivering large retinal genes, we sought to determine the transduction efficiency and tropism of HC-AdV particles with unmodified and modified capsids in human retinal organoids.
Historically, the testing of gene therapy vectors has been done in animal models, such as rodents and non-human primates [
11,
12]. Whilst these models offer a valuable platform for testing viral vectors
in vivo, there are substantial differences between the tissues and organs of these animal models and those of humans, including the retina. In recent years, the development of human iPSC-derived retinal organoids has allowed for the production of organoid models capable of closely mimicking the human retina with a high degree of accuracy that, for instance, yield laminated retinal tissue containing all major retinal cell types [
13,
14].
Commonly used adenovectors (AdV) are based on the prototypic human adenovirus type 5 from species C, which engages the coxsackievirus and adenovirus receptor (CAR) to enter cells. In contrast, species B AdVs interact with different primary receptors that, depending on the type, mostly involves desmoglein 2 or CD46. In this study, we utilised an iPSC-derived retinal organoid model to assess the transduction efficiency and tropism of two HC-AdV vectors based on the classical AdV type 5 (HC-AdV5.F5) and a capsid-modified version displaying CD46-binding fibre 50 domains (HC-AdV5.F50) capable of transducing CAR-negative cells [
15]. These two viral vectors contained sequences expressing a mCherry reporter gene under the control of the ubiquitous CAG promoter, and were applied to human retinal organoids at differentiation day (DD) 130. Strong transduction was observed throughout the retinal organoids, with efficient transduction of the photoreceptor layer and Müller glial cells being readily detected. Significantly, HC-AdV5.F5 and HC-AdV5.F50 vectors resulted in comparable transduction efficiencies and cell-type tropism profiles. Expression of the transgene was visible shortly following transduction and persisted until the termination of this study, namely, 110 days post-transduction. No detrimental effects were observed following HC-AdV transductions with organoid development and morphology aligning with non-transduced organoid controls.
Our results show the possibility of leveraging the HC-AdV system to deliver large genes in an efficient manner into both photoreceptors and Müller glial cells, and demonstrate the feasibility of using such vectors for the supplementation of large retinal genes which currently lack a suitable vector. A matter warranting further investigation, however, is the detection of swelling of the photoreceptor outer nuclear layer (ONL) at late timepoints after HC-AdV transduction of human retinal organoids.
3. Discussion
Viral vector delivered gene therapy is an increasingly utilised method for addressing diseases previously considered untreatable. Viral vector delivery of
RPE65 to the retina has met clinically successful endpoints [
19], with a number of other promising clinical trials currently underway [
20]. AAV has been the vehicle of choice in the first wave of viral vector delivered gene therapies. Although possessing a number of desirable traits, the limited cargo capacity of approximately 4.7 kb precludes the delivery of many large genes. This is of particular importance in the field of retinal gene therapy as a number of coding sequences of clinically relevant genes linked to the required regulatory sequences readily exceeds this limit
4. In response to this, alternative delivery methods should be explored, with one of these alternative delivery vehicles being high-capacity adenovectors, whose packaging capacity of 36 kb allows for the delivery of all known coding gene sequences found in the human genome [
9,
21].
To determine transduction endpoints (i.e., spatiotemporal transgene expression profiles) and morphological effects of these vectors with conventional or tropism-modified capsids in the context of the human retina, we utilised human iPSC-derived retinal organoids as a testing platform. Retinal organoids faithfully recapitulate many aspects of the human retina, including the formation of all major retinal cell types arranged in a highly organised stratified architecture [
13,
14]. HC-AdV vectors have previously been tested ex vivo on cultured human cadaver retinal explants analysed at 7 days post-transduction, and in vivo in the rat retina [
22]. Effective HC-AdV5-CMVp-
eGFP transduction of human retinal explants was demonstrated, resulting in transgene expression throughout the retina including in the photoreceptors [
22]. The 7-day culturing period of the human retinal explants did not allow, however, for intensive analysis of morphological structures. Conversely, in vivo subretinal transduction of HC-AdV on the rat retina revealed structural changes due to an early acute innate inflammatory response to the capsid proteins, including significant upregulation of proinflammatory cytokines and chemokines potentially from Müller glial cells and immune effector cells. Interestingly, inflammatory responses upon HC-AdV exposure were, however, not reported following transduction experiments in the mouse retina [
23,
24,
25,
26].
In this study, we investigated for up to 110 days post-transduction in human retinal organoids, the efficiency, tropism and cellular responses of HC-AdV vectors displaying either classical adenovirus type 5 fibres or apical fibre motifs from adenovirus type 50 [
15]. The former and latter vector utilize CAR and CD46 as primary receptors, respectively. Of note, CD46-binding vectors bypass the absence of CAR on various human cell types with therapeutic relevant or potential, namely, hematopoietic stem cells [
27,
28], mesenchymal stromal cells [
29] and muscle progenitor cells [
15]. In this study, using HC-AdV5.F5 and HC-AdV5.F50 vectors containing the same genome backbone and mCherry reporter, we found that these CAR- and CD46-binding vectors, respectively, are similarly effective gene delivery vehicles in human retinal tissue. The mCherry signals were detected shortly after transduction with live-cell imaging revealing reporter expression as early as 1 day post-transduction. Of note, these initial signals were primarily localised in the RPE, a cell type known for its high phagocytotic activity and ability to readily uptake adenovector and AAV particles [
18,
30]. The mCherry fluorescence became stronger and more evenly distributed in the retinal organoid shortly thereafter and, critically, it persisted for up to DD240 (110 days post-transduction) (
Figure 2D). Indeed, the transgene expression may have persisted in a sustained fashion even beyond the termination timepoint selected for our experiments.
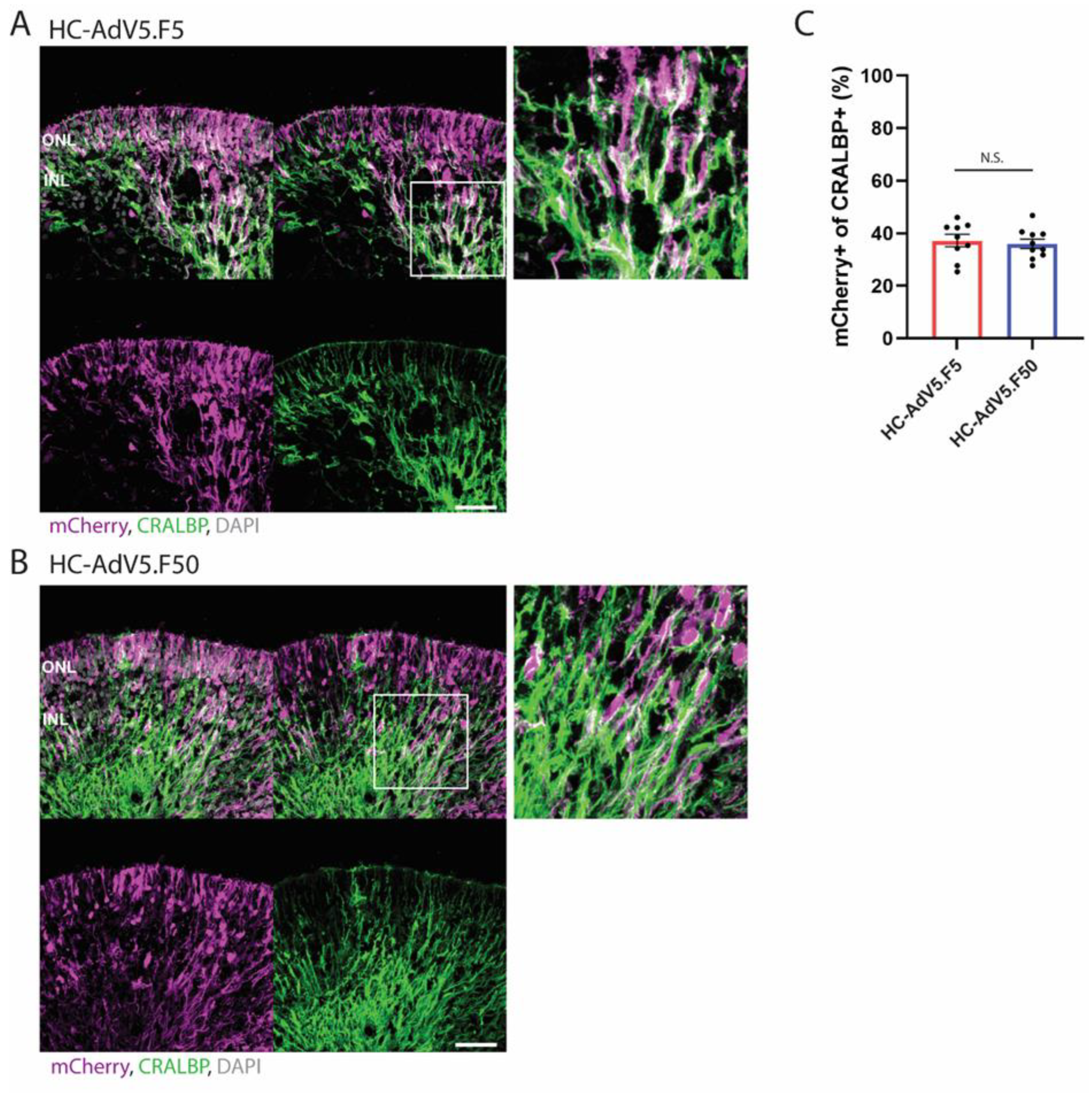
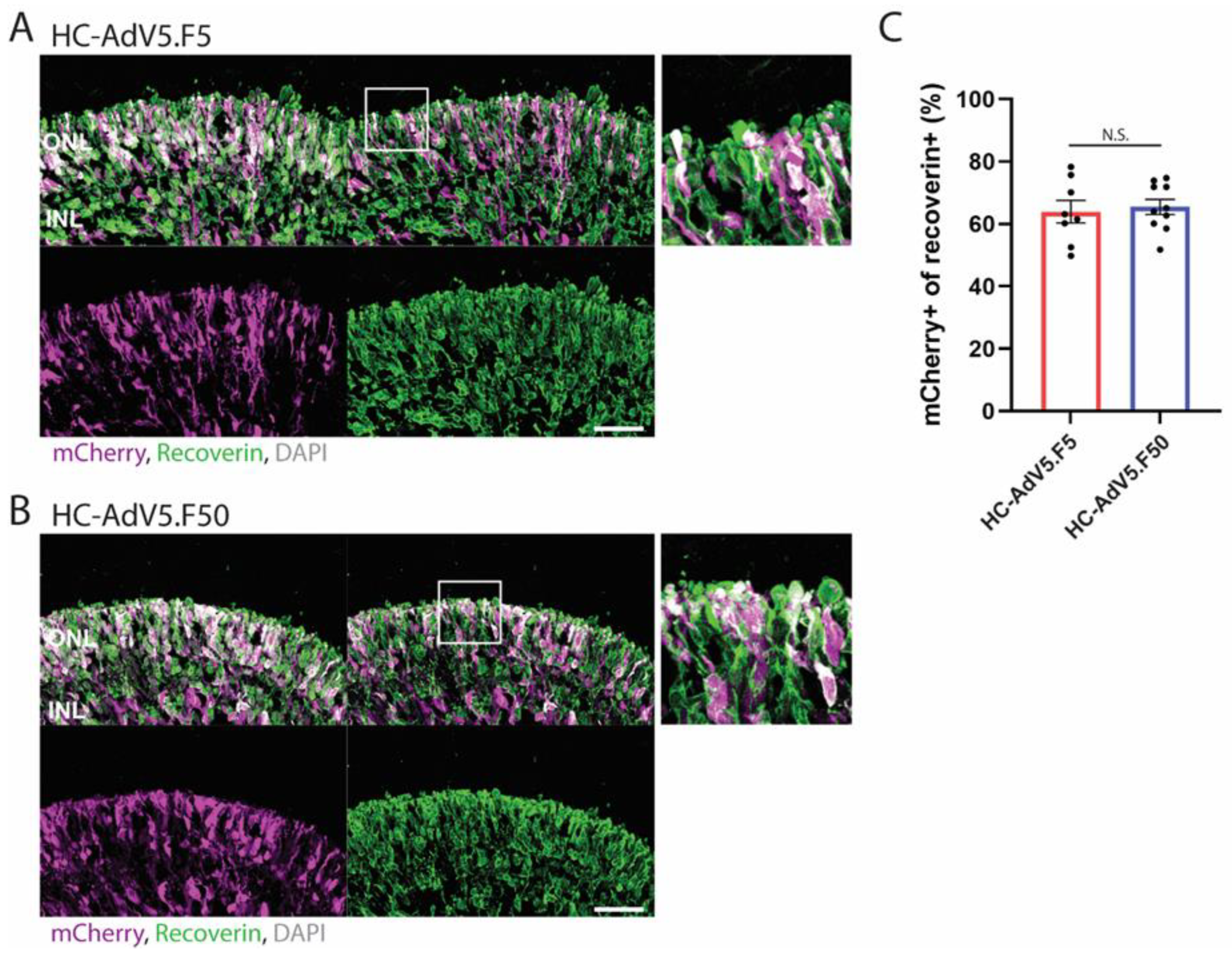
In addition to general organoid-level transduction analysis, we have also investigated the transduction of retinal cell subtypes, namely, Müller glial and photoreceptor cells, both of which are highly desirable targets for gene therapy. We determined the vector transduction profiles in these cell populations by measuring the colocalization of the mCherry reporter with established cell markers, i.e., CRALBP for Müller glial cells and recoverin for photoreceptors. Our results show a roughly equal capacity of regular and capsid-modified HC-AdV vectors to transduce Müller glial cells when measured at both DD210 and DD240 timepoints. Further, these two adenovector types performed also largely equally well in transducing photoreceptors with approximately 60% of photoreceptors co-expressing the mCherry reporter. Of note, it was observed that several cells within the inner retina also expressed recoverin, which may represent ON-bipolar cells [
31] or misplaced photoreceptors. Yet, as we could not define these cells with certainty as being photoreceptors, we limited photoreceptor quantifications to recoverin-positive areas exclusively located in the ONL, where photoreceptor nuclei typically reside.
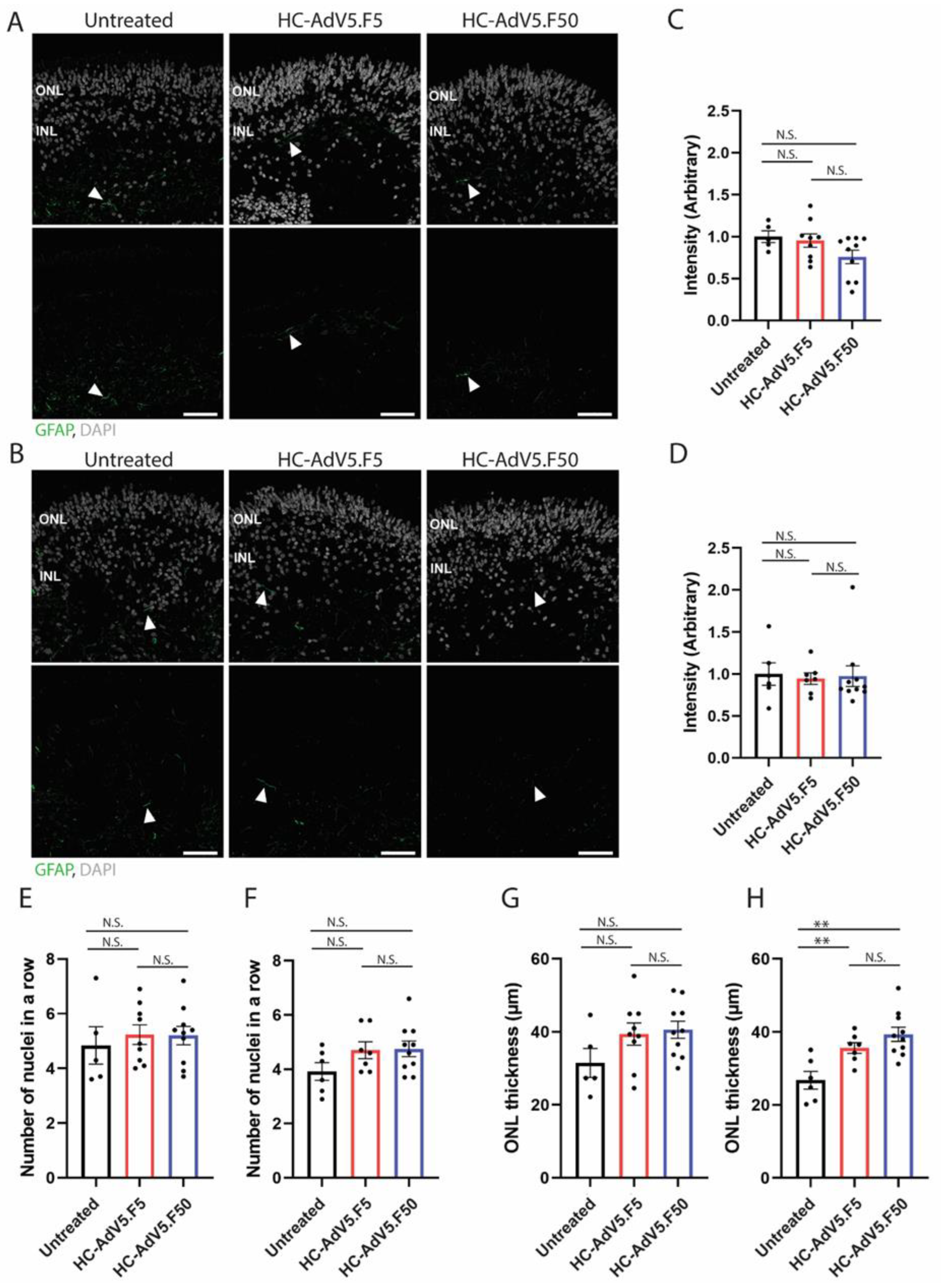
To address whether high adenovector loads trigger stress responses and degeneration in the retinal organoids, we investigated reactive gliosis which is evidenced by a strong upregulation of the intermediate filament protein GFAP [
32]. We compared GFAP levels in untreated organoids versus organoids transduced with HC-AdV5.F50 or HC-AdV5.F50. We found no statistically significant increase in GFAP levels nor did we observe the typical gliosis-associated cell morphology, noting only low GFAP expression in the end feet of Müller glial cells. Additionally, a common sign of retinal degeneration is the loss of photoreceptor cells which can be measured by a thinning of the ONL. Hence, we selected two parameters, the number of ONL nuclei in a row and the length of the ONL thickness in µm. By either metric we did not observe a thinning of the ONL suggesting no increased loss of photoreceptors following transductions with the two HC-AdV constructs. However, at DD240 we did observe a significant increase in ONL thickness following HC-AdV transduction. The swelling of the ONL is not due to changes in the number of photoreceptor nuclei (
Figure 6F). Further investigation is needed to reveal if the swelling is due to increased photoreceptor cell volume, increased Müller glial cell volume, extracellular matrix expansion, or expression of mCherry. Regarding the latter, it is of note the existence of experimental evidence indicating that long-term mCherry overexpression is associated with lysosomal accumulation and cytotoxicity in human cells and with abnormal eye development in
Xenopus laevis [
33]. Hence, further research will be necessary to determine whether the ONL thickening at late timepoints post-transduction is the result of HC-AdV exposure per se, long-term mCherry overexpression or a combination of both.
Activated Müller glial cells release inflammatory molecules such as cytokines and chemokines similar to canonical immune effector cells. Inflammatory responses can come from direct Müller glial cell activation or from indirect activation of Müller glial cells via photoreceptor cell damage [
34]. Toll-like receptor 9 (TLR9) triggers an early innate immune response to HC-AdV vectors as they sense unmethylated CpG motifs in incoming double-stranded DNA (dsDNA) genomes, which potentially cause cytokine secretion from Müller glial cells in the retinal organoids [
35,
36]. Besides this TLR9-dependent innate immunity mechanism, there are other early innate immunity restriction factors that could be involved in HC-AdV sensing [
37,
38]. Other restriction factors include cyclic GMP-AMP synthase (cGAS), which detects cytosolic dsDNA and triggers interferon responses through the interferon regulatory factor-3 (IRF3)–stimulator of interferon genes (STING) pathway. These cellular changes in photoreceptors and Müller glial cells may contribute to the increased thickness of the photoreceptor ONL at later stages.
Follow-up work will involve studying innate immune responses directed to DNA and protein components of incoming HC-AdV particles in retinal organoids. A better understanding of these responses might ultimately allow their modulation in the context of clinical HC-AdV delivery of large retinal transgenes. At present, we cannot exclude late-onset effects on the ONL due to toxicity by high levels of mCherry or contaminants in the adenovector preparations. Despite the significant ONL enlargement detected at DD240, transduced organoids developed otherwise normally resulting in a well-structured laminated inner retina, outer plexiform layer, and ONL with visible photoreceptor inner/outer segments arranged in a typical brush border configuration. When performing initial HC-AdV dose-response experiments to determine the optimal vector amount, we observed strong and broad transduction of human retinal organoids at 3.7x10
7 HTU when using vectors with either of the ubiquitous promoters tested, i.e., CAG or PGK. In contrast, the use of human cell type-specific promoters such as those of the rhodopsin kinase gene (
GRK1) and of the retinaldehyde binding protein 1 gene (
RLBP1) is expected to result in a more defined expression profile in photoreceptors and Müller glial cells, respectively, as previously observed in several studies using AAV vectors [
39]. Follow-up work should also exploit the large size of HC-AdV particles to accommodate extensive
cis-acting regulatory sequences to assure robust and cell type-specific expression of therapeutic transgenes in the retina. Finally, in addition to gene supplementation, the HC-AdV platform can also be in principle directed for delivering large gene-editing tools, such as, prime editing and base editing constructs which, in their full-length formats, cannot be delivered in a single AAV vector [
40]. Alternative methods exist for large gene delivery, mostly involving dual AAV vectors encoding transgene halves that, upon co-transduction, lead to mRNA trans-splicing, ribozyme-activated mRNA trans-ligation, Cre-lox DNA sequence-specific and near-unidirectional recombination, or full-length protein assembly via intein-mediated ligation [
41,
42,
43,
44]. However, in addition to issues related to the designing complexity and performance of dual AAV vectors, the need to produce at sufficient scale and qualify two independent drug substances increases the challenges and costs of such clinical trials.
In conclusion, here we report that conventional and capsid-modified HC-AdV vectors are capable of achieving robust and persistent transgene expression in human retinal organoids, effectively transducing both Müller glial cells and photoreceptors. Although the observed swelling of the ONL following HC-AdV transduction is a point of concern warranting further research, HC-AdV systems hold promise for the delivery of large genetic payloads to the human retina.
Author Contributions
Conceptualization, A.McD. and J.W.; formal analysis, A.McD., C.G., C.A., M.O., M.A.F.V.G. and J.W.; funding acquisition, J.W.; investigation, A.McD., C.G., C.A., and M.O., M.A.F.V.G. and J.W.; methodology, A.McD., C.G., C.A., M.O.; project administration, J.W.; resources, A.McD., C.G., C.A., and M.O; supervision, M.A.F.V.G. and J.W.; validation, A.McD., C.G., C.A., M.O., M.A.F.V.G. and J.W.; visualization, A.McD., C.G. and C.A.; writing—original draft, A.McD; writing—review and editing, A.McD., C.G., C.A., M.O., M.A.F.V.G. and J.W. All authors have read and agreed to the published version of the manuscript.
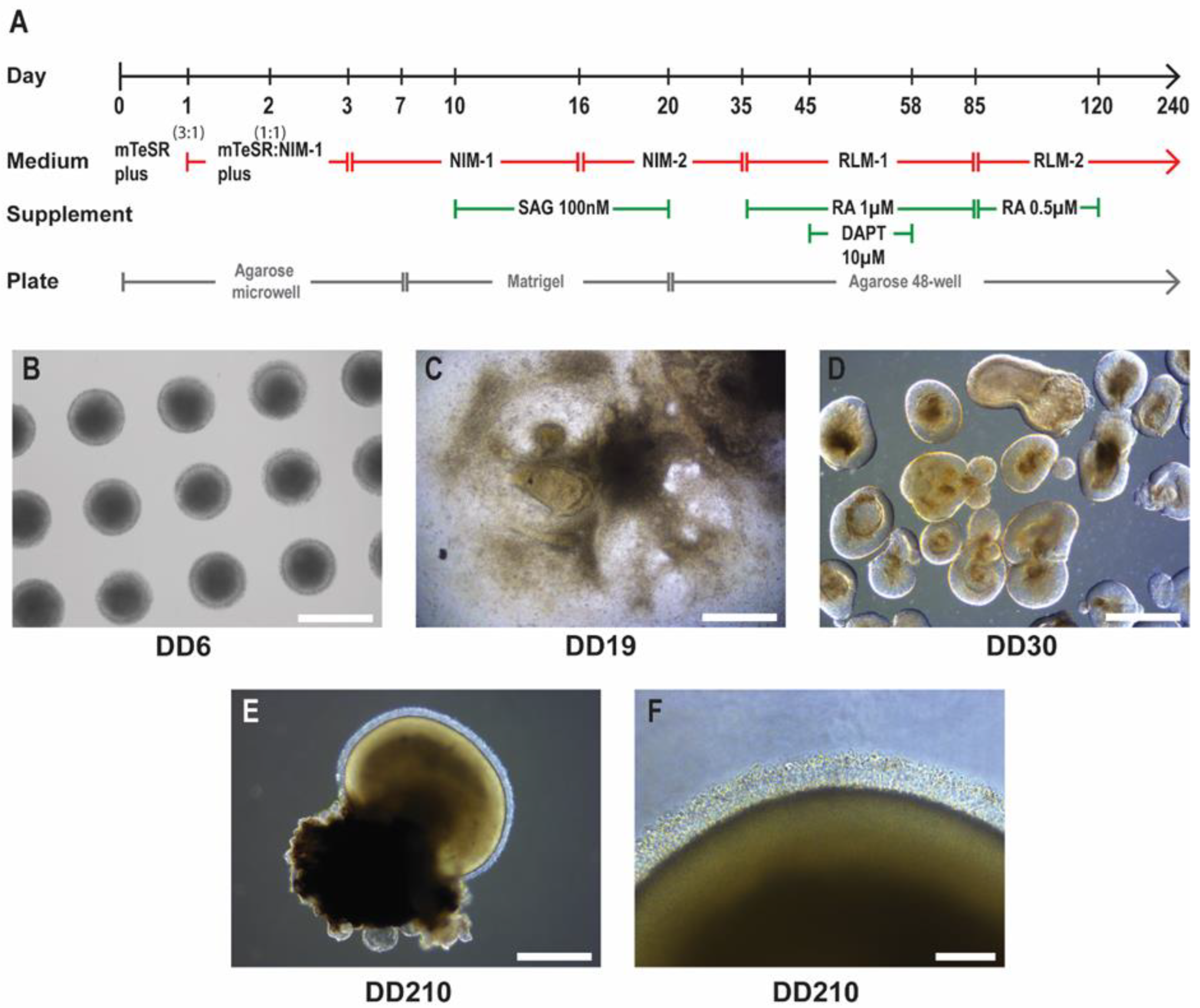
Figure 1.
iPSC-derived retinal organoid differentiation. A) Schematic of the retinal organoid differentiation timeline and culture conditions. Neural-induction medium 1 (NIM-1), neural-induction medium 2 (NIM-2), retinal lamination medium 1 (RLM-1), retinal lamination medium 2 (RLM-2), smoothened agonist (SAG), retinoic acid (RA), gamma secretase inhibitor IX (DAPT). B) Embryoid bodies formed from iPSCs in agarose microwells at DD6. Scale bar 500 µm. C) Neuroepithelium formation on Matrigel at DD19. Scale bar 1 mm. D) Isolated promising early-stage retinal organoid structures in floating culture at DD30. Scale bar 500 µm. E) Mature retinal organoid at DD210. Scale bar 500 µm. F) Higher magnification image of inner/outer segment-like structures at DD210. Scale bar 100 µm.
Figure 1.
iPSC-derived retinal organoid differentiation. A) Schematic of the retinal organoid differentiation timeline and culture conditions. Neural-induction medium 1 (NIM-1), neural-induction medium 2 (NIM-2), retinal lamination medium 1 (RLM-1), retinal lamination medium 2 (RLM-2), smoothened agonist (SAG), retinoic acid (RA), gamma secretase inhibitor IX (DAPT). B) Embryoid bodies formed from iPSCs in agarose microwells at DD6. Scale bar 500 µm. C) Neuroepithelium formation on Matrigel at DD19. Scale bar 1 mm. D) Isolated promising early-stage retinal organoid structures in floating culture at DD30. Scale bar 500 µm. E) Mature retinal organoid at DD210. Scale bar 500 µm. F) Higher magnification image of inner/outer segment-like structures at DD210. Scale bar 100 µm.
Figure 2.
Adenoviral vector transduction of iPSC-derived retinal organoids. A) A schematic representation of the HC-AdV vector genomes. Both HC-AdV5.F5 and HC-AdV5.F50 contain “stuffer” DNA and the reporter mCherry under the control of the hybrid CAG promoter. ITR and Ψ, adenoviral cis-acting inverted terminal repeat (origins of replication) and packaging signal elements necessary for, respectively, vector DNA replication and encapsidation in producer cells. B) Schematic detailing the adenoviral vector transduction procedure in retinal organoids. Transduction occurs at DD130 with either HC-AdV5.F5 or HC-AdV50.F50 with collection and further analysis of the retinal organoids occurring at DD210 and DD240. Created with Biorender.com. C) Representative live-cell fluorescence microscopy images of retinal organoids at 1 day and 80 days post-transduction. The mCherry reporter is visible in purple areas in the brightfield images (white arrows). Scale bars 500 µm. D) Live-cell fluorescence microscopy analysis for mCherry expression in human retinal organoids. The retinal organoids were transduced with the indicated adenoviral vectors at DD130. Three representative images per time point are depicted. DPT = days post-transduction. Scale bars 500 µm.
Figure 2.
Adenoviral vector transduction of iPSC-derived retinal organoids. A) A schematic representation of the HC-AdV vector genomes. Both HC-AdV5.F5 and HC-AdV5.F50 contain “stuffer” DNA and the reporter mCherry under the control of the hybrid CAG promoter. ITR and Ψ, adenoviral cis-acting inverted terminal repeat (origins of replication) and packaging signal elements necessary for, respectively, vector DNA replication and encapsidation in producer cells. B) Schematic detailing the adenoviral vector transduction procedure in retinal organoids. Transduction occurs at DD130 with either HC-AdV5.F5 or HC-AdV50.F50 with collection and further analysis of the retinal organoids occurring at DD210 and DD240. Created with Biorender.com. C) Representative live-cell fluorescence microscopy images of retinal organoids at 1 day and 80 days post-transduction. The mCherry reporter is visible in purple areas in the brightfield images (white arrows). Scale bars 500 µm. D) Live-cell fluorescence microscopy analysis for mCherry expression in human retinal organoids. The retinal organoids were transduced with the indicated adenoviral vectors at DD130. Three representative images per time point are depicted. DPT = days post-transduction. Scale bars 500 µm.
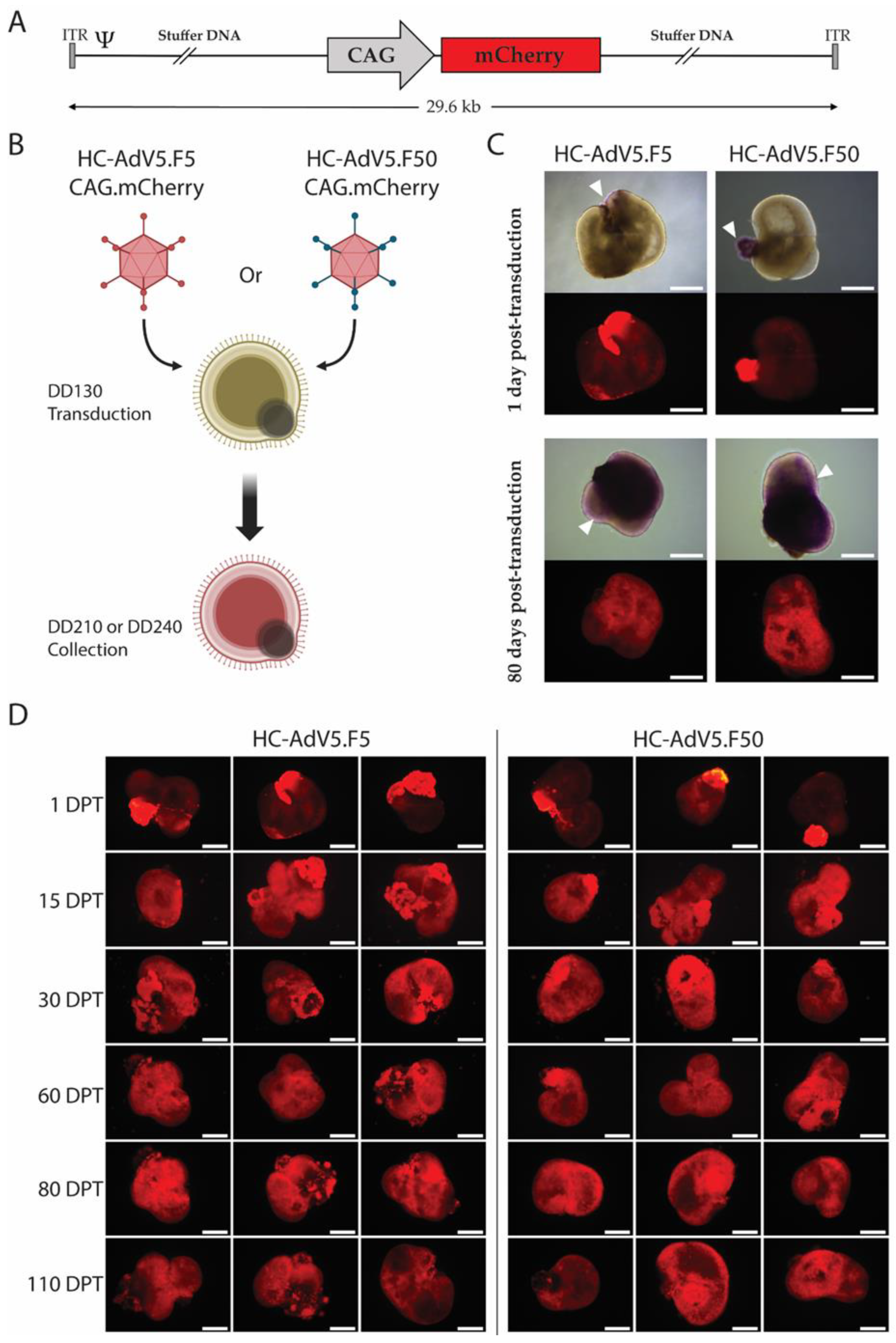
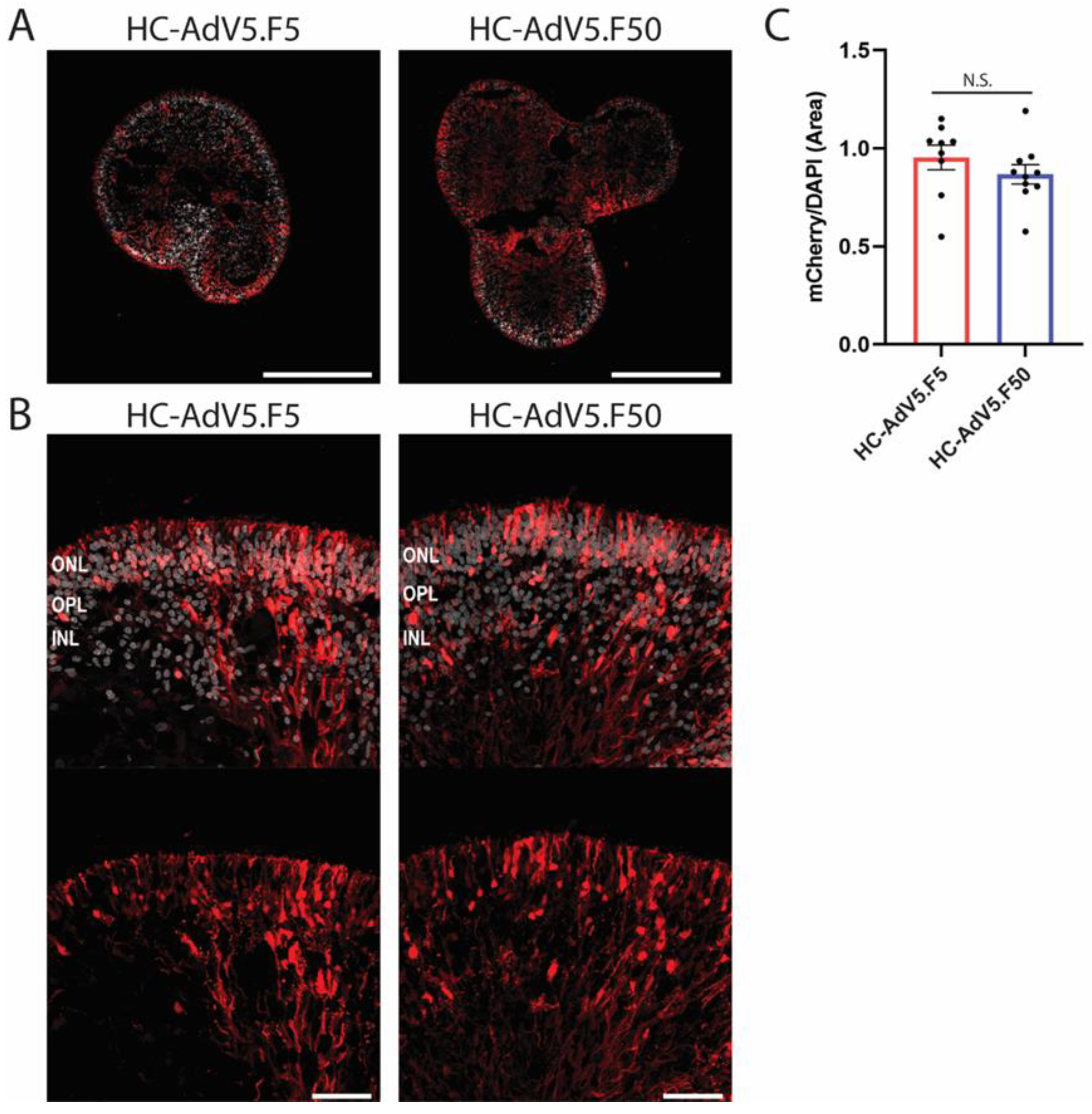
Figure 3.
Transduction efficiency of HC-AdV5.F5 and HC-AdV5.F50 in human retinal organoids. A) Representative images (10× magnification) of retinal organoids (DD210) following transduction at DD130 with either HC-AdV5.F5 or HC-AdV5.F50. Scale bars 500 µm. B) Representative images (40× magnification) of retinal organoids (DD210) upon transduction at DD130 with either HC-AdV5.F5 or HC-AdV5.F50. Outer nuclear layer (ONL), outer plexiform layer (OPL), and inner nuclear layer (INL). Scale bars 50 µm. C) Quantification of transduction efficiency in human retinal organoids using HC-AdV5.F5 and HC-AdV5.F50 calculated by mCherry positive area normalized to DAPI positive area. Each datapoint of the graph represents a single organoid, the value for each organoid is generated from the average value of 3 independent images at 40× magnification. Unpaired t-test. Error bars represent standard error of the mean (SEM). Number of individual organoids per condition: HC-AdV5.F5 n=9, HC-AdV5.F50 n=10.
Figure 3.
Transduction efficiency of HC-AdV5.F5 and HC-AdV5.F50 in human retinal organoids. A) Representative images (10× magnification) of retinal organoids (DD210) following transduction at DD130 with either HC-AdV5.F5 or HC-AdV5.F50. Scale bars 500 µm. B) Representative images (40× magnification) of retinal organoids (DD210) upon transduction at DD130 with either HC-AdV5.F5 or HC-AdV5.F50. Outer nuclear layer (ONL), outer plexiform layer (OPL), and inner nuclear layer (INL). Scale bars 50 µm. C) Quantification of transduction efficiency in human retinal organoids using HC-AdV5.F5 and HC-AdV5.F50 calculated by mCherry positive area normalized to DAPI positive area. Each datapoint of the graph represents a single organoid, the value for each organoid is generated from the average value of 3 independent images at 40× magnification. Unpaired t-test. Error bars represent standard error of the mean (SEM). Number of individual organoids per condition: HC-AdV5.F5 n=9, HC-AdV5.F50 n=10.
Figure 4.
Adenovector transduction of Müller glial cells. (A and B) Representative images of retinal organoids (DD210) following transduction with HC-AdV5.F5 and HC-AdV5.F50, respectively. Colocalization between the mCherry reporter and the Müller glial cell marker CRALBP is identified by white coloured regions. Outer nuclear layer (ONL) and inner nuclear layer (INL). Scale bars 50 µm. C) Quantification of transduced Müller glial cells, measured by the percentage of CRALBP-positive areas also expressing mCherry. Each datapoint of the graph represents a single organoid, the value for each organoid is generated from the average value of 3 independent images at 40× magnification. Unpaired t-test. Error bars represent standard error of the mean (SEM). Number of individual organoids per condition: HC-AdV5.F5 n=9, HC-AdV5.F50 n=10.
Figure 4.
Adenovector transduction of Müller glial cells. (A and B) Representative images of retinal organoids (DD210) following transduction with HC-AdV5.F5 and HC-AdV5.F50, respectively. Colocalization between the mCherry reporter and the Müller glial cell marker CRALBP is identified by white coloured regions. Outer nuclear layer (ONL) and inner nuclear layer (INL). Scale bars 50 µm. C) Quantification of transduced Müller glial cells, measured by the percentage of CRALBP-positive areas also expressing mCherry. Each datapoint of the graph represents a single organoid, the value for each organoid is generated from the average value of 3 independent images at 40× magnification. Unpaired t-test. Error bars represent standard error of the mean (SEM). Number of individual organoids per condition: HC-AdV5.F5 n=9, HC-AdV5.F50 n=10.
Figure 5.
Adenovector transduction of photoreceptors. (A and B) Representative images of retinal organoids (DD210) following transduction at DD130 with HC-AdV5.F5 and HC-AdV5.F50, respectively. Colocalization between the mCherry reporter and the photoreceptor marker recoverin is identified by white coloured regions. Outer nuclear layer (ONL) and inner nuclear layer (INL). Scale bars 50 µm. C) Quantification of transduced photoreceptors, measured by the percentage of the recoverin-positive area also expressing mCherry. Each datapoint of the graph represents a single organoid, the value for each organoid is generated from the average value of 3 independent images at 40× magnification. Unpaired t-test. Error bars represent standard error of the mean (SEM). Number of individual organoids per condition: HC-AdV5.F5 n=8, HC-AdV5.F50 n=10.
Figure 5.
Adenovector transduction of photoreceptors. (A and B) Representative images of retinal organoids (DD210) following transduction at DD130 with HC-AdV5.F5 and HC-AdV5.F50, respectively. Colocalization between the mCherry reporter and the photoreceptor marker recoverin is identified by white coloured regions. Outer nuclear layer (ONL) and inner nuclear layer (INL). Scale bars 50 µm. C) Quantification of transduced photoreceptors, measured by the percentage of the recoverin-positive area also expressing mCherry. Each datapoint of the graph represents a single organoid, the value for each organoid is generated from the average value of 3 independent images at 40× magnification. Unpaired t-test. Error bars represent standard error of the mean (SEM). Number of individual organoids per condition: HC-AdV5.F5 n=8, HC-AdV5.F50 n=10.
Figure 6.
Increased thickness of the ONL observed at DD240 following HC-AdV transduction. A) Representative images of GFAP expression in organoids at DD210 and (B) DD240 following adenoviral vector transduction (DD130). White arrows show GFAP positive regions. Scale bars 50 µm. C). Quantification of GFAP expression in DD210 and (D) DD240 organoids calculated by GFAP intensity normalized to DAPI area. E) Quantification of the number of photoreceptor nuclei in a row in transduced and non-transduced organoids at DD210 and (F) DD240. G) Quantification of ONL thickness in transduced and non-transduced organoids at DD210 and (H) DD240. Each datapoint of the graph represents a single organoid, the value for each organoid is generated from the average value of 3 independent images at 40× magnification. Error bars represent standard error of the mean. Unpaired t-test, p < 0.01 (∗∗). Number of individual organoids per condition: DD210 - Untreated n=5; HC-AdV5.F5 n=9; HC-AdV5.F50 n=10; DD240 - Untreated n=6; HC-AdV5.F5 n=7; HC-AdV5.F50 n=10.
Figure 6.
Increased thickness of the ONL observed at DD240 following HC-AdV transduction. A) Representative images of GFAP expression in organoids at DD210 and (B) DD240 following adenoviral vector transduction (DD130). White arrows show GFAP positive regions. Scale bars 50 µm. C). Quantification of GFAP expression in DD210 and (D) DD240 organoids calculated by GFAP intensity normalized to DAPI area. E) Quantification of the number of photoreceptor nuclei in a row in transduced and non-transduced organoids at DD210 and (F) DD240. G) Quantification of ONL thickness in transduced and non-transduced organoids at DD210 and (H) DD240. Each datapoint of the graph represents a single organoid, the value for each organoid is generated from the average value of 3 independent images at 40× magnification. Error bars represent standard error of the mean. Unpaired t-test, p < 0.01 (∗∗). Number of individual organoids per condition: DD210 - Untreated n=5; HC-AdV5.F5 n=9; HC-AdV5.F50 n=10; DD240 - Untreated n=6; HC-AdV5.F5 n=7; HC-AdV5.F50 n=10.