Submitted:
01 November 2024
Posted:
04 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Outbreaks
2.1.1. Free-Living Trahiras
2.1.2. Farmed Arapaima
2.2. Bacteriological Examinations
2.3. DNA Extraction
2.4. Capsular Typing
2.5. Repetitive Extragenic Palindromic-PCR (REP-PCR)
2.6. Multilocus Sequence Typing (MLST)
2.7. Whole-Genome Sequencing and Phylogenomic Analysis
2.8. Antimicrobial Susceptibility
2.9. Fish and Challenge Assay
2.10. Histopathological Analysis
3. Results
3.1. Clinical Signs at Field Conditions and Identification of Streptococcus Agalactiae Isolates
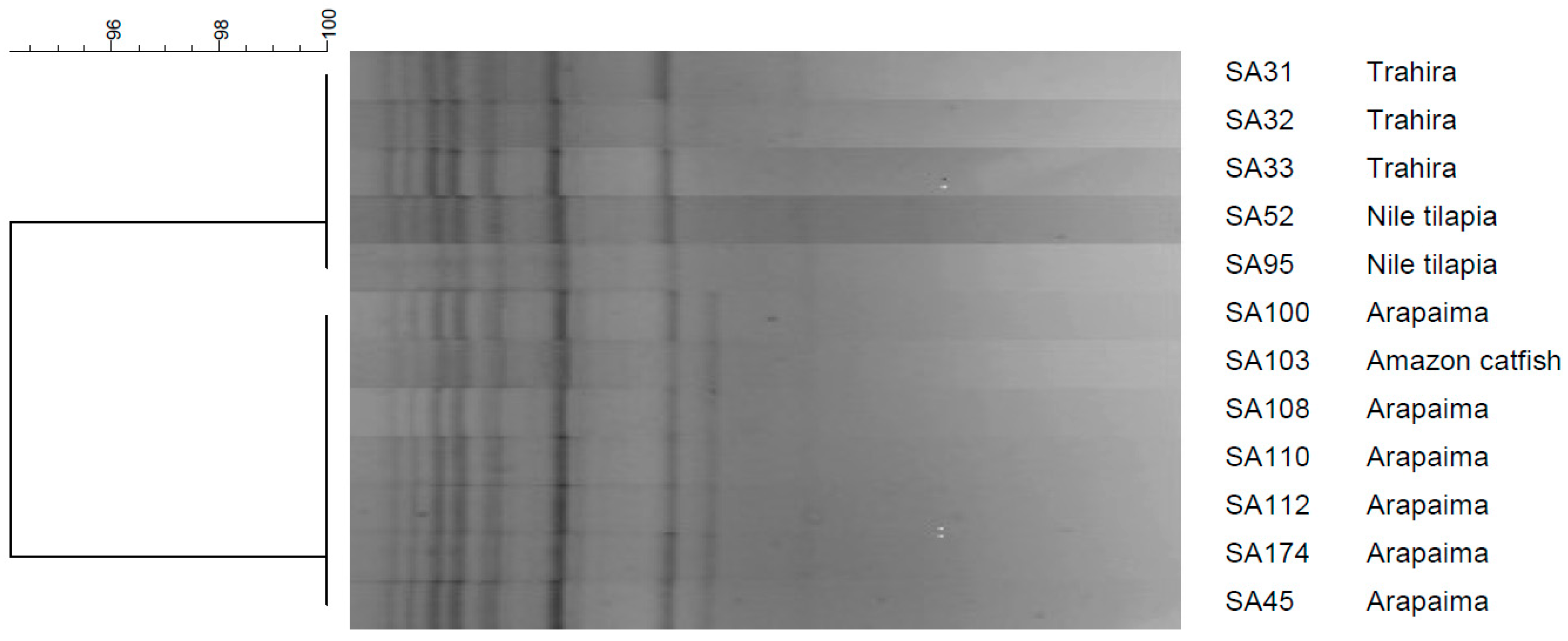
3.2. Genetic Diversity and Relationship Among the Isolates
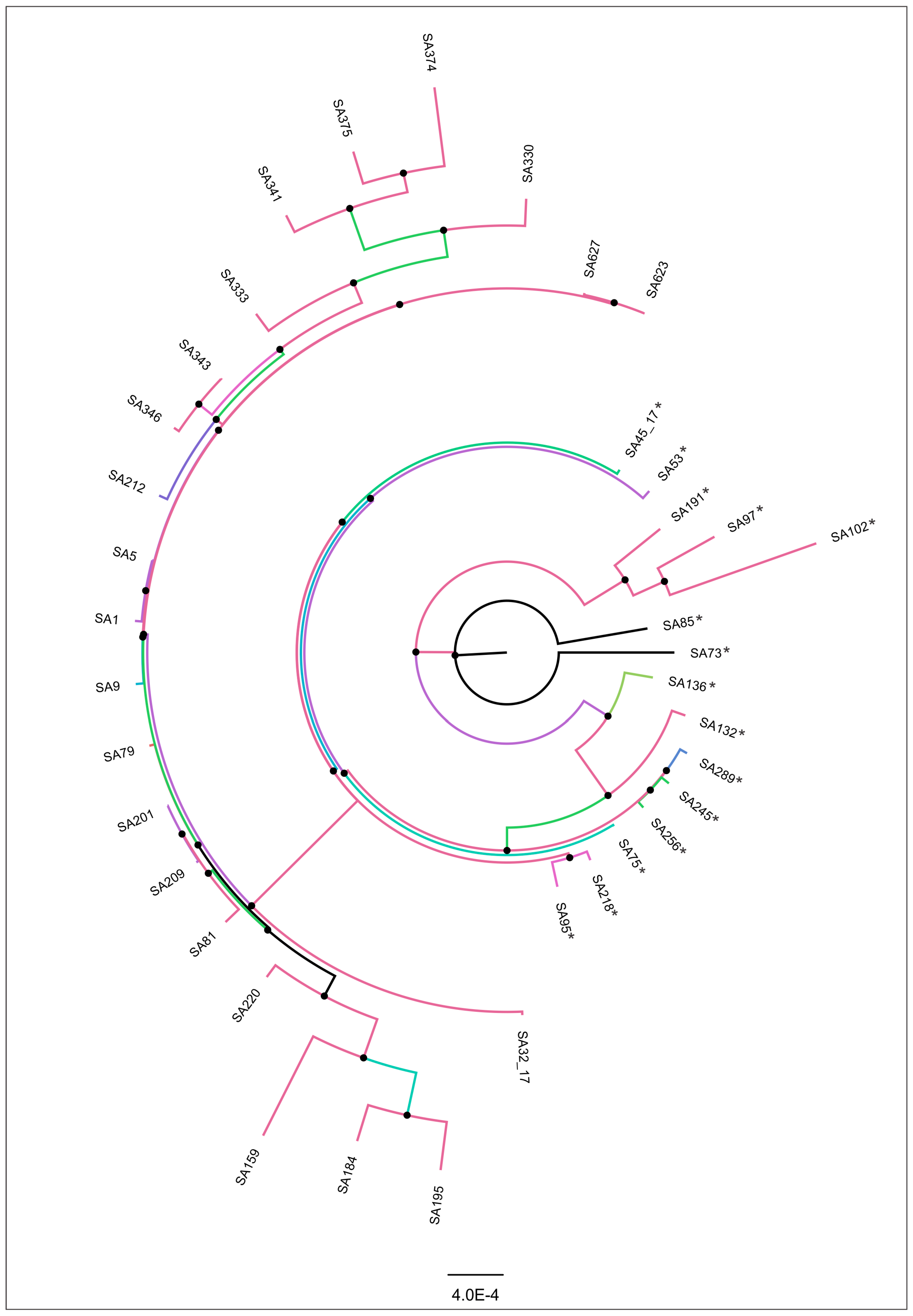
3.3. Genomic Similarity and Phylogenomic Analysis
3.4. Antimicrobial Susceptibility and In Silico Identification of AMR Genes
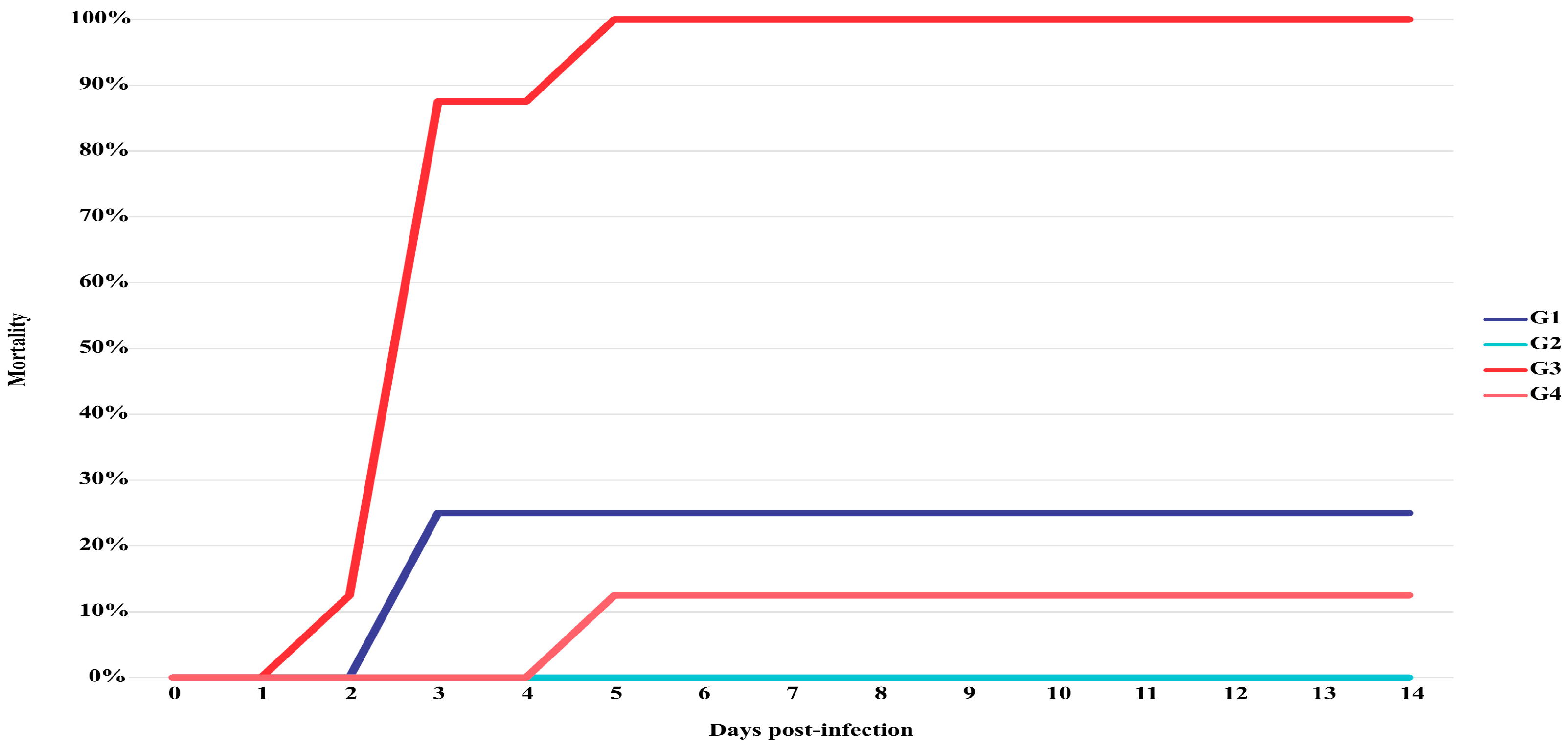
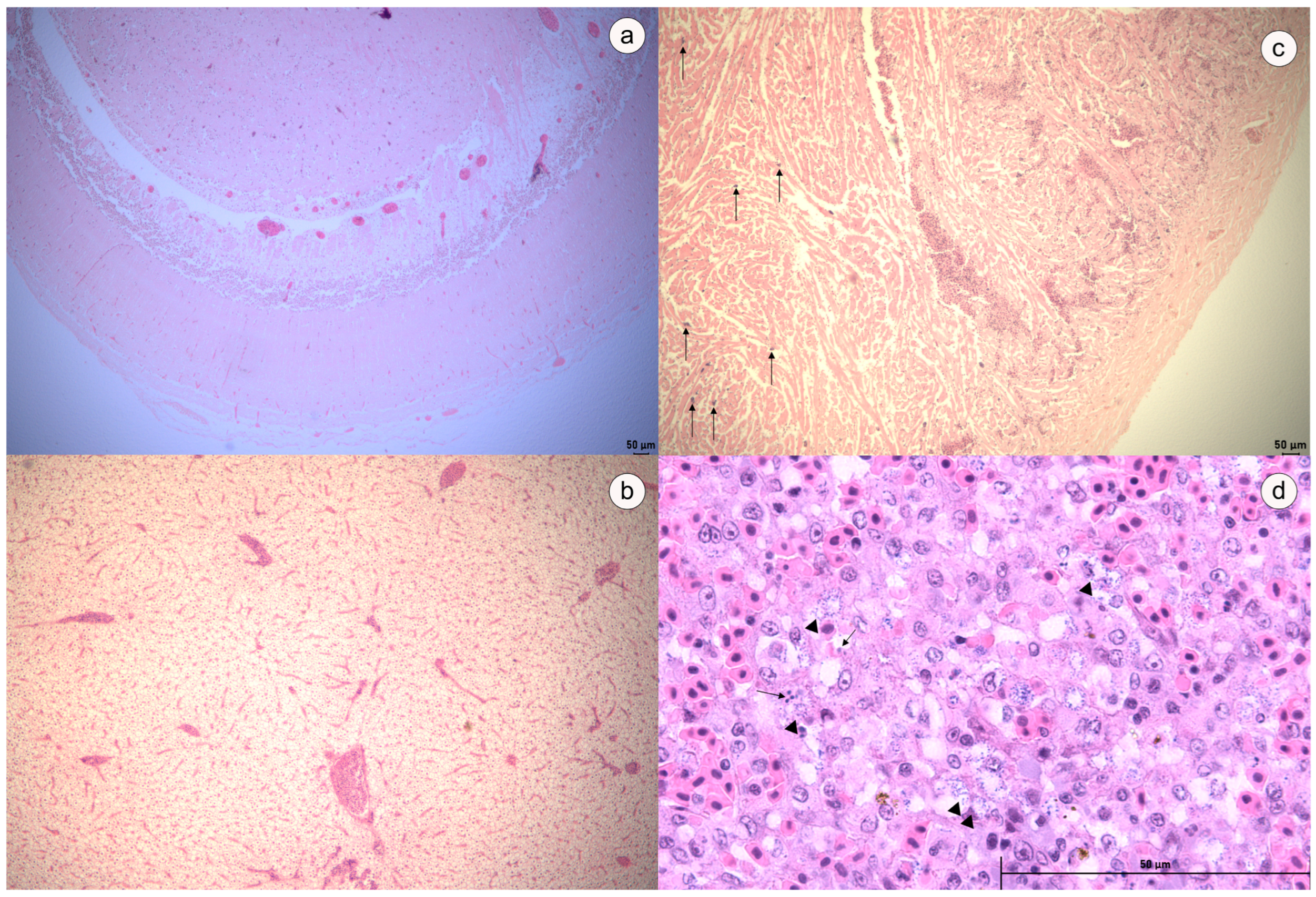
3.5. Challenge Assay and Histopathological Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Ethical Approval
References
- Lusiastuti, A.M.; Textor, M.; Seeger, H.; Akineden, Ö.; Zschöck, M. The Occurrence of Streptococcus agalactiae Sequence Type 261 from Fish Disease Outbreaks of Tilapia Oreochromis niloticus in Indonesia. Aquac. Res. 2014, 45, 1260–1263. [Google Scholar] [CrossRef]
- Johri, A.K.; Paoletti, L.C.; Glaser, P.; Dua, M.; Sharma, P.K.; Grandi, G.; Rappuoli, R. Group B Streptococcus: Global Incidence and Vaccine Development. Nat. Rev. Microbiol. 2006, 4, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Keefe, G.P. Streptococcus agalactiae Mastitis: A Review. Can. Vet. J. = La Rev. Vet. Can. 1997, 38, 429–437. [Google Scholar]
- Mian, G.F.; Godoy, D.T.; Leal, C.A.G.; Yuhara, T.Y.; Costa, G.M.; Figueiredo, H.C.P. Aspects of the Natural History and Virulence of S. agalactiae Infection in Nile Tilapia. Vet. Microbiol. 2009, 136, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Bowater, R.O.; Forbes-Faulkner, J.; Anderson, I.G.; Condon, K.; Robinson, B.; Kong, F.; Gilbert, G.L.; Reynolds, a.; Hyland, S.; McPherson, G.; et al. Natural Outbreak of Streptococcus agalactiae (GBS) Infection in Wild Giant Queensland Grouper, Epinephelus lanceolatus (Bloch), and Other Wild Fish in Northern Queensland, Australia. J. Fish Dis. 2012, 35, 173–186. [Google Scholar] [CrossRef]
- Evans, J.J.; Klesius, P.H.; Gilbert, P.M.; Shoemaker, C.A.; Sarawi, M.A. Al; Landsberg, J. Characterization of B-Haemolytic Group B Streptococcus agalactiae in Cultured Seabream, Sparus auratus L., and Wild Mullet, Liza klunzingeri ( Day ), in Kuwait. J. Fish Biol. 2002, 25, 505–513. [Google Scholar] [CrossRef]
- Leira, M.H.; Botelho, H.A.; Lago, A.A.; De Freitas, R.T.F.; Garcia, A.M.; De Azevedo Soares Dos Santos, H.C. Identification of Pathogens in Fish Polyculture Systems in Southern Minas Gerais, Brazil. Acta Vet. Bras. 2019, 13, 13–17. [Google Scholar] [CrossRef]
- Sebastião, F.A.; Furlan, L.R.; Hashimoto, D.T.; Pilarski, F. Identification of Bacterial Fish Pathogens in Brazil by Direct Colony PCR and 16S rRNA Gene Sequencing. Adv. Microbiol. 2015, 05, 409–424. [Google Scholar] [CrossRef]
- Tavares, G.C.; de Queiroz, G.A.; Assis, G.B.N.; Leibowitz, M.P.; Teixeira, J.P.; Figueiredo, H.C.P.; Leal, C.A.G. Disease Outbreaks in Farmed Amazon Catfish (Leiarius marmoratus x Pseudoplatystoma corruscans) Caused by Streptococcus agalactiae, S. iniae, and S. dysgalactiae. Aquaculture 2018, 495, 384–392. [Google Scholar] [CrossRef]
- Bialetzki, A.; Nakatani, K.; Sanches, P. V.; Baumgartner, G. Spatial and Temporal Distribution of Larvae and Juveniles of Hoplias malabaricus (Characiformes, Erythrinidae) in the Upper Paraná River Floodplain, Brazil. Brazilian J. Biol. 2002, 62, 211–222. [Google Scholar] [CrossRef]
- da Silva, C.A.; Oba, E.T.; Ramsdorf, W.A.; Magalhães, V.F.; Cestari, M.M.; Oliveira Ribeiro, C.A.; Silva de Assis, H.C. First Report about Saxitoxins in Freshwater Fish Hoplias malabaricus through Trophic Exposure. Toxicon 2011, 57, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, L.C.; Santos, A.C.G.; Ferreira, E.M.; Teófilo, T.S.; Pereira, D.M.; Costa, F.N. Aspectos Parasitológicos Da Traíra (Hoplias malabaricus) Proveniente Da Cidade de São Bento, MA. Arq. Bras. Med. Vet. e Zootec. 2017, 69, 264–268. [Google Scholar] [CrossRef]
- Benigno, R.N.M.; Knoff, M.; Matos, E.R.; Gomes, D.C.; Pinto, R.M.; Clemente, S.C.S. Morphological Aspects of Clinostomidae Metacercariae (Trematoda: Digenea) in Hoplerytrinus unitaeniatus and Hoplias malabaricus (Pisces: Erythrinidae) of the Neotropical Region, Brazil. An. Acad. Bras. Cienc. 2014, 86, 733–744. [Google Scholar] [CrossRef] [PubMed]
- IBGE Produção Da Pecuária Municipal, 50th ed.; Instituto Brasileiro de Geografia e Estatística: Rio de Janeiro, Brasil, 2023.
- Baia, R.R.J.; Florentino, A.C.; Silva, L.M.A.; Tavares-Dias, M. Patterns of the Parasite Communities in a Fish Assemblage of a River in the Brazilian Amazon Region. Acta Parasitol. 2018, 63, 304–316. [Google Scholar] [CrossRef]
- Meneguetti, D.U. de O.; Laray, M.P. de O.; Camargo, L.M.A. Primeiro Relato de Larvas de Eustrongylides Sp. (Nematoda: Dioctophymatidae) Em Hoplias malabaricus (Characiformes: Erythrinidae) No Estado de Rondônia, Amazônia Ocidental, Brasil. Rev. Pan-Amazônica Saúde 2013, 4, 55–58. [Google Scholar] [CrossRef]
- Oliveira, M.S.B.; Corrêa, L.L.; Oliveira Ferreira, D.; Neves, L.R.; Tavares-Dias, M. Records of New Localities and Hosts for Crustacean Parasites in Fish from the Eastern Amazon in Northern Brazil. J. Parasit. Dis. 2017, 41, 565–570. [Google Scholar] [CrossRef]
- Guimarães, L.; Santos, A.C. dos; Ferreira, E.; Pereira, D.; Costa, F. Microbiological Quality of Trahira Fish (Hoplias malabaricus) from Baixada Maranhense, Municipality of São Bento, MA. Arq. Inst. Biol. (Sao. Paulo). 2018, 84, 1–7. [Google Scholar] [CrossRef]
- Honczaryk, A.; Inoue, L.A.K.A. Anesthesia in Pirarucu by Benzocaine Sprays in the Gills. Ciência Rural 2010, 40, 204–207. [Google Scholar] [CrossRef]
- Imbiriba, E.P. Potencial de Criação de Pirarucu, Arapaima gigas,Em Cativeiro. Acta Amaz. 2001, 31, 299. [Google Scholar] [CrossRef]
- Fazzi-Gomes, P.F.; Melo, N.; Palheta, G.; Guerreiro, S.; Amador, M.; Ribeiro-Dos-santos, A.K.; Santos, S.; Hamoy, I. Genetic Diversity and Differentiation in Natural Populations of Arapaima gigas from Lower Amazon Revealed by Microsatellites. Genet. Mol. Res. 2017, 16, 1–6. [Google Scholar] [CrossRef]
- Ferraris Jr., C.J. Family Arapaimatidae (Bonytongues). In Check list of the freshwater fishes of South and Central America; Reis, R.E., Kullander, S.O., Ferraris, C.J., Jr., Eds.; EDIPUCRS: Porto Alegre, Brazil, 2003; pp. 32–33. [Google Scholar]
- Pereira-Filho, M.; Roubach, R. Pirarucu (Arapaima gigas). In Espécies nativas para piscicultura no Brasil; Baldisserotto, B., de C. Gomes, L., Eds.; Editora UFSM: Santa Maria, Brazil, 2010; pp. 27–56. [Google Scholar]
- Carvalho, F.R.; Casatti, L.; Manzotti, A.R.; Ravazzi, D.C.W. First Record of Arapaima gigas (Schinz, 1822) (Teleostei: Osteoglossomorpha), the “Pirarucu”, in the Upper Paraná River Basin, Southeast Brazil. Check List 2015, 11, 1–4. [Google Scholar] [CrossRef]
- Lawson, L.L.; Tuckett, Q.M.; Lawson, K.M.; Watson, C.A.; Hill, J.E. Lower Lethal Temperature for Arapaima Arapaima gigas: Potential Implications for Culture and Establishment in Florida. N. Am. J. Aquac. 2015, 77, 497–502. [Google Scholar] [CrossRef]
- Miranda-Chumacero, G.; Wallace, R.; Calderón, H.; Calderón, G.; Willink, P.; Guerrero, M.; Siles, T.M.; Lara, K.; Chuqui, D. Distribution of Arapaima (Arapaima gigas) (Pisces: Arapaimatidae) in Bolivia: Implications in the Control and Management of a Non-Native Population. BioInvasions Rec. 2012, 1, 129–138. [Google Scholar] [CrossRef]
- Ohs, C.; Hill, J.; Wright, S.; Giddings, H.M.; Durland Donahou, A. Candidate Species for Florida Aquaculture: Arapaima Arapaima gigas. Edis 2021, 2021, 1–13. [Google Scholar] [CrossRef]
- Wyman-Grothem, K.; Castello, L.; DTBS, C.; CRC, D.; ALB, M.; Patoka, J.; Stewart, D.; Watson, C. Invasion Risk to the United States from Arapaima spp. Hinges on Climate Suitability. Aquac. Environ. Interact. 2024, 16, 175–188. [Google Scholar] [CrossRef]
- Drumond, G.V.F.; Caixeiro, A.P. de A.; Tavares-Dias, M.; Marcon, J.L.; Affonso, E.G. Características Bioquímicas e Hematológicas Do Pirarucu Arapaima gigas Schinz, 1822 (Arapaimidae) de Cultivo Semi-Intensivo Na Amazônia. Acta Amaz. 2010, 40, 591–595. [Google Scholar] [CrossRef]
- Cruz, M.G. da; Jerônimo, G.T.; Bentes, S.P.C.; Gonçalves, L.U. Trichlorfon Is Effective against Dawestrema cycloancistrium and Does Not Alter the Physiological Parameters of Arapaima (Arapaima gigas): A Large Neotropical Fish from the Amazon. J. Fish Dis. 2022, 45, 203–212. [Google Scholar] [CrossRef]
- Dias, M.K.R.; Sampaio, L.S.; Proietti-Junior, A.A.; Yoshioka, E.T.O.; Rodrigues, D.P.; Rodriguez, A.F.R.; Ribeiro, R.A.; Faria, F.S.E.D.V.; Ozório, R.O.A.; Tavares-Dias, M. Lethal Dose and Clinical Signs of Aeromonas hydrophila in Arapaima gigas (Arapaimidae), the Giant Fish from Amazon. Vet. Microbiol. 2016, 188, 12–15. [Google Scholar] [CrossRef]
- Ribeiro, M. da S.; da Fonseca, F.A.L.; de Queiroz, M.N.; Affonso, E.G.; da Conceição, L.E.C.; Gonçalves, L.U. Fish Protein Hydrolysate as an Ingredient in Diets for Arapaima gigas Juveniles. Bol. do Inst. Pesca 2017, 43, 85–92. [Google Scholar] [CrossRef]
- Marinho, R.G.B.; Tavares-Dias, M.; Dias-Grigório, M.K.R.; Neves, L.R.; Yoshioka, E.T.O.; Boijink, C.L.; Takemoto, R.M. Helminthes and Protozoan of Farmed Pirarucu (Arapaima gigas) in Eastern Amazon and Host-Parasite Relationship. Arq. Bras. Med. Vet. e Zootec. 2013, 65, 1192–1202. [Google Scholar] [CrossRef]
- Andrade-Porto, S.M.; Cárdenas, M.Q.; Martins, M.L.; Oliveira, J.K.Q.; Pereira, J.N.; Araújo, C.S.O.; Malta, J.C.O. First Record of Larvae of Hysterothylacium (Nematoda: Anisakidae) with Zoonotic Potential in the Pirarucu Arapaima gigas (Osteichthyes: Arapaimidae) from South America. Brazilian J. Biol. 2015, 75, 790–795. [Google Scholar] [CrossRef] [PubMed]
- Proietti-Junior, A.A.; Lima, L.S.; Roges, E.M.; Rodrigues, Y.C.; Lima, K.V.B.; Rodrigues, D.P.; Tavares-Dias, M. Experimental Co-Infection by Aeromonas hydrophila and Aeromonas jandaei in Pirarucu Arapaima gigas (Pisces: Arapaimidae). Aquac. Res. 2021, 52, 1688–1696. [Google Scholar] [CrossRef]
- Choresca Jr., C.H.; Gomez, D.K.; Shin, S.P.; Kim, J.H.; Han, J.E.; Jun, J.W.; Park, S.C. Molecular Detection of Edwardsiella tarda with gyrB Gene Isolated from Pirarucu, Arapaima gigas Which Is Exhibited in an Indoor Private Commercial Aquarium. African J. Biotechnol. 2011, 10, 848–850. [Google Scholar]
- Kodama, H.; Nakanishi, Y.; Yamamoto, F.; Mikami, T.; Izawa, H.; Imagawa, T.; Hashimoto, Y.; Kudo, N. Salmonella arizonae Isolated from a Pirarucu, Arapaima gigas Cuvier, with Septicaemia. J. Fish Dis. 1987, 10, 509–512. [Google Scholar] [CrossRef]
- Choresca Jr., C.H.; Kim, J.H.; Gomez, D.K.; Jang, H.; Joh, S.J.; Park, S.C. Isolation of Serratia fonticola from Pirarucu Arapaima gigas. Korean J. Vet. Res. 2008, 48, 89–92. [Google Scholar]
- Barbanti, A.C.C.; do Rosário, A.E.C.; da Silva Maia, C.R.M.; Rocha, V.P.; Costa, H.L.; Trindade, J.M.; Nogueira, L.F.F.; Rosa, J.C.C.; Ranzani-Paiva, M.J.T.; Pilarski, F.; et al. Genetic Characterization of Lactococcosis-Causing Bacteria Isolated from Brazilian Native Fish Species. Aquaculture 2024, 593, 741305. [Google Scholar] [CrossRef]
- Serrano-Martínez, E.; Verónica, C.P.; Marco, Q.H.; Gina, C. V.; Jorge, L.Q. Isolation of Bacteria and Fungi in Tissues of Paiche (Arapaima gigas) Reared in Captivity. Rev. Investig. Vet. del Peru 2014, 25, 117–122. [Google Scholar]
- Assis, G.B.N.; Pereira, F.L.; Zegarra, A.U.; Tavares, G.C.; Leal, C.A.; Figueiredo, H.C.P. Use of MALDI-TOF Mass Spectrometry for the Fast Identification of Gram-Positive Fish Pathogens. Front. Microbiol. 2017, 8, 1492. [Google Scholar] [CrossRef]
- Poyart, C.; Tazi, A.; Réglier-Poupet, H.; Billoët, A.; Tavares, N.; Raymond, J.; Trieu-Cuot, P. Multiplex PCR Assay for Rapid and Accurate Capsular Typing of Group B Streptococci. J. Clin. Microbiol. 2007, 45, 1985–1988. [Google Scholar] [CrossRef]
- Costa, F.A.A.; Leal, C.A.G.; Leite, R.C.; Figueiredo, H.C.P. Genotyping of Streptococcus dysgalactiae Strains Isolated from Nile Tilapia, Oreochromis niloticus (L.). J. Fish Dis. 2014, 37, 463–469. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the Amount of Ecologic Association between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Ishii, S.; Sadowsky, M.J. Applications of the Rep-PCR DNA Fingerprinting Technique to Study Microbial Diversity, Ecology and Evolution. Environ. Microbiol. 2009, 11, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Hunter, P.R.; Gaston, M.A. Numerical Index of the Discriminatory Ability of Typing Systems: An Application of Simpson’s Index of Diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef] [PubMed]
- Pinto, F.R.; Melo-Cristino, J.; Ramirez, M. A Confidence Interval for the Wallace Coefficient of Concordance and Its Application to Microbial Typing Methods. PLoS One 2008, 3, e3696. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.; Bohnsack, J.F.; Takahashi, S.; Oliver, K.A.; Chan, M.-S.; Kunst, F.; Glaser, P.; Rusniok, C.; Crook, D.W.M.; Harding, R.M.; et al. Multilocus Sequence Typing System for Group B Streptococcus. J. Clin. Microbiol. 2003, 41, 2530–2536. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.Org Website and Their Applications. Wellcome Open Res. 2018, 3, 1–20. [Google Scholar] [CrossRef]
- Page, A.J.; De Silva, N.; Hunt, M.; Quail, M.A.; Parkhill, J.; Harris, S.R.; Otto, T.D.; Keane, J.A. Robust High-Throughput Prokaryote de Novo Assembly and Improvement Pipeline for Illumina Data. Microb. genomics 2016, 2, e000083. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de Novo Short Read Assembly Using de Bruijn Graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Boetzer, M.; Henkel, C. V; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding Pre-Assembled Contigs Using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef]
- Boetzer, M.; Pirovano, W. Toward Almost Closed Genomes with GapFiller. Genome Biol. 2012, 13, R56. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Brown, G.R.; Maglott, D.R. NCBI Reference Sequences (RefSeq): Current Status, New Features and Genome Annotation Policy. Nucleic Acids Res. 2012, 40, D130–5. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.L.N.; Ariute, J.C.; Rodrigues da Costa, F.M.; Benko-Iseppon, A.M.; Barh, D.; Azevedo, V.; Aburjaile, F. PanViTa: Pan Virulence and ResisTance Analysis. Front. Bioinforma. 2023, 3. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- CLSI VET03: Methods for Antimicrobial Broth Dilution and Disk Diffusion Susceptibility Testing of Bacteria Isolated from Aquatic Animals; Clinical and Laboratory Standards Institute: Wayne, USA, 2020.
- Leal, C.A.G.; Silva, B.A.; Colombo, S.A. Susceptibility Profile and Epidemiological Cut-Off Values Are Influenced by Serotype in Fish Pathogenic Streptococcus agalactiae. Antibiotics 2023, 12. [Google Scholar] [CrossRef]
- Kronvall, G.; Smith, P. Normalized Resistance Interpretation, the NRI Method. APMIS 2016, 124, 1023–1030. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Luna, L.G. Manual of Histologic Staining Methods of the Armed Forces Institute of Pathology, 3rd ed. Blakiston Division: New York, 1968.
- Barony, G.M.; Tavares, G.C.; Pereira, F.L.; Carvalho, A.F.; Dorella, F.A.; Leal, C.A.G.; Figueiredo, H.C.P. Large-Scale Genomic Analyses Reveal the Population Structure and Evolutionary Trends of Streptococcus agalactiae Strains in Brazilian Fish Farms. Sci. Rep. 2017, 7, 13538. [Google Scholar] [CrossRef]
- Laith, A.A.; Ambak, M.A.; Hassan, M.; Sheriff, S.M.; Nadirah, M.; Draman, A.S.; Wahab, W.; Ibrahim, W.N.W.; Aznan, A.S.; Jabar, A.; et al. Molecular Identification and Histopathological Study of Natural Streptococcus agalactiae Infection in Hybrid Tilapia (Oreochromis niloticus). Vet. world 2017, 10, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Delamare-Deboutteville, J.; Bowater, R.; Condon, K.; Reynolds, A.; Fisk, A.; Aviles, F.; Barnes, A.C. Infection and Pathology in Queensland Grouper, Epinephelus lanceolatus, (Bloch), Caused by Exposure to Streptococcus agalactiae via Different Routes. J. Fish Dis. 2015, 38, 1021–1035. [Google Scholar] [CrossRef] [PubMed]
- Ortega Asencios, Y.; Barreiro Sánchez, F.; Bueno Mendizábal, H.; Huancaré Pusari, K.; Ostos Alfonso, H.; Manchego Sayán, A.; Pereira Figueiredo, M.A.; Gómez Manrique, W.; de Andrade Belo, M.A.; Sandoval Chaupe, N. First Report of Streptococcus agalactiae Isolated from Oreochromis niloticus in Piura, Peru: Molecular Identification and Histopathological Lesions. Aquac. Reports 2016, 4, 74–79. [Google Scholar] [CrossRef]
- Gonzalez-Callirgos, L.; da Costa, J.I.; Yunis-Aguinaga, J. Economic Evaluation of Arapaima gigas Production in Earth Ponds: Case Study of a Small Fish Farm at San Martin-Peru. Bol. do Inst. Pesca 2023, 49, 1–8. [Google Scholar] [CrossRef]
- Venturieri, R.; Bernardino, G. Pirarucu: Espécie Ameaçada Pode Ser Salva Através Do Cultivo. Panor. da Aquicultura 1999, 9. [Google Scholar]
- Iregui, C.A.; Comas, J.; Vásquez, G.M.; Verján, N. Experimental Early Pathogenesis of Streptococcus agalactiae Infection in Red Tilapia Oreochromis spp. J. Fish Dis. 2016, 39, 205–215. [Google Scholar] [CrossRef]
- Chideroli, R.T.; Amoroso, N.; Mainardi, R.M.; Suphoronski, S.A.; de Padua, S.B.; Alfieri, A.F.; Alfieri, A.A.; Mosela, M.; Moralez, A.T.P.; de Oliveira, A.G.; et al. Emergence of a New Multidrug-Resistant and Highly Virulent Serotype of Streptococcus agalactiae in Fish Farms from Brazil. Aquaculture 2017, 479, 45–51. [Google Scholar] [CrossRef]
- de Oliveira, T.F.; Queiroz, G.A.; Teixeira, J.P.; Figueiredo, H.C.P.; Leal, C.A.G. Recurrent Streptoccoccus agalactiae Infection in Nile Tilapia (Oreochromis niloticus) Treated with Florfenicol. Aquaculture 2018, 493, 51–60. [Google Scholar] [CrossRef]
- Faria, F.C.; Leal, C. a G.; Carvalho-Castro, G. a; Leite, R.C.; Figueiredo, H.C.P. Carrier State Induced by Oxytetracycline Therapy against Streptococcosis in Nile Tilapia, Oreochromis niloticus (L.). J. Fish Dis. 2014, 37, 853–857. [Google Scholar] [CrossRef]

| Genes | Reaction mixture | PCR conditions |
|---|---|---|
|
atr glnA tkt |
Buffer 10X: 2.5 μL 25 mM MgCl2: 1.5 μL 10 mM dNTP: 0.5 μL 10 mM each primer: 0.5 μL Taq DNA Polymerase: 0.25 μL |
Initial step: 95 ℃ for 5 min Cycling: 35 cycles of 95 ℃ for 1 min, 56 ℃ for 45 s, and 72 ℃ for 1 min; Final elongation: 72 ℃ for 15 min |
|
glcK pheS |
Buffer 10X: 2.5 μL 25 mM MgCl2: 1.5 μL 10 mM dNTP: 0.5 μL 10 mM each primer: 0.5 μL Taq DNA Polymerase: 0.25 μL |
Initial step: 95 ℃ for 15 min Cycling: 35 cycles of 95 ℃ for 1 min, 56 ℃ for 1 min, and 72 ℃ for 1 min; Final elongation: 72 ℃ for 10 min |
|
adhP sdhA |
Buffer 10X: 2.5 μL 25 mM MgCl2: 1.0 μL 10 mM dNTP: 0.25 μL 10 mM each primer: 0.2 μL Taq DNA Polymerase: 0.05 μL |
Initial step: 95 ℃ for 5 min Cycling: 35 cycles of 95 ℃ for 1 min, 56 ℃ for 40 s, and 72 ℃ for 1 min; Final elongation: 72 ℃ for 10 min |
| SA32-17 | SA45-17 | |
|---|---|---|
| Genome statistics by QUAST tool | ||
| #contigs (>= 0 bp) | 29 | 23 |
| #contigs (>= 1000 bp) | 28 | 23 |
| Total length (>= 0 bp) | 1,824,566 | 1,817,806 |
| Total length (>= 1000 bp) | 1,823,865 | 1,817,806 |
| #contigs | 29 | 23 |
| Largest contig | 368,081 | 283,920 |
| Total length | 1,824,566 | 1,817,806 |
| G+C (%) | 35.34 | 35.32 |
| N50 | 136,190 | 124,698 |
| N90 | 42,369 | 56,228 |
| auN | 192,272.4 | 144,941.0 |
| L50 | 4 | 5 |
| L90 | 13 | 15 |
| # N’s per 100 kbp | 1.32 | 0.00 |
| Attributes | ||
| Genes (total) | 1,957 | 1,922 |
| Coding sequences (total) | 1,852 | 1,834 |
| Pseudogenes | 29 | 26 |
| rRNA | 11 | 10 |
| tRNA | 64 | 51 |
| tmRNA | 1 | 1 |
| AMR gene detection using PanViTa tool | ||
| arlR (%) | 50.4 | 52.2 |
| RlmA(II) (%) | 53.2 | 53.2 |
| lmrD (%) | 61.2 | 61.2 |
| efrA (%) | 52.8 | 52.6 |
| norB (%) | 59.7 | 59.5 |
| pmrA (%) | 57.5 | 57.3 |
| mreA (%) | 99.7 | 99.4 |
| efrB (%) | 0.0 | 53.3 |
| lmrP (%) | 54.4 | 54.4 |
| Saga_mprF (%) | 99.4 | 99.1 |
| rpoB2 (%) | 52.2 | 52.2 |
| qacJ (%) | 58.9 | 0.0 |
| Antimicrobial agent* | ||||||
|---|---|---|---|---|---|---|
| Strain ID | SXTa | OXYb | FLOb | AMOb | NORa | ERYa |
| SA31 | 20 (NWT) | 28 (WT) | 30 (WT) | 28 (NWT) | 30 (WT) | 25 (WT) |
| SA32 | 19 (NWT) | 28 (WT) | 30 (WT) | 28 (NWT) | 20 (WT) | 17 (NWT) |
| SA33 | 18 (NWT) | 30 (WT) | 32 (WT) | 32 (WT) | 10 (NWT) | 32 (WT) |
| SA45 | 6 (NWT) | 25 (WT) | 24 (WT) | 27 (NWT) | 16 (WT) | 27 (WT) |
| SA100 | 6 (NWT) | 27 (WT) | 25 (WT) | 27 (NWT) | 6 (NWT) | 27 (WT) |
| SA108 | 6 (NWT) | 25 (WT) | 26 (WT) | 31 (WT) | 6 (NWT) | 24 (NWT) |
| SA110 | 6 (NWT) | 23 (WT) | 26 (WT) | 34 (WT) | 6 (NWT) | 27 (WT) |
| SA112 | 6 (NWT) | 25 (WT) | 25 (WT) | 27 (NWT) | 6 (NWT) | 28 (WT) |
| SA174 | 6 (NWT) | 25 (WT) | 24 (WT) | 28 (NWT) | 6 (NWT) | 22 (NWT) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





