Submitted:
17 October 2024
Posted:
17 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
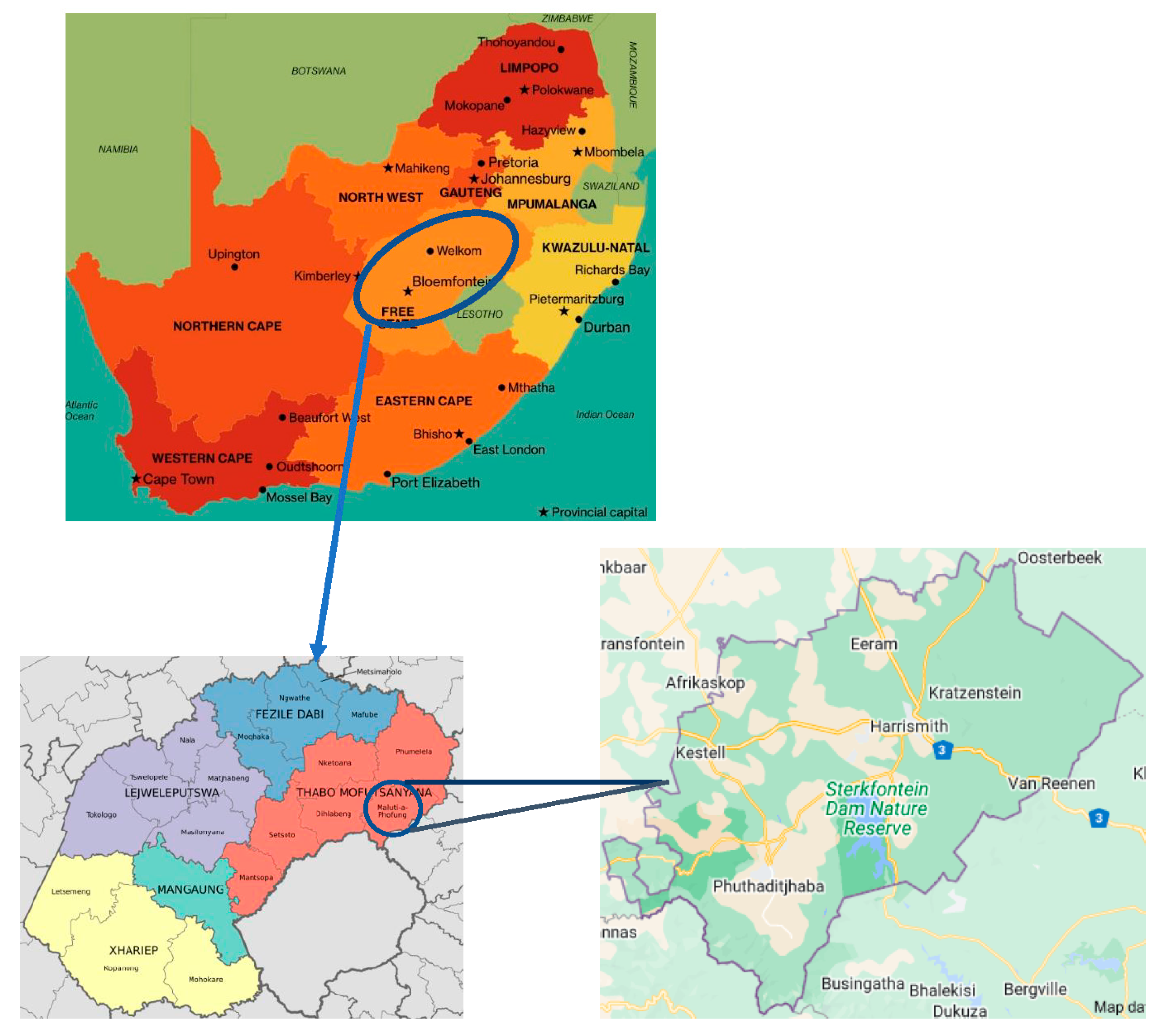
2.1. Soil Sampling
2.2. Soil Microbiology, Physical and Chemical Analysis
2.3. Mapping of Artemisia Afra Distribution and Potential Regions for Cultivation
- GFDL-CM2.0 from the National Oceanic and Atmospheric Administration (NOAA)
- GFDL-CM2.1 of NOAA
- ECHAM5/MPI-Ocean Model from Germany
- UKMO-HadCM3 from the United Kingdom
- MIROC3.2-medres from the Japanese Agency for Marine-Earth Science and Technology (JAMSTEC)
- CSIRO Mark3.5 from Australia
3. Results
3.1. Soil Physical and Chemical Properties
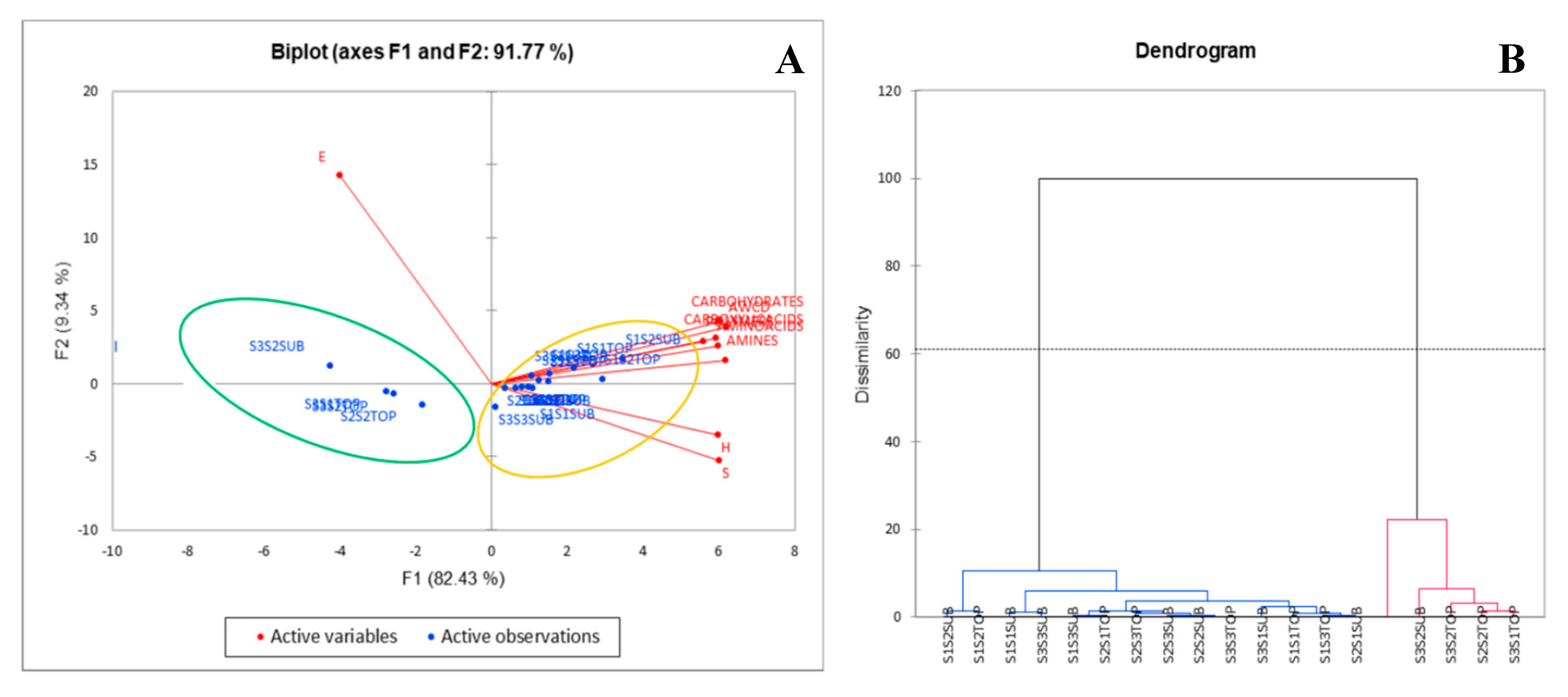
3.2. Soil Microbiology
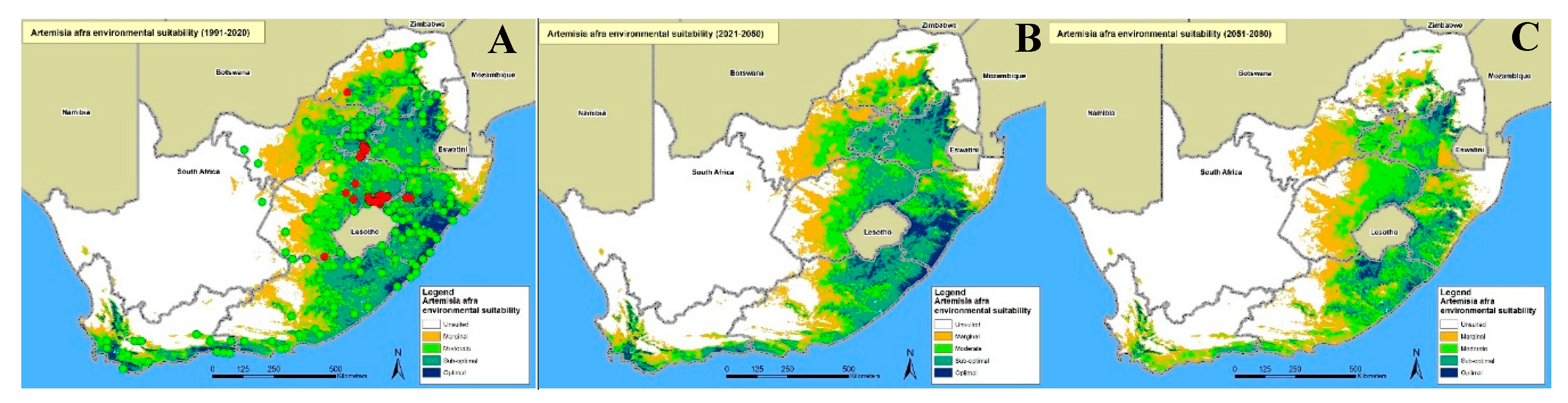
3.3. Mapping of Potentially Suitable Areas for Artemisia Afra Cultivation
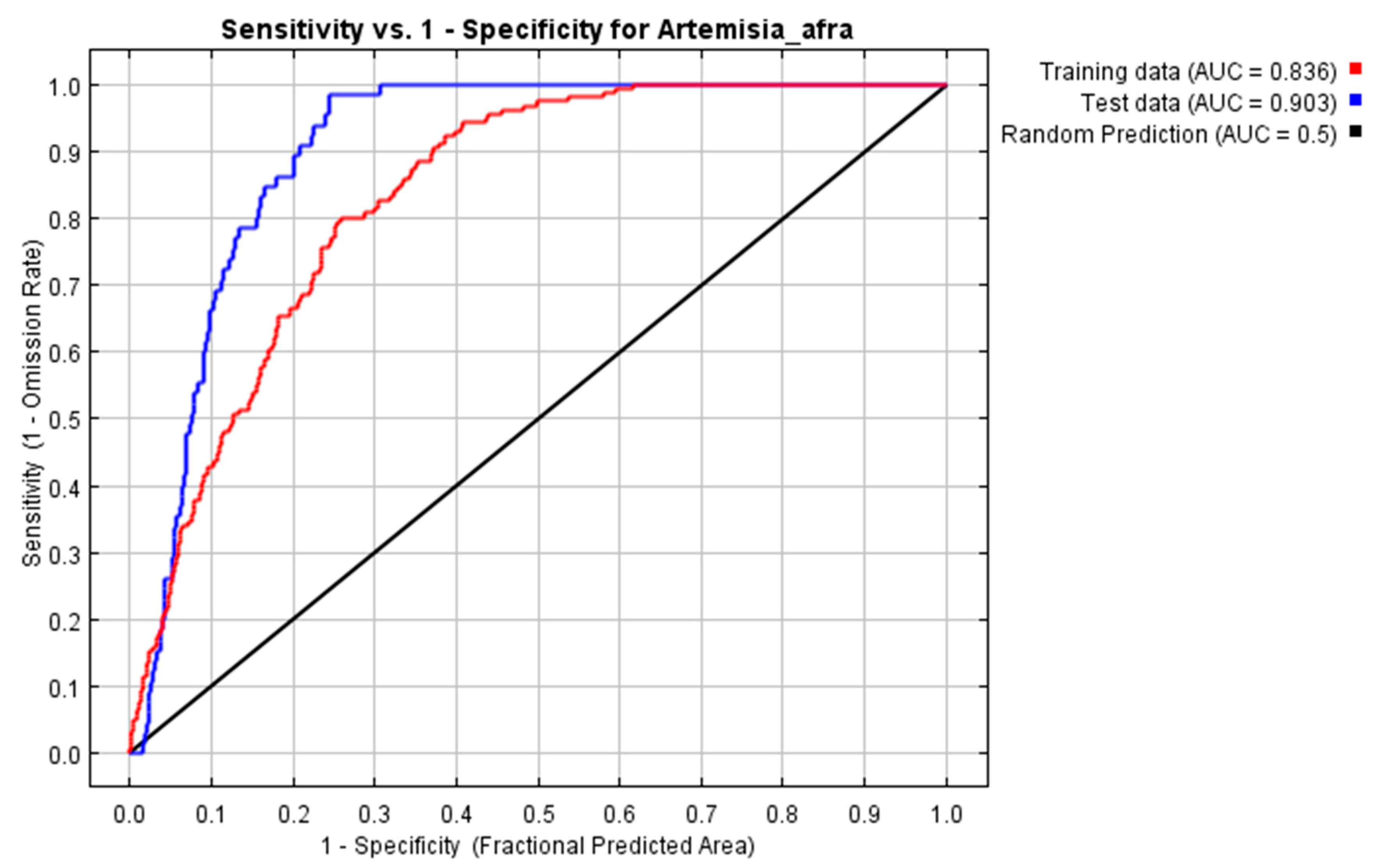
3.3.1. Mapping Using the Maxent Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Martini, C.M. , Zhang, T., Williams, J.T., Abramovitch, R.B., Weathers, P.J., Shell, S.S. Artemisia annua and Artemisia afra extracts exhibit strong bactericidal activity against Mycobacterium tuberculosis. J. Ethnopharmacol. 2020, 262, 113191. [Google Scholar] [CrossRef] [PubMed]
- Daddy, B. , Lutgen, P., Gisenya, P. Breakthrough against tuberculosis: high efficacy Artemisia afra infusions. Pharm. Pharmacol. Int. J. 2021, 9, 58–62. [Google Scholar]
- Nie, C. , Trimpert, J., Moon, S., Haag, R., Gilmore, K., Kaufer, B.B., Seeberger, P.H. In vitro efficacy of Artemisia extracts against SARS-CoV-2. Virol. J. 2021, 18, 182. [Google Scholar] [CrossRef]
- Spies, L. , Koekemoer, T.C., Sowemimo, A.A., Goosen, E.D., Van de Venter, M. Caspase-dependent apoptosis is induced by Artemisia afra Jacq. ex Willd in a mitochondria-dependent manner after G2/M arrest. S. Afr. J. Bot. 2013, 84, 104–109. [Google Scholar]
- Shinyuy, L.M. , Loe, G.E., Jansen, O., Mamede, L., Ledoux, A., Noukimi, S.F., Abenwie, S.N., Ghogomu, S.M., Souopgui, J., Robert, A., Deyemer, K., Frederich, M. Secondary metabolites isolated from Artemisia afra and Artemisia annua and their anti-malarial, anti-inflammatory and immunomodulating properties—pharmacokinetics and pharmacodynamics: a review. Metabolites 2023, 13, 613. [Google Scholar] [CrossRef]
- Van Noorden, R. Demand for malarial drug soars. Nature 2010, 466, 672–673. [Google Scholar]
- Zhang, L. , Cao, B., Bai, C., Li, G. & Mao, M. Predicting suitable cultivation regions of medicinal plants with Maxent modeling and fuzzy logics: a case study of Scutellaria baicalensis in China. Environ. Earth Sci. 2016, 75, 361. [Google Scholar] [CrossRef]
- Bariotakis, M. , Georgescu, L., Laina, D., Oikonomou, J., Ntagounakis, G., Koufaki, M.-I., Souma, M., Choreftakis, M., Zormpa, O.G., Smykal, P., Sourvinos, G., Lionis, C., Castanas, E., Karousou, R., Pirintos, S.A. From wild harvest towards precision agriculture: use of ecological niche modelling to direct potential cultivation of wild medicinal plants in Crete. Sci. Total Environ. 2019, 694, 133681. [Google Scholar] [CrossRef]
- Maharjan, S.K. , Sterck, F.J., Raes, N., Poorter, L. Temperature and soils predict the distribution of plant species along the Himalayan elevation gradient. J. Trop. Ecol. 2022, 38, 58–70. [Google Scholar]
- Rahman, I.U. , Hart, R.E., Ijaz, F., Afzal, A., Iqbal, Z., Calixto, E.S., Abd_Allah, E.F., Alqawari, A.A., Hashem, A., Al-Arjani, A.B.F., Kausar, R., Haq, S.M. Environmental variables drive plant species composition and distribution in the moist temperate forests of Northwestern Himalaya, Pakistan. Plos One 2022, 17, e0260687. [Google Scholar] [CrossRef]
- Chauvier, Y. , Thuiller, W., Brun, P., Lavergne, S., Descombes, P., Karger, D.N., Renaud, J., Zimmermann, N.E. Influence of climate, soil, and land cover on plant species distribution in the European Alps. Ecol. Monogr. 2021, 91, e01433. [Google Scholar]
- Chen, G. , Wang, S., Huang, X., Hong, J., Du, L., Zhang, L., Ye, L. Environmental factors affecting growth and development of Banlangen (Radix isatidis) in China. Afr. J. Plant Sci. 2015, 9, 421–426. [Google Scholar]
- Ncube, B. , Finnie, J.F., Van Staden, J. Quality from the field: the impact of environmental factors as quality determinants in medicinal plants. S. Afr. J. Bot. 2012, 82, 11–20. [Google Scholar]
- Wu, J. , Li, X., Huang, L., Meng, X., Hu, H., Luo, L., Chen, S. A new GIS model for ecologically suitable distributions of medicinal plants. Chin. Med. 2019, 14, 4. [Google Scholar] [CrossRef]
- Tshwene-Mauchaza, B. , Aguirre-Gutiérrez, J. Climatic drivers of plant species distribution across spatial grains in southern Africa tropical forests. Front. For. Glob. Change 2019, 2, 69. [Google Scholar] [CrossRef]
- Phillips, S.J. , Anderson, R.P., Schapire, R.E. Maximum entropy modelling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar]
- Tshabalala, T. , Mutanga, O., Abdel-Rahman, E.M. Predicting the geographical distribution shift of medicinal plants in South Africa due to climate change. Conservation 2022, 2, 694–708. [Google Scholar] [CrossRef]
- Zhao, Y. , Zhao, M., Zhang, L., Wang, C., Xu, Y. Predicting possible distribution of tea (Camellia sinensis L.) under climate change scenarios using MaxEnt Model in China. Agriculture 2021, 11, 1122. [Google Scholar] [CrossRef]
- Sullivan, T.S., Barth, V., Lewis, R.W. Soil acidity impacts beneficial soil microorganisms. Soil acidification series, Washington State University 2017, FS247E, www.ext.wsu.edu.
- Ntalo, M. , Ravhuhali, K.E., Moyo, B., Mmbi, N.E., Mokoboki, K.H. Physical and chemical properties of the soils in selected communal property associations of South Africa. PeerJ 2022, 10, e13960. [Google Scholar] [CrossRef]
- Mofokeng, M.M. , Habig, J., Amoo, S.O., du Plooy, C.P., Mashela, P.W., Moeletsi, M.E., Venter, S., Araya, H. Differences in soil microbial communities and enzyme activity due to the application of bioslurry under cultivation. S. Afr. J. Plant Soil 2020, 37, 283–291. [Google Scholar] [CrossRef]
- Engelbrecht, F.A. , Bopape, M. J. High-resolution projected climate futures for southern Africa. In proceedings of the 27th Annual Conference of the South African Society for Atmospheric Sciences, Hartebeeshoek, South Africa, September 2011, ISBN 978-0-620-47333-0.
- Yan, H. , He, J., Xu, X., Yao, X., Wang, G., Tang, L., Feng, L., Zou, L., Gu, X., Qu, Y., Qu, L. Prediction of potentially suitable distributions of Codonopsis pilosula in China based on an optimized MaxEnt Model. Front. Ecol. Evol. 2021, 9, 773396. [Google Scholar] [CrossRef]
- Viljoen, A.M. , Van Vuuren, S.F., Gwebu, T., Demirci, B., Bașer, K.H.C. The geographical variation and antimicrobial activity of African wormwood (Artemisia afra Jacq.) essential oil. J. Essent. Oil Res. 2006, 18, 19–25. [Google Scholar] [CrossRef]
- Kane, N.F. , Kyama, M.C., Nganga, J.K., Hassanali, A., Diallo, M., Kimani, F.T. Comparison of phytochemical profiles and antimalarial activities of Artemisia afra plant collected from five countries in Africa. S. Afr. J. Bot. 2019, 125, 126–133. [Google Scholar]
- Oyedeji, A.O. , Afolayan, A.J., Hutchings, A. Compositional variation of the essential oils of Artemisia afra Jacq. from three provinces in South Africa – a case study of its safety. Nat. Prod. Commun. 2009, 4, 849–852. [Google Scholar] [PubMed]
- Du Preez, C.C. , Van Huyssteen, C.W., Mnkeni, P.N.S. Land use and soil organic matter in South Africa 1: a review on spatial variability and the influence of rangeland stock production. S. Afr. J. Sci. 2011, 107, Art. #354. [Google Scholar]
- Zhou, W. , Han, G., Liu, M., Li, X. Effects of soil pH and texture on soil carbon and nitrogen in soil profiles under different land uses in Mun River Basin, Northeast Thailand. PeerJ 2019, 7, e7880. [Google Scholar] [CrossRef]
- Omer, E.A. , Abou Hussein, E.A, Hendawy, S.F., El-din, E., Azza, A., El-Gendy, A.G. Effect of soil type and seasonal variation on growth, yield, essential oil and artemisinin content of Artemisia annua L. Int. Res. J. Hortic. 2013, 1, 15–27. [Google Scholar] [CrossRef]
- Neina, D. The role of soil pH in plant nutrition and soil remediation. Appl. Environ. Soil Sci. 2019, Article ID 5794869. [CrossRef]
- Hansen-Quartey, J.H. Soil properties as influenced by cultivation of the aromatic shrub Artemisia afra. S. Afr. J. Plant Soil 1998, 15, 14–18. [Google Scholar]
- Sukitprapanon, T.-S., Jantamenchai, M., Tulaphikat, D., Vityakon, P. Nutrient composition of diverse organic residues and their long-term effects on available nutrients in a tropical sandy soil. Heliyon 2020, 6, e05601. [CrossRef]
- Koehorst, R. , Laubscher, C.P., Ndakidemi, P.A. Growth response of Artemisia afra Jacq. to different pH levels in a closed hydroponics system. J. Med. Plants Res. 2010, 4, 1617–1623. [Google Scholar]
- Sinsabaugh, R.L. , Lauber, C., Weintraub, M.N., Ahmed, B., Allison, S.D., Crenshaw, C., Contosta, A.R., Cusack, D., Frey, S., Gallo, M.E., Gartner, T.B., Hobbie, S.E., Holland, K., Keeler, B.L., Powers, J.S., Stursova, M., Takacs-Vesbach, C., Waldrop, M.P., Wallenstein, M.D., Zak, D.R., Zeglin, L.H. Stoichiometry of soil enzyme activity at global scale. Ecol. Lett. 2008, 11, 1252–1264. [Google Scholar]
- Turner, B.L. Variation in pH optima of hydrolytic enzyme activities in tropical rain forest soils. Appl. Environ. Microbiol. 2010, 76, 6485–6493. [Google Scholar] [PubMed]
- Fierer, N. , Jackson, R.B. The diversity and biogeography of soil bacterial communities. PNAS 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Hartman, W.H. , Richardson, C.J., Vilgalys, R., Bruland, G.L. Environmental and anthropogenic controls over bacterial communities in wetland soils. PNAS 2008, 105, 17842–17847. [Google Scholar] [CrossRef]
- Will, C. , Thürmer, A., Wollherr, A., Nacke, H., Herold, N., Schrumpf, M., Gutknecht, J., Wubet, T., Buscot, F., Daniel, R. Horizon-specific bacterial community composition of German grassland soils, as revealed by pyrosequencing-based analysis of 16S rRNA genes. Appl. Environ. Microbiol. 2010, 76, 6751–6759. [Google Scholar] [CrossRef]
- Rousk, J. , Bååth, E., Brookes, P.C., Lauber, C.L., Lozupone, C., Caporaso, J.G., Knight, R., Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. The ISME J. 2010, 4, 1340–1351. [Google Scholar]
- Conradie, D.C.U. South Africa’s climatic zones: today, tomorrow. In proceeding of the International Green Building Conference and Exhibition, Sandton, South Africa, July 25-26 2012. [Google Scholar]
- Cole, M.J. , Bailey, R.M., Cullis, J.D.S., New, M.G. Spatial inequality in water access and water use in South Africa. Water Policy 2018, 20, 37–52. [Google Scholar] [CrossRef]
| Sand (%) | Silt (%) | Clay (%) | |||||
|---|---|---|---|---|---|---|---|
| Sample ID | Coarse 2-0.5 |
Medium 0.5-0.25 |
Fine 0.25-0.106 |
Very Fine 0.106-0.05 |
Coarse 0.05-0.02 |
Fine 0.02-0.002 |
<0.002 |
| mm | |||||||
| Pp1 SP1 top | 3.1 | 5.0 | 20.1 | 12.1 | 12.8 | 14.6 | 31.3 |
| Pp1 SP1 sub | 0.6 | 2.9 | 19.0 | 11.9 | 11.4 | 14.9 | 38.1 |
| Pp1 SP2 top | 16.1 | 12.6 | 22.7 | 15.1 | 9.4 | 10.0 | 13.2 |
| Pp1 SP2 sub | 22.3 | 9.6 | 20.2 | 14.6 | 12.2 | 9.4 | 10.7 |
| Pp1 SP3 top | 6.3 | 5.6 | 22.6 | 11.2 | 11.0 | 18.8 | 23.7 |
| Pp1 SP3 sub | 6.4 | 3.2 | 17.1 | 10.3 | 10.2 | 20.8 | 30.7 |
| Pp2 SP1 top | 11.6 | 5.3 | 11.9 | 12.7 | 13.5 | 14.9 | 28.6 |
| Pp2 SP1 sub | 2.7 | 7.7 | 20.2 | 20.6 | 12.8 | 12.6 | 22.1 |
| Pp2 SP2 top | 8.0 | 8.2 | 13.8 | 15.3 | 12.1 | 14.3 | 27.3 |
| Pp2 SP2 sub | 5.6 | 6.6 | 12.6 | 16.0 | 13.1 | 16.4 | 28.4 |
| Pp2 SP3 top | 22.0 | 10.2 | 17.7 | 13.4 | 12.3 | 9.3 | 13.9 |
| Pp2 SP3 sub | 12.2 | 7.9 | 14.8 | 15.0 | 11.1 | 13.4 | 24.4 |
| Pp3 SP1 top | 3.6 | 4.1 | 18.4 | 13.4 | 10.1 | 17.2 | 32.0 |
| Pp3 SP1 sub | 3.9 | 3.0 | 17.7 | 12.8 | 11.2 | 19.9 | 30.5 |
| Pp3 SP2 top | 4.6 | 6.1 | 21.2 | 14.5 | 11.6 | 14.4 | 26.6 |
| Pp3 SP2 sub | 4.3 | 6.4 | 22.9 | 14.3 | 10.4 | 14.1 | 26.4 |
| Pp3 SP3 top | 4.2 | 6.0 | 18.7 | 12.9 | 12.1 | 17.7 | 27.6 |
| Pp3 SP3 sub | 4.6 | 6.6 | 19.0 | 13.1 | 10.1 | 15.2 | 30.3 |
| Sample ID | Total P | Total N | N-NO3 | N-NH4 | P | K | Na | Ca | Mg | CEC | pH | OM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ppm | mg/kg | cmol(+)/kg | H2O | % | ||||||||
| Pp1 SP1 top | 4.67 | 0.131 | 0.70 | 6.67 | 1.79 | 277 | 15.6 | 2300 | 532 | 23.19 | 6.66 | 0.050 |
| Pp1 SP1 sub | 4.44 | 0.120 | 0.71 | 5.07 | 0.42 | 210 | 44.4 | 2300 | 651 | 33.04 | 5.95 | 0.074 |
| Pp1 SP2 top | 5.06 | 0.120 | 1.03 | 4.60 | 1.63 | 223 | 15.3 | 1720 | 502 | 25.35 | 5.3 | 1.264 |
| Pp1 SP2 sub | 5.46 | 0.113 | 1.67 | 4.84 | 2.06 | 265 | 23.9 | 1560 | 428 | 21.02 | 5.91 | 0.098 |
| Pp1 SP3 top | 5.08 | 0.162 | 0.59 | 3.75 | 4.02 | 458 | 7.92 | 2130 | 525 | 21.89 | 6.72 | 0.068 |
| Pp1 SP3 sub | 3.79 | 0.096 | 0.83 | 3.36 | 1.28 | 263 | 14.6 | 2560 | 700 | 26.42 | 6.29 | 0.049 |
| Pp2 SP1 top | 4.32 | 0.067 | 0.63 | 2.19 | 0.38 | 390 | 14.6 | 3340 | 1240 | 36.79 | 6.48 | 0.059 |
| Pp2 SP1 sub | 3.74 | 0.067 | 1.18 | 6.91 | 0.06 | 140 | 19.3 | 2700 | 988 | 35.52 | 6.70 | 0.048 |
| Pp2 SP2 top | 4.13 | 0.089 | 0.64 | 3.47 | 1.36 | 317 | 18.7 | 3060 | 1080 | 33.61 | 6.22 | 0.060 |
| Pp2 SP2 sub | 3.94 | 0.071 | 0.73 | 5.11 | 0.70 | 251 | 22.2 | 3470 | 1110 | 34.86 | 6.59 | 0.057 |
| Pp2 SP3 top | 5.43 | 0.090 | 1.60 | 8.25 | 1.29 | 196 | 44.9 | 2090 | 653 | 23.14 | 6.66 | 0.841 |
| Pp2 SP3 sub | 3.92 | 0.067 | 0.84 | 5.20 | 0.13 | 147 | 25.4 | 2900 | 990 | 33.11 | 6.86 | 0.055 |
| Pp3 SP1 top | 4.32 | 0.152 | 1.33 | 3.43 | 0.67 | 409 | 8.16 | 1300 | 648 | 23.35 | 6.15 | 0.062 |
| Pp3 SP1 sub | 4.33 | 0.151 | 1.67 | 3.27 | 0.44 | 334 | 10.0 | 1310 | 660 | 17.99 | 6.04 | 0.058 |
| Pp3 SP2 top | 4.32 | 0.115 | 1.71 | 1.60 | 0.79 | 284 | 20.9 | 1220 | 586 | 21.70 | 6.50 | 0.047 |
| Pp3 SP2 sub | 4.02 | 0.096 | 1.47 | 2.61 | 0.49 | 190 | 22.4 | 982 | 529 | 20.39 | 6.24 | 0.042 |
| Pp3 SP3 top | 4.31 | 0.155 | 2.25 | 5.40 | 1.09 | 405 | 7.8 | 1320 | 638 | 20.54 | 6.21 | 0.065 |
| Pp3 SP3 sub | 3.96 | 0.121 | 0.74 | 4.94 | 0.26 | 189 | 12.3 | 1320 | 747 | 26.61 | 6.06 | 0.049 |
| Sample ID | Carbon Source Utilization (Functional diversity) | Enzyme Activities | ||||
|---|---|---|---|---|---|---|
| zAWCD | Shannon Weaver Index (H’) | Richness (S) | Evenness | β-glucosidase | Phosphatase | |
| Pp1 SP1 top | 1.547abc | 3.36a | 29.33ab | 0.994abc | 19.51 | 92.13 |
| Pp1 SP1 sub | 1.316cde | 3.33abc | 29.66ab | 0.982c | 16.79 | 99.56 |
| Pp1 SP2 top | 1.614ab | 3.38a | 30.66a | 0.988bc | 36.58 | 93.46 |
| Pp1 SP2 sub | 1.753a | 3.36a | 30.33a | 0.995abc | 30.35 | 99.86 |
| Pp1 SP3 top | 1.453bcd | 3.34abc | 29.00ab | 0.993abc | 25.76 | 93.15 |
| Pp1 SP3 sub | 1.279de | 3.35ab | 29.66ab | 0.991abc | 14.59 | 91.11 |
| Pp2 SP1 top | 1.296de | 3.35ab | 29.66ab | 0.991abc | 19.99 | 106.20 |
| Pp2 SP1 sub | 1.385b-e | 3.34abc | 29.00ab | 0.991abc | 13.44 | 102.81 |
| Pp2 SP2 top | 0.941g | 3.26cd | 27.33abc | 0.988bc | 17.05 | 98.19 |
| Pp2 SP2 sub | 1.355cde | 3.34abc | 29.33ab | 0.988bc | 25.31 | 96.94 |
| Pp2 SP3 top | 1.444bcd | 3.33abc | 29.00ab | 0.984abc | 25.54 | 99.73 |
| Pp2 SP3 sub | 1.284de | 3.29bcd | 28.00ab | 0.988abc | 11.97 | 87.55 |
| Pp3 SP1 top | 0.841g | 3.25d | 26.00bc | 0.996abc | 21.70 | 101.82 |
| Pp3 SP1 sub | 1.425bcd | 3.28bcd | 27.33abc | 0.992abc | 18.33 | 92.16 |
| Pp3 SP2 top | 0.995fg | 3.05e | 23.66c | 0.989abc | 18.53 | 90.31 |
| Pp3 SP2 sub | 0.847g | 3.02e | 19.33d | 1.004ab | 14.84 | 88.24 |
| Pp3 SP3 top | 1.359cde | 3.33abc | 29.00ab | 0.988abc | 15.28 | 90.04 |
| Pp3 SP3 sub | 1.186ef | 3.28bcd | 28.33ab | 0.982c | 17.36 | 95.81 |
| LSD0.05 | 0.236 | 0.081 | 3.69 | 0.019 | ||
| Sample ID | Amino acids | Carbohydrates | Carboxylic acids | Polymers | Amines | Avg. No. of colonies | Colony forming units (CFU g-1) |
|---|---|---|---|---|---|---|---|
| Pp1 SP1 top | 1.628abc | 1.579a-d | 1.381abc | 1.689ab | 1.609ab | 17(10)6 | 1.7 x 108 |
| Pp1 SP1 sub | 1.502bcd | 1.299cde | 1.189c-f | 1.313b-e | 1.417a-c | 5(10)6 | 5.0 x 108 |
| Pp1 SP2 top | 1.778ab | 1.626ab | 1.521ab | 1.547a-d | 1.619a | 95(10)6 | 9.5 x 106 |
| Pp1 SP2 sub | 1.912a | 1.766a | 1.626a | 1.858a | 1.586ab | 19(10)6 | 1.9 x 108 |
| Pp1 SP3 top | 1.576a-d | 1.434def | 1.338a-d | 1.604ab | 1.402a-c | 13(10)6 | 1.3 x 108 |
| Pp1 SP3 sub | 1.445bcd | 1.249ef | 1.219b-e | 1.230b-e | 1.276a-c | 27(10)6 | 2.7 x 108 |
| Pp2 SP1 top | 1.385cd | 1.265def | 1.171c-f | 1.527a-d | 1.294a-c | 55(10)4 | 5.5 x 106 |
| Pp2 SP1 sub | 1.647abc | 1.347b-f | 1.176c-f | 1.566a-c | 1.372a-c | 300(10)7 | 3 x 1010 |
| Pp2 SP2 top | 0.957ef | 0.888gh | 0.906fg | 1.211b-e | 0.768fg | 13(10)6 | 1.3 x 108 |
| Pp2 SP2 sub | 1.542bcd | 1.461a-e | 1.063d-f | 1.526a-d | 1.240a-e | 39(10)7 | 3.9 x 109 |
| Pp2 SP3 top | 1.487bcd | 1.580a-d | 1.134c-f | 1.835a | 1.259a-d | 13(10)6 | 1.3 x 108 |
| Pp2 SP3 sub | 1.439bcd | 1.336b-f | 1.079c-f | 1.503a-d | 1.045c-f | 12(10)6 | 1.2 x 108 |
| Pp3 SP1 top | 0.981ef | 0.769hi | 0.721g | 1.093c-e | 0.805efg | 45(10)4 | 4.5 x 106 |
| Pp3 SP1 sub | 1.708abc | 1.616abc | 1.159c-f | 1.508de | 1.542ab | 300(10)7 | 3.0 x 1010 |
| Pp3 SP2 top | 1.273de | 0.740hi | 0.975e-g | 0.859ef | 0.823d-g | 33(10)4 | 3.3 x 106 |
| Pp3 SP2 sub | 1.032e | 0.698hi | 0.948e-g | 0.840ef | 1.542ab | 24(10)4 | 2.4 x 106 |
| Pp3 SP3 top | 1.481bc | 1.444a-e | 1.161c-f | 1.498a-d | 1.191a-f | 14(10)7 | 1.4 x 109 |
| Pp3 SP3 sub | 1.560bcd | 1.110fg | 1.128c-f | 0.961e | 1.161b-f | 10(10)4 | 1.0 x 106 |
| LSD0.05 | 0.339 | 0.342 | 0.308 | 0.500 | 0.451 |
| Environmental variable | Percentage contribution | Permutation importance |
|---|---|---|
| Long term average annual rainfall | 62.0% | 41.0% |
| Highest long-term average monthly maximum temperature | 21.5% | 26.1% |
| Average soil depth | 7.5% | 11.1% |
| Lowest long-term average monthly maximum temperature | 3.6% | 5.3% |
| Long-term average monthly minimum relative humidity | 2.0% | 4.2% |
| Lowest long-term average monthly minimum temperature | 1.1% | 1.9% |
| Highest long-term average monthly minimum temperature | 1.0% | 6.7% |
| Average soil clay% | 1.0% | 2.8% |
| Long-term average monthly maximum relative humidity | 0.4% | 0.7% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





