Submitted:
16 October 2024
Posted:
17 October 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Materials & Methods
Plant Materials and Experiment Design
Measurement of Phenotypic Parameters
RNA Extraction and Sequencing
Transcriptome Data Analysis
Gene Functional Annotation Analyses
Data Processing, Visualization, and Statistical Analysis
Results
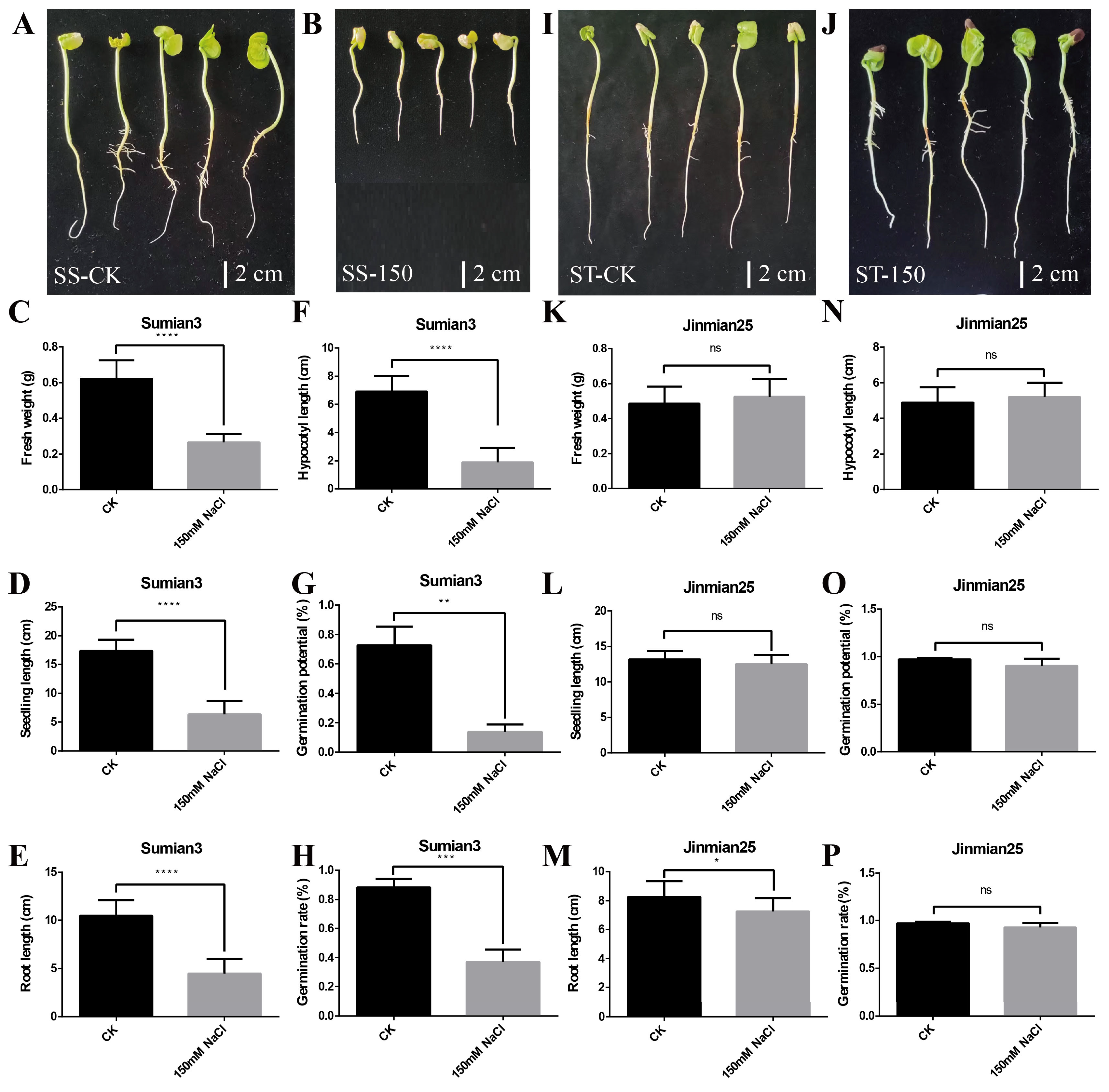
Salt Sensitivity Assessment of Su-Mian 3 and Jin-Mian 25 Cotton Cultivars
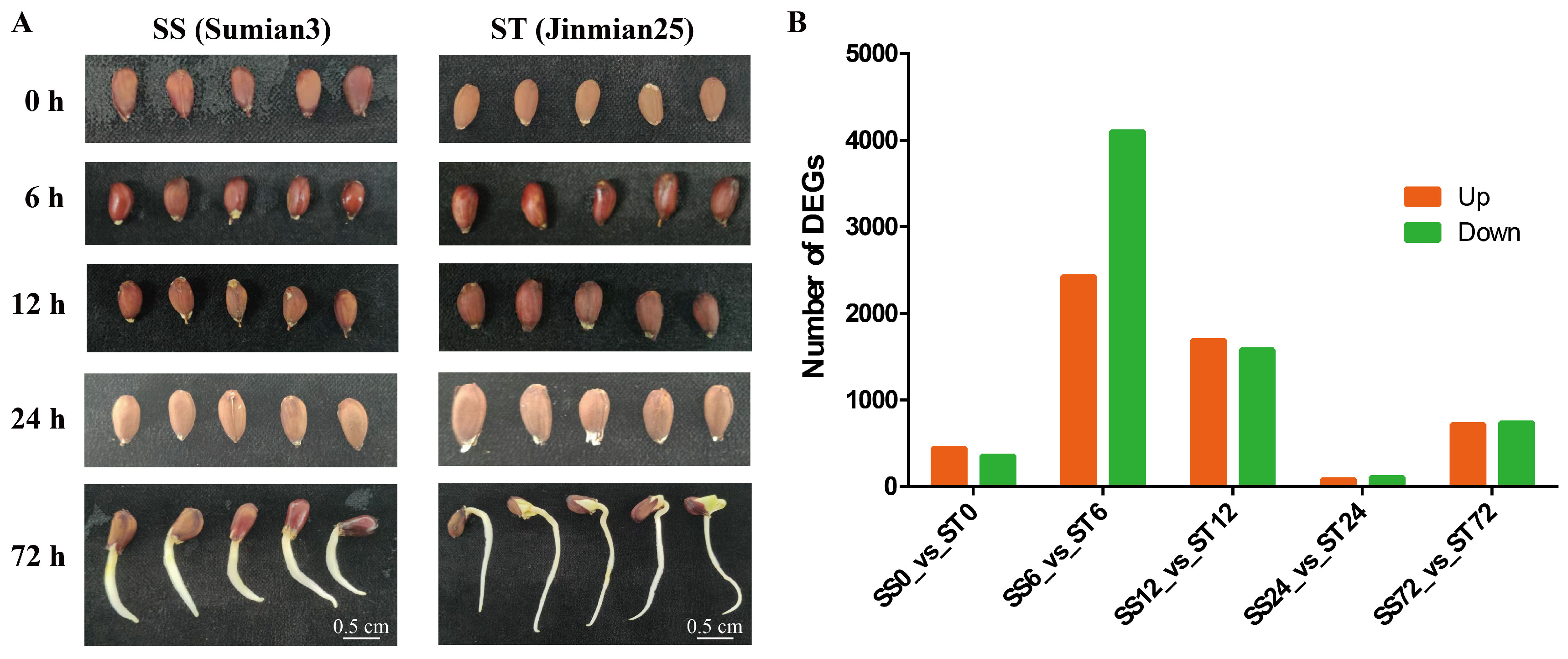
Transcriptome Profiles of Su-Mian 3 and Jin-Mian 25 Cotton Plants Exposed to Salt Stress
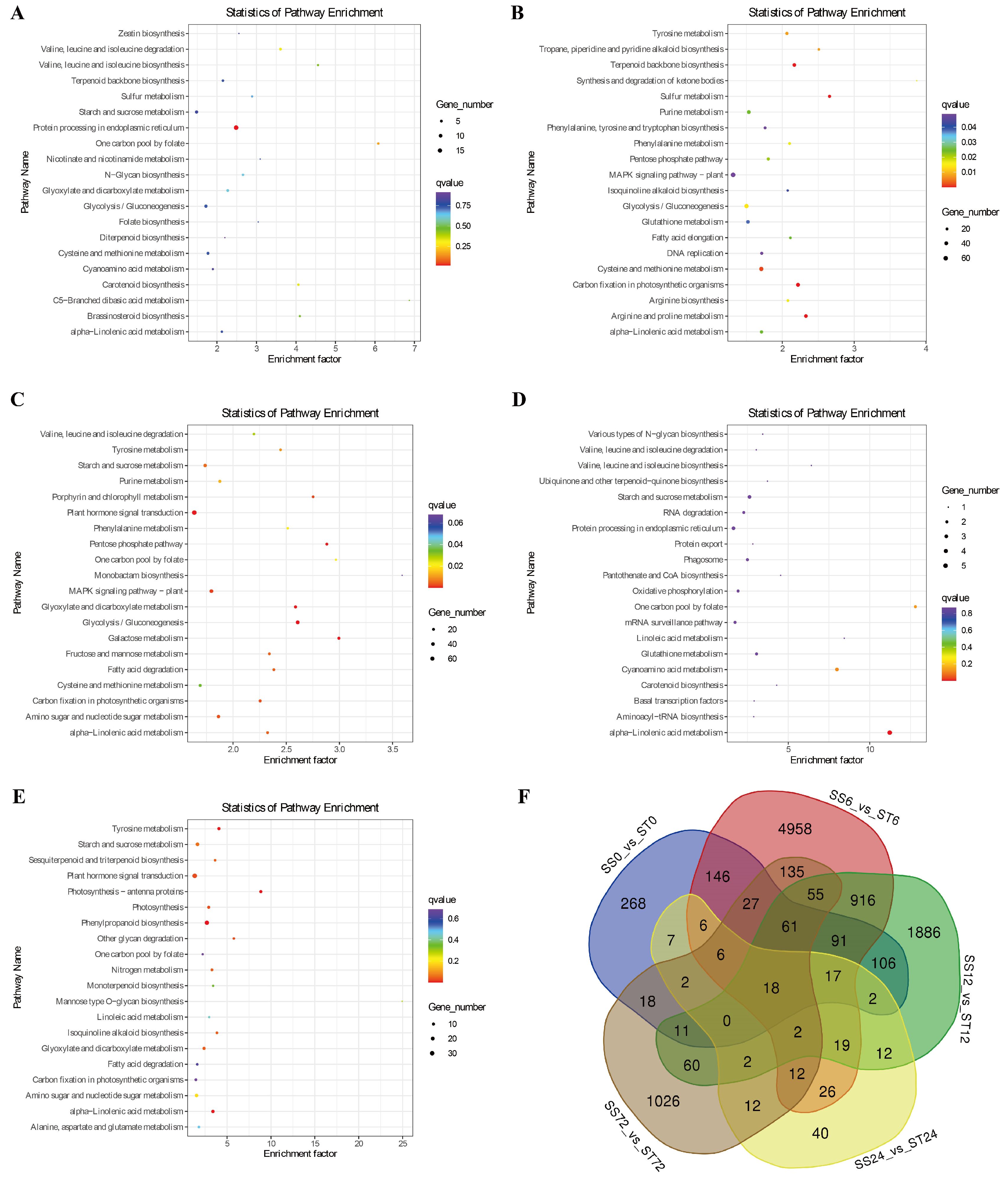
Pathway Enrichment of DEGs
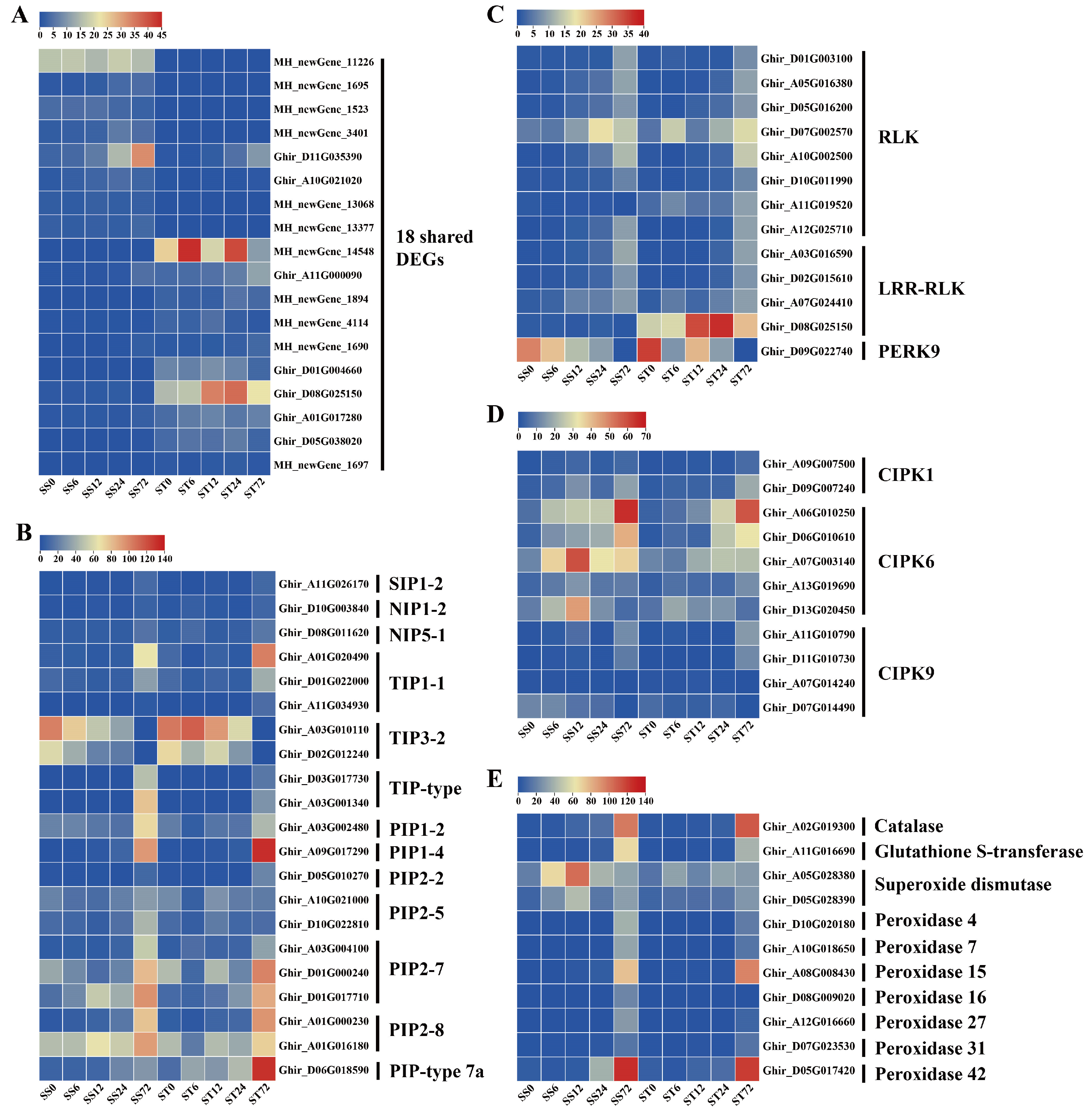
Expression Patterns of Genes Related to Aquaporins, Kinases, and ROS Scavenging during Salt Stress in Both Genotypes
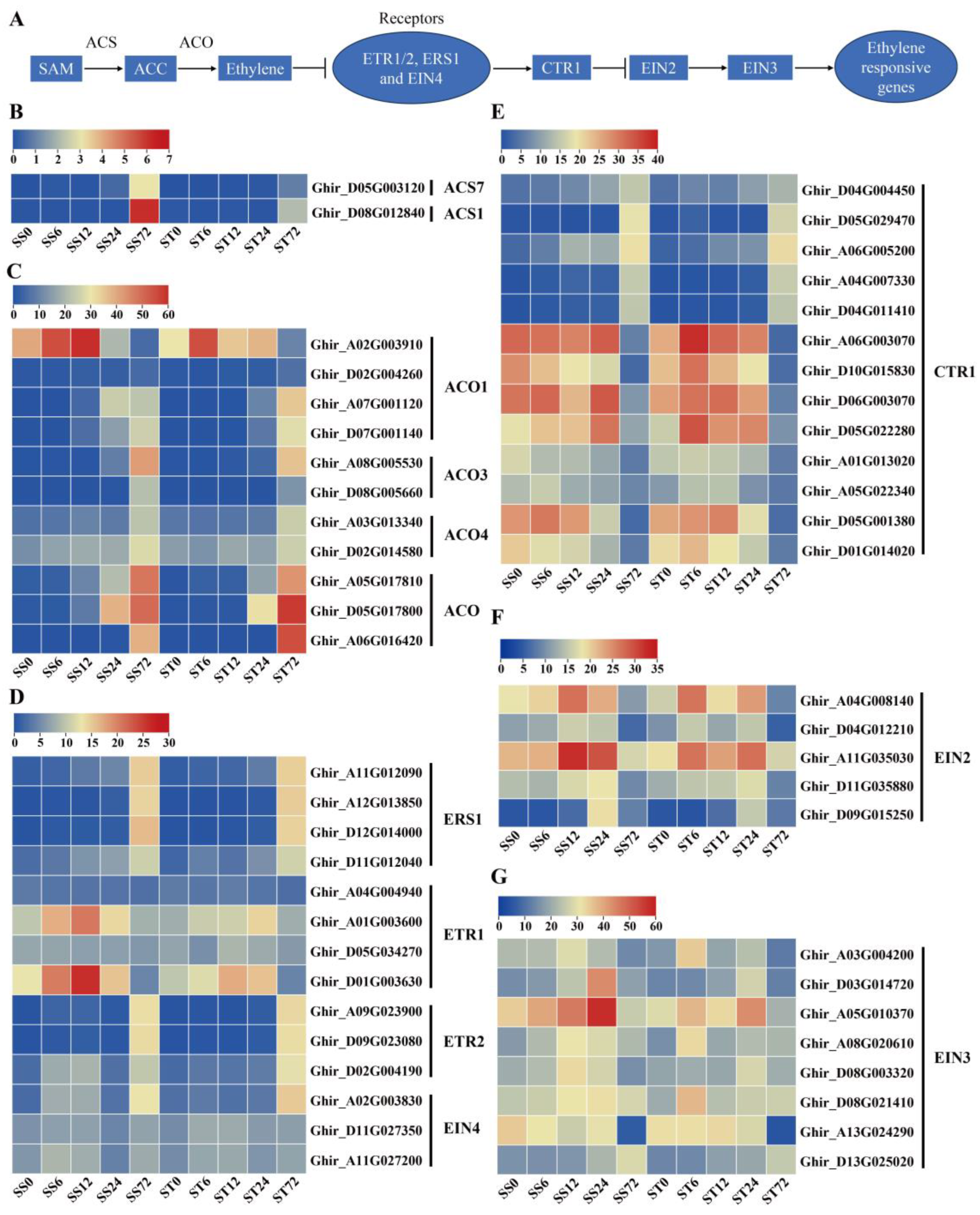
Expression Patterns of Genes Related to Ethylene Biosynthesis and Signaling during Salt Stress in Both Genotypes
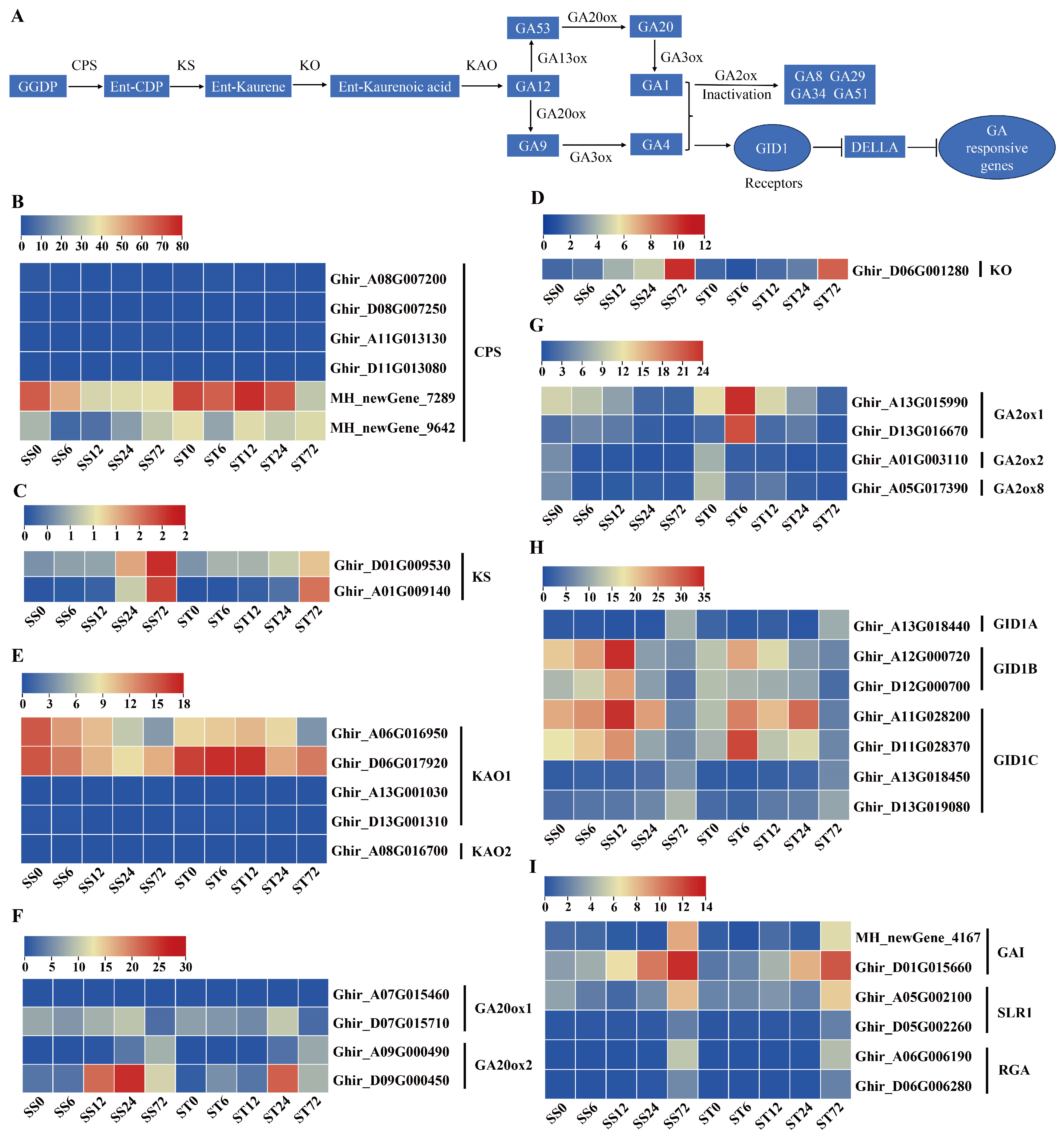
Expression Patterns of Genes Related to Gibberellin (GA) Biosynthesis and Signaling during Salt Stress in Both Genotypes
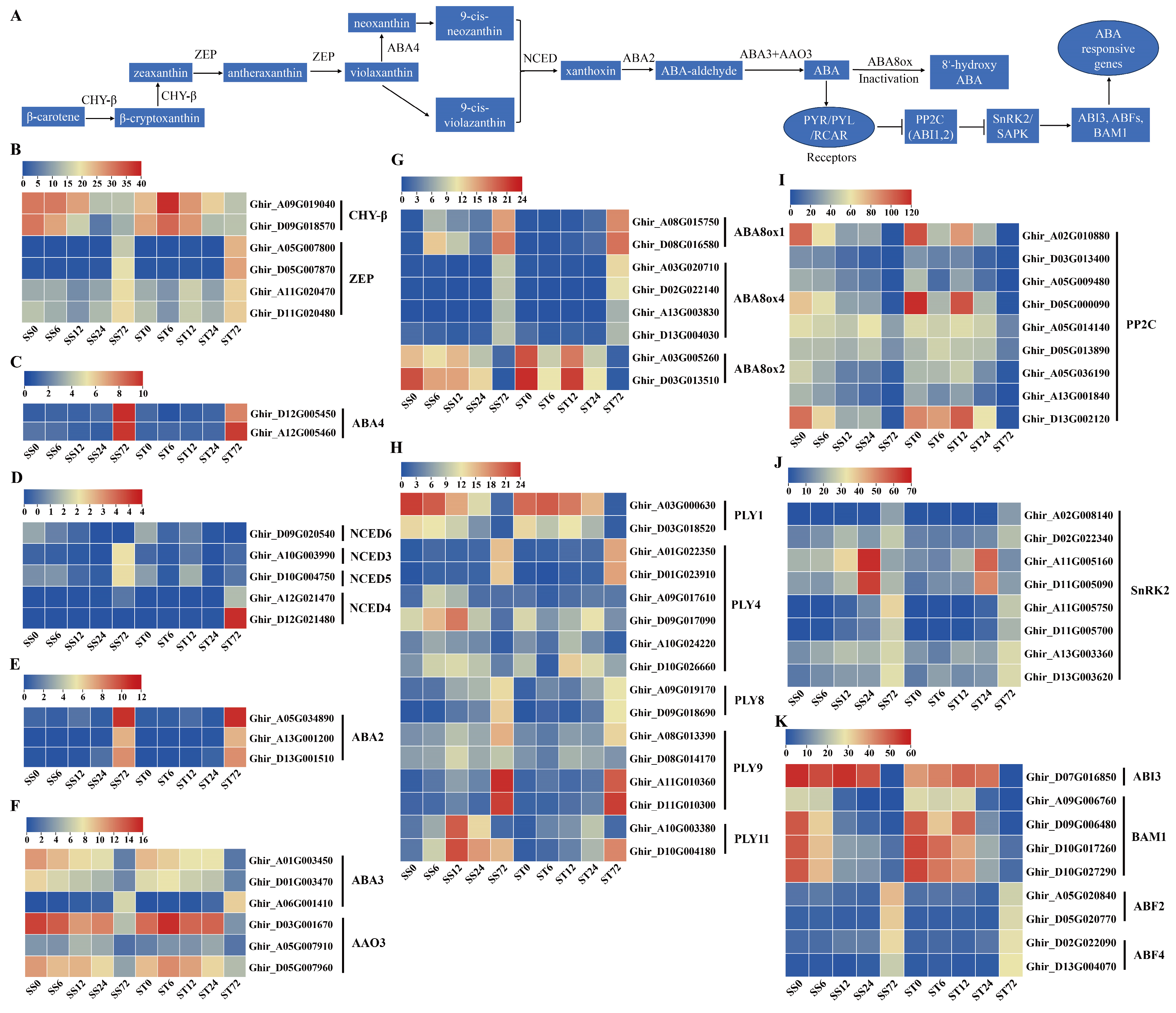
Expression Patterns of Genes Related to Abscisic Acid (ABA) Biosynthesis and Signaling during Salt Stress in Both Genotypes
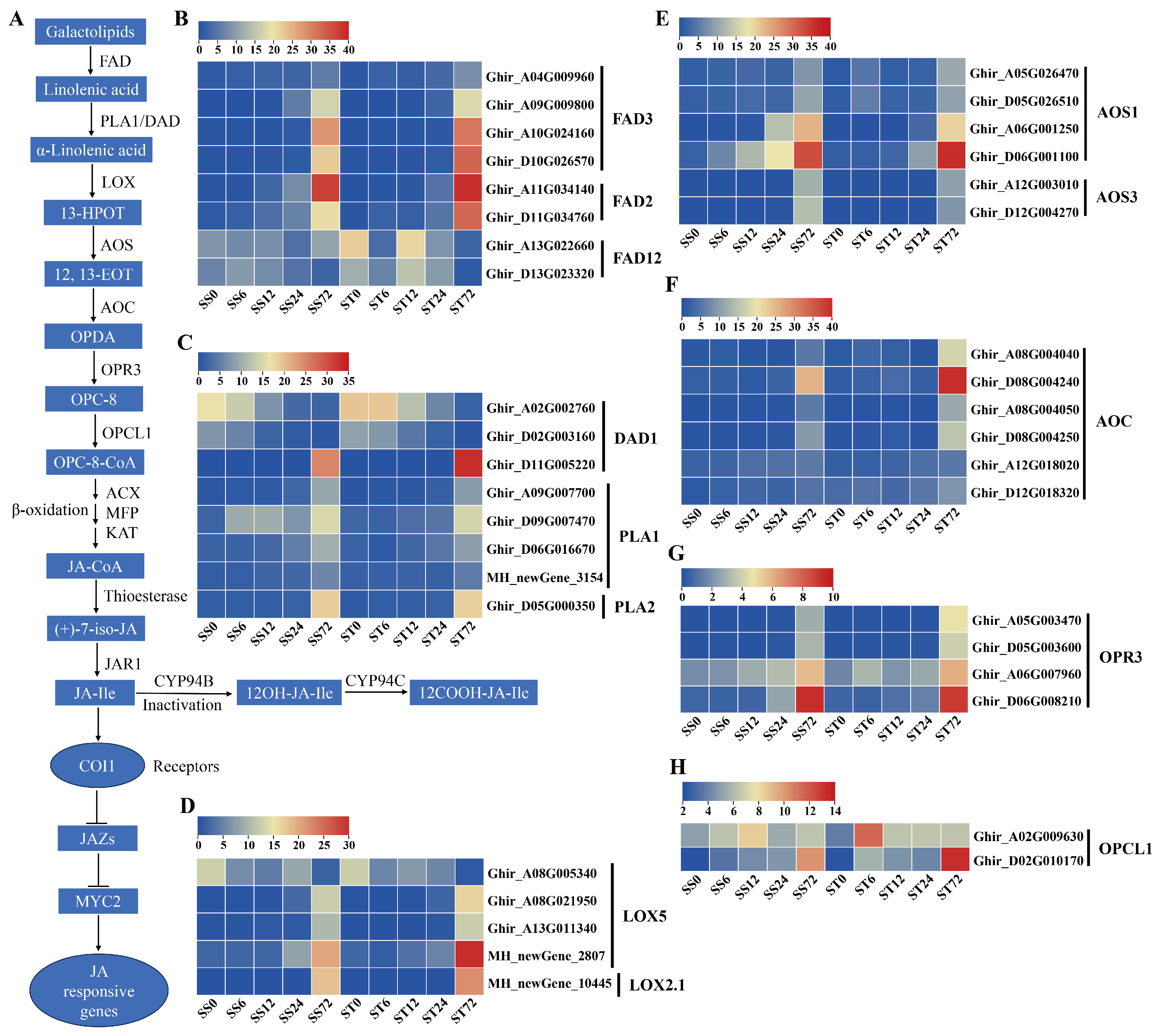
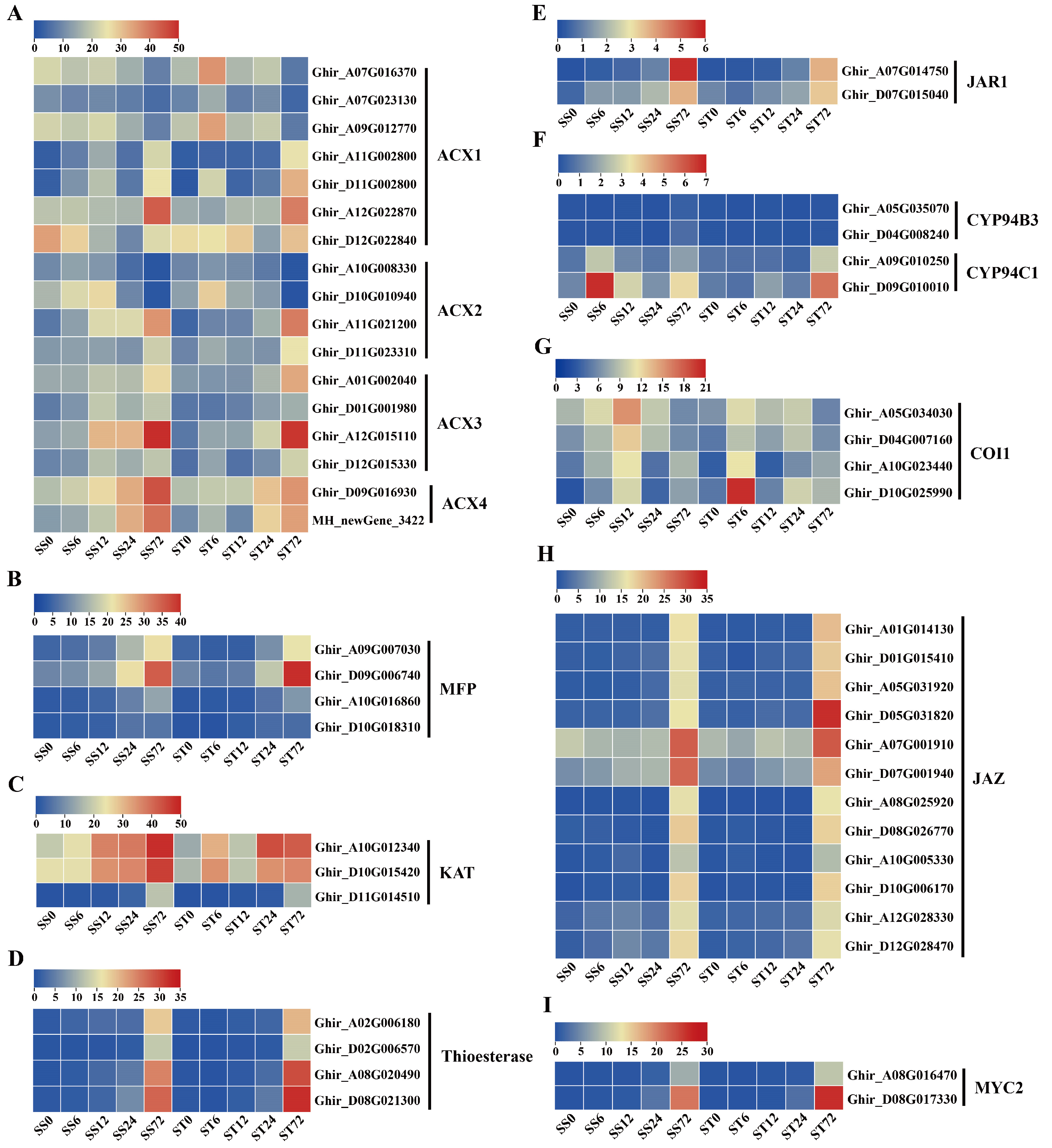
Expression Patterns of Genes Related to Jasmonic Acid (JA) Biosynthesis and Signaling during Salt Stress in Both Genotypes
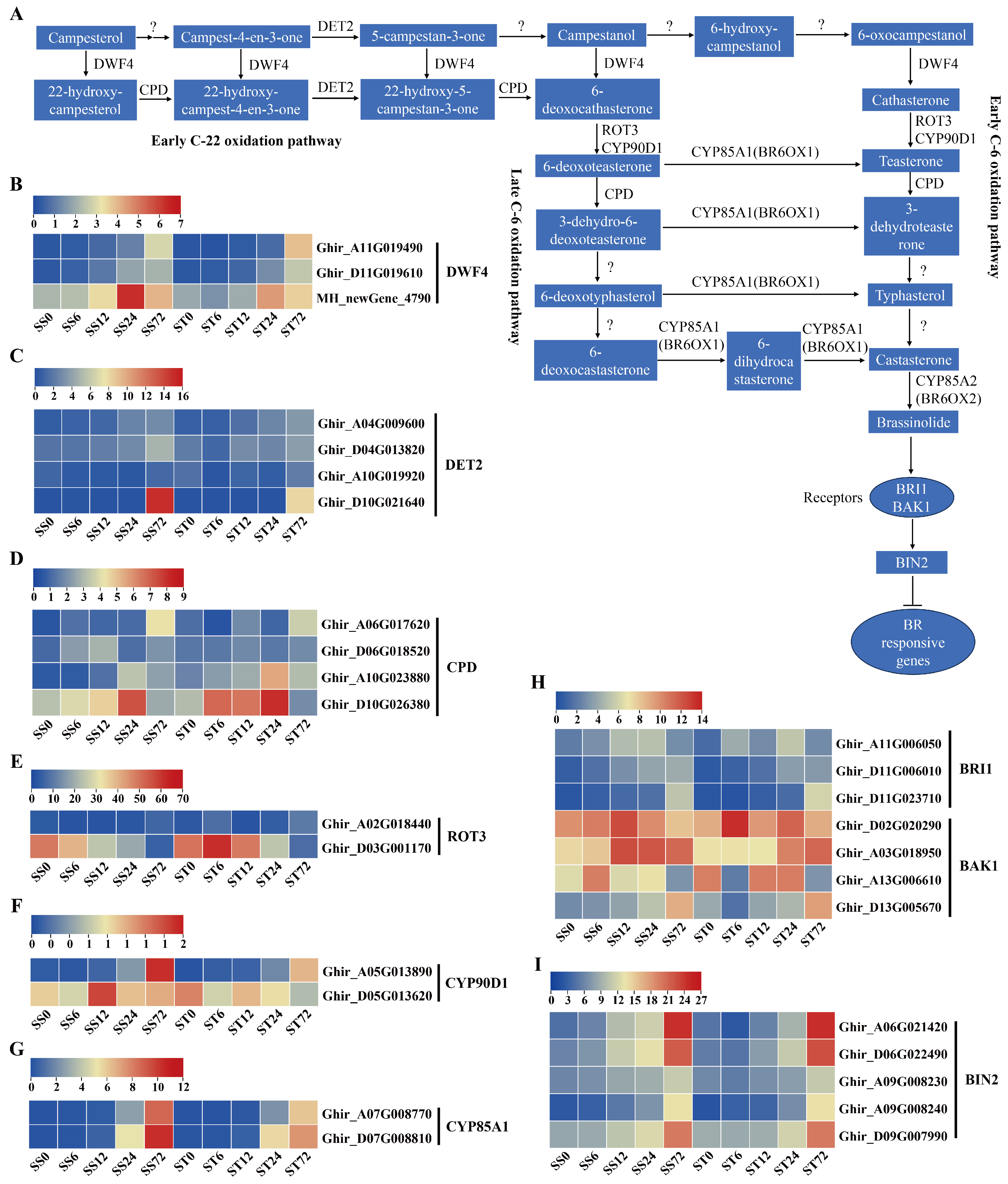
Expression Patterns of Genes Related to Brassinosteroids (BRs) Biosynthesis and Signaling during Salt Stress in Both Genotypes
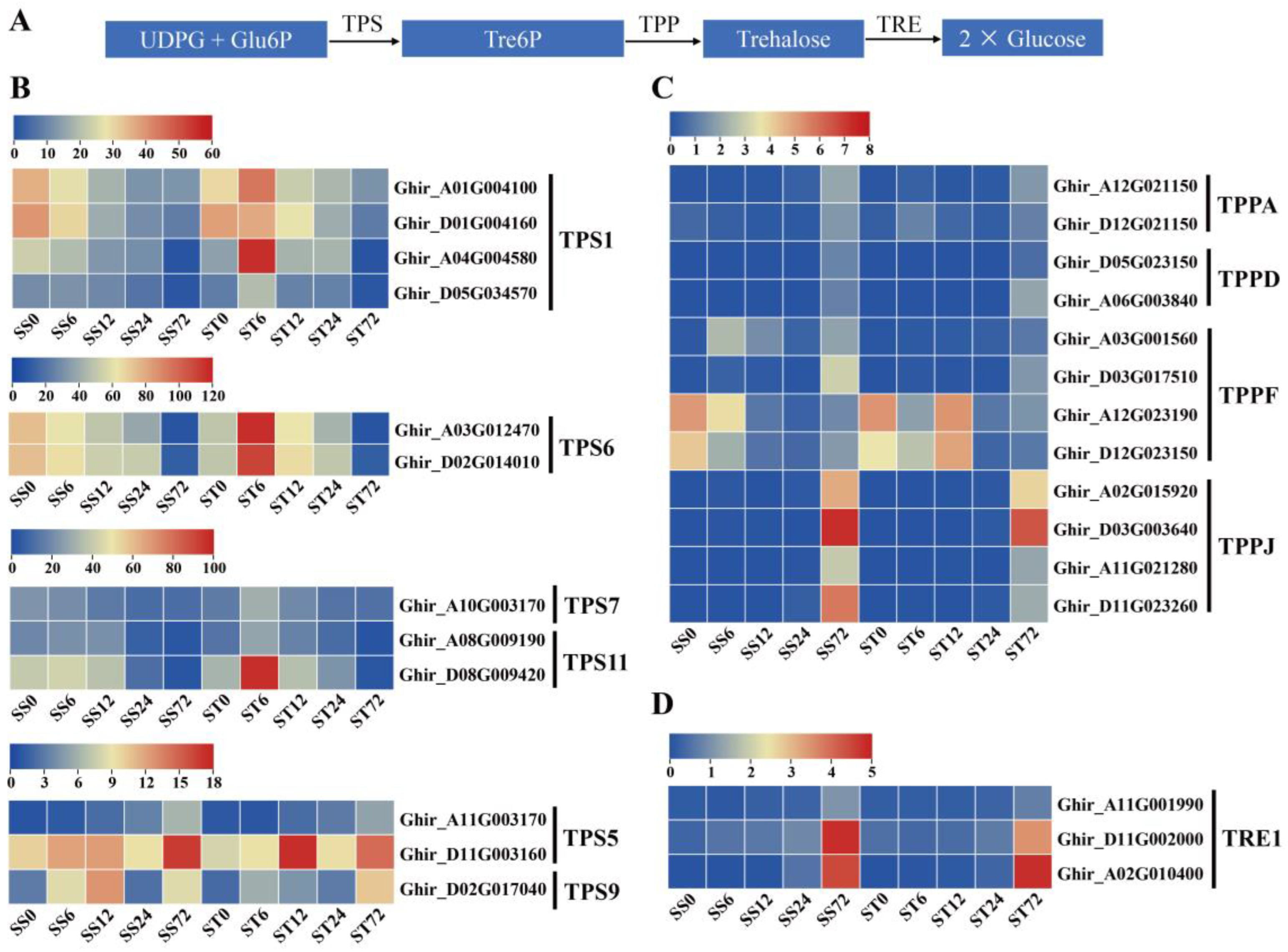
Expression Patterns of Genes Related to Trehalose Metabolism Pathway during Salt Stress in Both Genotypes
Discussion
Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Abbreviations
References
- Wen, X.; Chen, Z.; Yang, Z.; Wang, M.; Jin, S.; Wang, G.; Zhang, L.; Wang, L.; Li, J.; Saeed, S.; He, S.; Wang, Z.; Wang, K.; Kong, Z.; Li, F.; Zhang, X.; Chen, X.; Zhu, Y. A comprehensive overview of cotton genomics, biotechnology and molecular biological studies. Sci China Life Sci 2023, 66((10)), 2214–2256. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, D.; Hua, J.; Kong, X.; Wang, X.; Wang, J.; Si, A.; Zhao, F.; Liu, W.; Yu, Y.; Chen, Z. Genetic and morpho-physiological differences among transgenic and no-transgenic cotton cultivars. Plants 2023, 12((19)), 3437. [Google Scholar] [CrossRef]
- Huang, W.; Wu, F.; Han, W.; Li, Q.; Han, Y.; Wang, G.; Feng, L.; Li, X.; Yang, B.; Lei, Y.; Fan, Z.; Xiong, S.; Xin, M.; Li, Y.; Wang, Z. Carbon footprint of cotton production in China: Composition, spatiotemporal changes and driving factors. Science of The Total Environment 2022, 821, 153407. [Google Scholar] [CrossRef]
- Hassani, A.; Azapagic, A.; Shokri, N. Global predictions of primary soil salinization under changing climate in the 21st century. Nature Communications 2021, 12((1)), 6663. [Google Scholar] [CrossRef]
- Li, D.; Yang, Y.; Zhao, Y.; Zhou, X.; Han, Q.; Liu, H.; Li, M. Optimizing cotton yield and soil salinity management: Integrating brackish water leaching and freshwater drip irrigation with subsurface drainage. Field Crops Research 2024, 314, 109454. [Google Scholar] [CrossRef]
- Sun, H.; Meng, M.; Yan, Z.; Lin, Z.; Nie, X.; Yang, X. Genome-wide association mapping of stress-tolerance traits in cotton. The Crop Journal 2019, 7((1)), 77–88. [Google Scholar] [CrossRef]
- Ashraf, M.; Ahmad, S. Influence of sodium chloride on ion accumulation, yield components and fibre characteristics in salt-tolerant and salt-sensitive lines of cotton (Gossypium hirsutum L.). Field Crops Research 2000, 66, 115–127. [Google Scholar] [CrossRef]
- Ibrahim, W.; Qiu, C. W.; Zhang, C.; Cao, F.; Shuijin, Z.; Wu, F. Comparative physiological analysis in the tolerance to salinity and drought individual and combination in two cotton genotypes with contrasting salt tolerance. Physiol Plant 2019, 165((2)), 155–168. [Google Scholar] [CrossRef]
- Ibrahim, W.; Zhu, Y. M.; Chen, Y.; Qiu, C. W.; Zhu, S.; Wu, F. Genotypic differences in leaf secondary metabolism, plant hormones and yield under alone and combined stress of drought and salinity in cotton genotypes. Physiol Plant 2019, 165((2)), 343–355. [Google Scholar] [CrossRef]
- Wang, N.; Qi, H.; Qiao, W.; Shi, J.; Xu, Q.; Zhou, H.-W.; Yan, G.; Huang, Q. Cotton (Gossypium hirsutum L.) genotypes with contrasting K+/Na+ ion homeostasis: implications for salinity tolerance. Acta Physiologiae Plantarum 2017, 39, 1-10.
- Zhang, L.; Zhang, G.; Wang, Y.; Zhou, Z.; Meng, Y.; Chen, B. Effect of soil salinity on physiological characteristics of functional leaves of cotton plants. J Plant Res 2013, 126((2)), 293–304. [Google Scholar] [CrossRef]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; Ye, Z.; Huang, H.; Yan, F.; Ma, Y.; Zhang, L.; Liu, M.; You, J.; Yang, Y.; Liu, Z.; Huang, F.; Li, B.; Qiu, P.; Zhang, Q.; Zhu, L.; Jin, S.; Yang, X.; Min, L.; Li, G.; Chen, L.-L.; Zheng, H.; Lindsey, K.; Lin, Z.; Udall, J. A.; Zhang, X. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nature Genetics 2019, 51((2)), 224–229. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, J.; Qi, Z.; Long, Y.; Pei, L.; Huang, X.; Grover, C. E.; Du, X.; Xia, C.; Wang, P.; Liu, Z.; You, J.; Tian, X.; Ma, Y.; Wang, R.; Chen, X.; He, X.; Fang, D. D.; Sun, Y.; Tu, L.; Jin, S.; Zhu, L.; Wendel, J. F.; Zhang, X. Genomic innovation and regulatory rewiring during evolution of the cotton genus Gossypium. Nature Genetics 2022, 54((12)), 1959–1971. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; Baruch, K.; Fang, D.; Liu, X.; Ruan, Y.-l.; Rahman, M.-u.; Han, J.; Wang, K.; Wang, Q.; Wu, H.; Mei, G.; Zang, Y.; Han, Z.; Xu, C.; Shen, W.; Yang, D.; Si, Z.; Dai, F.; Zou, L.; Huang, F.; Bai, Y.; Zhang, Y.; Brodt, A.; Ben-Hamo, H.; Zhu, X.; Zhou, B.; Guan, X.; Zhu, S.; Chen, X.; Zhang, T. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nature Genetics 2019, 51((4)), 739–748. [Google Scholar] [CrossRef]
- Huang, G.; Wu, Z.; Percy, R. G.; Bai, M.; Li, Y.; Frelichowski, J. E.; Hu, J.; Wang, K.; Yu, J. Z.; Zhu, Y. Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nature Genetics 2020, 52((5)), 516–524. [Google Scholar] [CrossRef]
- Chen, Z. J.; Sreedasyam, A.; Ando, A.; Song, Q.; De Santiago, L. M.; Hulse-Kemp, A. M.; Ding, M.; Ye, W.; Kirkbride, R. C.; Jenkins, J.; Plott, C.; Lovell, J.; Lin, Y.-M.; Vaughn, R.; Liu, B.; Simpson, S.; Scheffler, B. E.; Wen, L.; Saski, C. A.; Grover, C. E.; Hu, G.; Conover, J. L.; Carlson, J. W.; Shu, S.; Boston, L. B.; Williams, M.; Peterson, D. G.; McGee, K.; Jones, D. C.; Wendel, J. F.; Stelly, D. M.; Grimwood, J.; Schmutz, J. Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nature Genetics 2020, 52((5)), 525–533. [Google Scholar] [CrossRef]
- Peng, Z.; He, S.; Gong, W.; Sun, J.; Pan, Z.; Xu, F.; Lu, Y.; Du, X. Comprehensive analysis of differentially expressed genes and transcriptional regulation induced by salt stress in two contrasting cotton genotypes. BMC Genomics 2014, 15((1)), 760. [Google Scholar] [CrossRef]
- Yuan, Z.; Zhang, C.; Zhu, W.; Yan, G.; Chen, X.; Qiu, P.; Ruzimurod, B.; Ye, W.; Qaraevna, B. Z.; Yin, Z. Molecular mechanism that underlies cotton response to salt and drought stress revealed by complementary transcriptomic and iTRAQ analyses. Environmental and Experimental Botany 2023, 209, 105288. [Google Scholar] [CrossRef]
- Zhang, F.; Zhu, G.; Du, L.; Shang, X.; Cheng, C.; Yang, B.; Hu, Y.; Cai, C.; Guo, W. Genetic regulation of salt stress tolerance revealed by RNA-Seq in cotton diploid wild species, Gossypium davidsonii. Sci Rep 2016, 6, 20582. [Google Scholar] [CrossRef]
- Guo, J.; Shi, G.; Guo, X.; Zhang, L.; Xu, W.; Wang, Y.; Su, Z.; Hua, J. Transcriptome analysis reveals that distinct metabolic pathways operate in salt-tolerant and salt-sensitive upland cotton varieties subjected to salinity stress. Plant Sci 2015, 238, 33–45. [Google Scholar] [CrossRef]
- Ju, F.; Pang, J.; Sun, L.; Gu, J.; Wang, Z.; Wu, X.; Ali, S.; Wang, Y.; Zhao, W.; Wang, S.; Zhou, Z.; Chen, B. Integrative transcriptomic, metabolomic and physiological analyses revealed the physiological and molecular mechanisms by which potassium regulates the salt tolerance of cotton (Gossypium hirsutum L.) roots. Industrial Crops and Products 2023, 193, 116177.
- Li, P.; Liu, Q.; Wei, Y.; Xing, C.; Xu, Z.; Ding, F.; Liu, Y.; Lu, Q.; Hu, N.; Wang, T.; Zhu, X.; Cheng, S.; Li, Z.; Zhao, Z.; Li, Y.; Han, J.; Cai, X.; Zhou, Z.; Wang, K.; Zhang, B.; Liu, F.; Jin, S.; Peng, R. Transcriptional landscape of cotton roots in response to salt stress at single-cell resolution. Plant Communications 2024, 5((2)), 100740. [Google Scholar] [CrossRef]
- Hao, Y.; Lu, G.-q.; Wang, L.-h.; Wang, C.-l.; Guo, H.; Li, Y.-F.; Cheng, H. Overexpression of AmDUF1517 enhanced tolerance to salinity, drought, and cold stress in transgenic cotton. Journal of Integrative Agriculture 2018. [Google Scholar] [CrossRef]
- Liu, G.; Li, X.; Jin, S.; Liu, X.; Zhu, L.; Nie, Y.; Zhang, X. Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS One 2014, 9((1)), e86895. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wan, S.; Liu, H.; Fan, S.; Zhang, Y.; Wang, W.; Xia, M.; Yuan, R.; Deng, F.; Shen, F. Overexpression of an Apocynum venetum DEAD-box helicase gene (AvDH1) in cotton confers salinity tolerance and increases yield in a saline field. Front Plant Sci 2015, 6, 1227. [Google Scholar] [CrossRef]
- Su, Y.; Liang, W.; Liu, Z.; Wang, Y.; Zhao, Y.; Ijaz, B.; Hua, J.-p. Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in Gossypium hirsutum. Journal of plant physiology 2017, 218, 222–234. [Google Scholar] [CrossRef]
- Song, J.; Zhang, R.; Yue, D.; Chen, X.; Guo, Z.; Cheng, C.; Hu, M.; Zhang, J.; Zhang, K. Co-expression of ApGSMT2g and ApDMT2g in cotton enhances salt tolerance and increases seed cotton yield in saline fields. Plant Science 2018, 274, 369–382. [Google Scholar] [CrossRef]
- Liang, C.; Meng, Z.; Meng, Z.; Malik, W.; Yan, R.; Lwin, K. M.; Lin, F.; Wang, Y.; Sun, G.; Zhou, T.; Zhu, T.; Li, J.; Jin, S.; Guo, S.; Zhang, R. GhABF2, a bZIP transcription factor, confers drought and salinity tolerance in cotton (Gossypium hirsutum L.). Sci Rep 2016, 6, 35040.
- Zhang, H.; Mao, L.; Xin, M.; Xing, H.; Zhang, Y.; Wu, J.; Xu, D.; Wang, Y.; Shang, Y.; Wei, L.; Cui, M.; Zhuang, T.; Sun, X.; Song, X. Overexpression of GhABF3 increases cotton(Gossypium hirsutum L.) tolerance to salt and drought. BMC Plant Biology 2022, 22, (1), 313.
- Kim, D.; Langmead, B.; Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015, 12((4)), 357–60. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G. M.; Leek, J. T.; Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 2016, 11((9)), 1650–67. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, J.; Qiao, J.; Li, W.; Guan, Z.; Liu, Z.; Bai, X.; Xing, B.; Zhang, J.; Li, J.; Yin, W.; Zhu, H. Graphene-mediated antioxidant enzyme activity and respiration in plant roots. ACS Agricultural Science & Technology 2022, 2, (3), 646-660.
- Liu, L.; Grover, C. E.; Kong, X.; Jareczek, J.; Wang, X.; Si, A.; Wang, J.; Yu, Y.; Chen, Z. Expression profile analysis of cotton fiber secondary cell wall thickening stage. PeerJ 2024, 12, e17682. [Google Scholar] [CrossRef]
- Love, M. I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15((12)), 550. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J. G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21((19)), 3787–3793. [Google Scholar] [CrossRef]
- Young, M. D.; Wakefield, M. J.; Smyth, G. K.; Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 2010, 11((2)), R14. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H. R.; Frank, M. H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant 2020, 13((8)), 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, E. A. Seed priming to alleviate salinity stress in germinating seeds. Journal of Plant Physiology 2016, 192, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yu, F.; Xie, P.; Sun, S.; Qiao, X.; Tang, S.; Chen, C.; Yang, S.; Mei, C.; Yang, D.; Wu, Y.; Xia, R.; Li, X.; Lu, J.; Liu, Y.; Xie, X.; Ma, D.; Xu, X.; Liang, Z.; Feng, Z.; Huang, X.; Yu, H.; Liu, G.; Wang, Y.; Li, J.; Zhang, Q.; Chen, C.; Ouyang, Y.; Xie, Q. A Gγ protein regulates alkaline sensitivity in crops. Science 2023, 379((6638)), eade8416. [Google Scholar] [CrossRef]
- Ubersax, J. A.; Ferrell Jr, J. E. Mechanisms of specificity in protein phosphorylation. Nature Reviews Molecular Cell Biology 2007, 8((7)), 530–541. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Q. Q.; Zhou, L.; Ren, F.; Li, D. D.; Li, X. B. Arabidopsis CBL-interacting protein kinase (CIPK6) is involved in plant response to salt/osmotic stress and ABA. Mol Biol Rep 2013, 40((8)), 4759–67. [Google Scholar] [CrossRef]
- Su, Y.; Guo, A.; Huang, Y.; Wang, Y.; Hua, J. GhCIPK6a increases salt tolerance in transgenic upland cotton by involving in ROS scavenging and MAPK signaling pathways. BMC Plant Biology 2020, 20((1)), 421. [Google Scholar] [CrossRef]
- Tripathi, V.; Parasuraman, B.; Laxmi, A.; Chattopadhyay, D. CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. The Plant Journal 2009, 58((5)), 778–790. [Google Scholar] [CrossRef]
- Castro, B.; Citterico, M.; Kimura, S.; Stevens, D. M.; Wrzaczek, M.; Coaker, G. Stress-induced reactive oxygen species compartmentalization, perception and signalling. Nature Plants 2021, 7((4)), 403–412. [Google Scholar] [CrossRef]
- Kesawat, M. S.; Satheesh, N.; Kherawat, B. S.; Kumar, A.; Kim, H. U.; Chung, S. M.; Kumar, M. Regulation of reactive oxygen species during salt stress in plants and their crosstalk with other signaling molecules-current perspectives and future directions. Plants (Basel) 2023, 12, (4).
- Li, X.; Pang, Y.; Zhong, Y.; Cai, Z.; Ma, Q.; Wen, K.; Nian, H. GmGSTU23 encoding a Tau class glutathione s-transferase protein enhances the salt tolerance of Soybean (Glycine max L.). Int J Mol Sci 2023, 24, (6), 5547.
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci 2020, 25((11)), 1117–1130. [Google Scholar] [CrossRef]
- S F Yang, a.; Hoffman, N. E. Ethylene biosynthesis and its regulation in higher plants. Annual Review of Plant Physiology 1984, 35((1)), 155–189. [Google Scholar] [CrossRef]
- Wang, K. L.; Li, H.; Ecker, J. R. Ethylene biosynthesis and signaling networks. Plant Cell 2002, 14 Suppl (Suppl), S131–51. [Google Scholar] [CrossRef] [PubMed]
- Ruduś, I.; Sasiak, M.; Kępczyński, J. Regulation of ethylene biosynthesis at the level of 1-aminocyclopropane-1-carboxylate oxidase (ACO) gene. Acta Physiologiae Plantarum 2013, 35((2)), 295–307. [Google Scholar] [CrossRef]
- Pattyn, J.; Vaughan-Hirsch, J.; Van de Poel, B. The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytol 2021, 229((2)), 770–782. [Google Scholar] [CrossRef]
- Guo, H.; Ecker, J. R. The ethylene signaling pathway: new insights. Curr Opin Plant Biol 2004, 7((1)), 40–9. [Google Scholar] [CrossRef]
- Xiao, F.; Zhou, H. Plant salt response: Perception, signaling, and tolerance. Front Plant Sci 2022, 13, 1053699. [Google Scholar] [CrossRef]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. The Plant Journal 2004, 37((5)), 720–729. [Google Scholar] [CrossRef]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J 2008, 56((4)), 613–26. [Google Scholar] [CrossRef]
- Richards, D. E.; King, K. E.; Ait-Ali, T.; Harberd, N. P. How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 2001, 52, 67–88. [Google Scholar] [CrossRef]
- Magome, H.; Nomura, T.; Hanada, A.; Takeda-Kamiya, N.; Ohnishi, T.; Shinma, Y.; Katsumata, T.; Kawaide, H.; Kamiya, Y.; Yamaguchi, S. CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc Natl Acad Sci U S A 2013, 110((5)), 1947–52. [Google Scholar] [CrossRef]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol 2008, 59, 225–51. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Miura, K.; Itoh, H.; Tatsumi, T.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Agrawal, G. K.; Takeda, S.; Abe, K.; Miyao, A.; Hirochika, H.; Kitano, H.; Ashikari, M.; Matsuoka, M. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 2004, 134((4)), 1642–53. [Google Scholar] [CrossRef] [PubMed]
- Waśkiewicz, A.; Beszterda, M.; Goliński, P. ABA: role in plant signaling under salt stress. In Salt Stress in Plants: Signalling, Omics and Adaptations, Ahmad, P.; Azooz, M. M.; Prasad, M. N. V., Eds. Springer New York: New York, NY, 2013; pp 175-196.
- Jia, K.-P.; Mi, J.; Ali, S.; Ohyanagi, H.; Moreno, J. C.; Ablazov, A.; Balakrishna, A.; Berqdar, L.; Fiore, A.; Diretto, G.; Martínez, C.; de Lera, A. R.; Gojobori, T.; Al-Babili, S. An alternative, zeaxanthin epoxidase-independent abscisic acid biosynthetic pathway in plants. Molecular Plant 2022, 15((1)), 151–166. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Okamoto, M.; Koshiba, T. ABA Biosynthetic and Catabolic Pathways. In Abscisic Acid: Metabolism, Transport and Signaling, Zhang, D.-P., Ed. Springer Netherlands: Dordrecht, 2014; pp 21-45.
- Huang, Y.; Jiao, Y.; Xie, N.; Guo, Y.; Zhang, F.; Xiang, Z.; Wang, R.; Wang, F.; Gao, Q.; Tian, L.; Li, D.; Chen, L.; Liang, M. OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci 2019, 287, 110188. [Google Scholar] [CrossRef]
- Shiono, K.; Yoshikawa, M.; Kreszies, T.; Yamada, S.; Hojo, Y.; Matsuura, T.; Mori, I. C.; Schreiber, L.; Yoshioka, T. Abscisic acid is required for exodermal suberization to form a barrier to radial oxygen loss in the adventitious roots of rice (Oryza sativa). New Phytologist 2022, 233((2)), 655–669. [Google Scholar] [CrossRef]
- Zhu, M.; Liu, Y.; Cai, P.; Duan, X.; Sang, S.; Qiu, Z. Jasmonic acid pretreatment improves salt tolerance of wheat by regulating hormones biosynthesis and antioxidant capacity. Front Plant Sci 2022, 13, 968477. [Google Scholar] [CrossRef]
- Song, R. F.; Li, T. T.; Liu, W. C. Jasmonic acid impairs Arabidopsis seedling salt stress tolerance through MYC2-mediated repression of CAT2 expression. Front Plant Sci 2021, 12, 730228. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 2013, 111((6)), 1021–58. [Google Scholar] [CrossRef]
- Wasternack, C.; Song, S. Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J Exp Bot 2017, 68((6)), 1303–1321. [Google Scholar] [CrossRef]
- Koo, A. J. K.; Chung, H. S.; Kobayashi, Y.; Howe, G. A. Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis*♦. Journal of Biological Chemistry 2006, 281((44)), 33511–33520. [Google Scholar] [CrossRef]
- Zhang, H.-Y.; Wang, X.; Wang, X.-N.; Liu, H.-F.; Zhang, T.-T.; Wang, D.-R.; Liu, G.-D.; Liu, Y.-Q.; Song, X.-h.; Zhang, Z.; You, C. Brassinosteroids biosynthetic gene MdBR6OX2 regulates salt stress tolerance in both apple and Arabidopsis. Plant Physiology and Biochemistry 2024, 212, 108767. [Google Scholar] [CrossRef] [PubMed]
- Rattan, A.; Kapoor, D.; Ashish; Kapoor, N.; Bhardwaj, R.; Sharma, A., Chapter 12 - Involvement of brassinosteroids in plant response to salt stress. In Brassinosteroids in Plant Developmental Biology and Stress Tolerance, Ahammed, G. J.; Sharma, A.; Yu, J., Eds. Academic Press: 2022; pp 237-253.
- Zebosi, B.; Vollbrecht, E.; Best, N. B. Brassinosteroid biosynthesis and signaling: Conserved and diversified functions of core genes across multiple plant species. Plant Communications 2024, 100982. [Google Scholar] [CrossRef] [PubMed]
- Bajguz, A.; Chmur, M.; Gruszka, D. Comprehensive overview of the brassinosteroid biosynthesis pathways: substrates, products, inhibitors, and connections. Front Plant Sci 2020, 11, 1034. [Google Scholar] [CrossRef]
- Vriet, C.; Russinova, E.; Reuzeau, C. From squalene to brassinolide: the steroid metabolic and signaling pathways across the plant kingdom. Mol Plant 2013, 6((6)), 1738–57. [Google Scholar] [CrossRef]
- Zhao, B.; Li, J. Regulation of brassinosteroid biosynthesis and inactivation. J Integr Plant Biol 2012, 54((10)), 746–59. [Google Scholar] [CrossRef]
- Fujioka, S.; Yokota, T. Biosynthesis and metabolism of brassinosteroids. Annual Review of Plant Biology 2003, 54(2003), 137–164. [Google Scholar] [CrossRef]
- Lunn, J. E.; Delorge, I.; Figueroa, C. M.; Van Dijck, P.; Stitt, M. Trehalose metabolism in plants. Plant J 2014, 79((4)), 544–67. [Google Scholar] [CrossRef]
- Minami, A.; Yano, K.; Gamuyao, R.; Nagai, K.; Kuroha, T.; Ayano, M.; Nakamori, M.; Koike, M.; Kondo, Y.; Niimi, Y.; Kuwata, K.; Suzuki, T.; Higashiyama, T.; Takebayashi, Y.; Kojima, M.; Sakakibara, H.; Toyoda, A.; Fujiyama, A.; Kurata, N.; Ashikari, M.; Reuscher, S. Time-course transcriptomics analysis reveals key responses of submerged deepwater rice to flooding. Plant Physiol 2018, 176((4)), 3081–3102. [Google Scholar] [CrossRef]
- Yao, Y.; Yang, Y.; Pan, Y.; Liu, Z.; Hou, X.; Li, Y.; Zhang, H.; Wang, C.; Liao, W. Crucial roles of trehalose and 5-azacytidine in alleviating salt stress in tomato: Both synergistically and independently. Plant Physiology and Biochemistry 2023, 203, 108075. [Google Scholar] [CrossRef]
- Yang, Y.; Yao, Y.; Li, J.; Zhang, J.; Zhang, X.; Hu, L.; Ding, D.; Bakpa, E. P.; Xie, J. Trehalose alleviated salt stress in tomato by regulating ROS metabolism, photosynthesis, osmolyte synthesis, and trehalose metabolic pathways. Front Plant Sci 2022, 13, 772948. [Google Scholar] [CrossRef]
- Zhou, H.; Shi, H.; Yang, Y.; Feng, X.; Chen, X.; Xiao, F.; Lin, H.; Guo, Y. Insights into plant salt stress signaling and tolerance. Journal of Genetics and Genomics 2024, 51((1)), 16–34. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of plant responses to salt stress. Int J Mol Sci 2021, 22, (9).
- Cao, Y. R.; Chen, S. Y.; Zhang, J. S. Ethylene signaling regulates salt stress response: An overview. Plant Signal Behav 2008, 3((10)), 761–3. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytologist 2013, 199((3)), 639–649. [Google Scholar] [CrossRef] [PubMed]
- Cebrián, G.; Iglesias-Moya, J.; García, A.; Martínez, J.; Romero, J.; Regalado, J. J.; Martínez, C.; Valenzuela, J. L.; Jamilena, M. Involvement of ethylene receptors in the salt tolerance response of Cucurbita pepo. Horticulture Research 2021, 8((1)), 73. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Liu, J.; Lin, Z.; Sun, W.; Wu, Z.; Hu, H.; Zhang, Y. ERF transcription factors involved in salt response in tomato. Plant Growth Regulation 2018, 84((3)), 573–582. [Google Scholar] [CrossRef]
- Zhang, M.; Smith, J. A. C.; Harberd, N. P.; Jiang, C. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Molecular Biology 2016, 91((6)), 651–659. [Google Scholar] [CrossRef]
- Jiang, C.; Belfield, E. J.; Cao, Y.; Smith, J. A. C.; Harberd, N. P. An Arabidopsis soil-salinity–tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. The Plant Cell 2013, 25((9)), 3535–3552. [Google Scholar] [CrossRef]
- Peng, J.; Li, Z.; Wen, X.; Li, W.; Shi, H.; Yang, L.; Zhu, H.; Guo, H. Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet 2014, 10((10)), e1004664. [Google Scholar] [CrossRef]
- Wang, Y.; Diao, P.; Kong, L.; Yu, R.; Zhang, M.; Zuo, T.; Fan, Y.; Niu, Y.; Yan, F.; Wuriyanghan, H. Ethylene enhances seed germination and seedling growth under salinity by reducing oxidative stress and promoting chlorophyll content via ETR2 pathway. Front Plant Sci 2020, 11, 1066. [Google Scholar] [CrossRef]
- Li, H.; Sun, H.; Ping, W.; Liu, L.; Zhang, Y.; Zhang, K.; Bai, Z.; Li, A.; Zhu, J.; Li, C. Exogenous ethylene promotes the germination of cotton seeds under salt stress. Journal of Plant Growth Regulation 2023, 42((6)), 3923–3933. [Google Scholar] [CrossRef]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J 2004, 37((5)), 720–9. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Renou, J. P.; Berthomé, R.; Harberd, N. P.; Genschik, P. Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr Biol 2008, 18((9)), 656–60. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Rashotte, A. M.; Wysocka-Diller, J. W. ABA and GA signaling pathways interact and regulate seed germination and seedling development under salt stress. Acta Physiologiae Plantarum 2011, 33((2)), 261–271. [Google Scholar] [CrossRef]
- Zhu, J. K. Abiotic stress signaling and responses in plants. Cell 2016, 167((2)), 313–324. [Google Scholar] [CrossRef]
- Yang, G.; Yu, Z.; Gao, L.; Zheng, C. SnRK2s at the crossroads of growth and stress responses. Trends Plant Sci 2019, 24((8)), 672–676. [Google Scholar] [CrossRef]
- Szymańska, K. P.; Polkowska-Kowalczyk, L.; Lichocka, M.; Maszkowska, J.; Dobrowolska, G. SNF1-related protein kinases SnRK2.4 and SnRK2.10 modulate ROS homeostasis in plant response to salt stress. Int J Mol Sci 2019, 20, (1).
- Delgado, C.; Mora-Poblete, F.; Ahmar, S.; Chen, J. T.; Figueroa, C. R. Jasmonates and plant salt stress: molecular players, physiological effects, and improving tolerance by using genome-associated tools. Int J Mol Sci 2021, 22, (6).
- Gao, Z.; Gao, S.; Li, P.; Zhang, Y.; Ma, B.; Wang, Y. Exogenous methyl jasmonate promotes salt stress-induced growth inhibition and prioritizes defense response of Nitraria tangutorum Bobr. Physiologia Plantarum 2021, 172((1)), 162–175. [Google Scholar] [CrossRef]
- Nolan, T. M.; Vukašinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell 2020, 32((2)), 295–318. [Google Scholar] [CrossRef]
- Jia, C.; Zhao, S.; Bao, T.; Zhao, P.; Peng, K.; Guo, Q.; Gao, X.; Qin, J. Tomato BZR/BES transcription factor SlBZR1 positively regulates BR signaling and salt stress tolerance in tomato and Arabidopsis. Plant Sci 2021, 302, 110719. [Google Scholar] [CrossRef]
- Özdemir, F.; Bor, M.; Demiral, T.; Türkan, İ. Effects of 24-epibrassinolide on seed germination, seedling growth, lipid peroxidation, proline content and antioxidative system of rice (Oryza sativa L.) under salinity stress. Plant Growth Regulation 2004, 42, (3), 203-211.
- Wani, A. S.; Ahmad, A.; Hayat, S.; Tahir, I. Epibrassinolide and proline alleviate the photosynthetic and yield inhibition under salt stress by acting on antioxidant system in mustard. Plant Physiol Biochem 2019, 135, 385–394. [Google Scholar] [CrossRef]
- Su, Q.; Zheng, X.; Tian, Y.; Wang, C. Exogenous Brassinolide Alleviates Salt Stress in Malus hupehensis Rehd. by Regulating the Transcription of NHX-Type Na(+)(K(+))/H(+) Antiporters. Front Plant Sci 2020, 11, 38.
- Xia, X. J.; Chen, Z.; Yu, J. Q. ROS mediate brassinosteroids-induced plant stress responses. Plant Signal Behav 2010, 5((5)), 532–4. [Google Scholar] [CrossRef]
- Abdelraheem, A.; Esmaeili, N.; O’Connell, M.; Zhang, J. Progress and perspective on drought and salt stress tolerance in cotton. Industrial Crops and Products 2019, 130, 118–129. [Google Scholar] [CrossRef]
- Cao, J.-F.; Huang, J.-Q.; Liu, X.; Huang, C.-C.; Zheng, Z.-S.; Zhang, X.-F.; Shangguan, X.-X.; Wang, L.-J.; Zhang, Y.-G.; Wendel, J. F.; Grover, C. E.; Chen, Z.-W. Genome-wide characterization of the GRF family and their roles in response to salt stress in Gossypium. BMC Genomics 2020, 21((1)), 575. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.; Hao, J.; Su, Y.; Li, B.; Zhao, N.; Zhu, M.; Huang, Y.; Tian, B.; Shi, G.; Hua, J. Two aquaporin genes, GhPIP2;7 and GhTIP2;1, positively regulate the tolerance of upland cotton to salt and osmotic stresses. Front Plant Sci 2021, 12, 780486. [Google Scholar] [CrossRef]
- Yang, L.; Wang, X.; Zhao, F.; Zhang, X.; Li, W.; Huang, J.; Pei, X.; Ren, X.; Liu, Y.; He, K.; Zhang, F.; Ma, X.; Yang, D. Roles of S-adenosylmethionine and its derivatives in salt tolerance of cotton. International Journal of Molecular Sciences 2023, 24((11)), 9517. [Google Scholar] [CrossRef]
- Nawaz, M.; Hassan, M. U.; Chattha, M. U.; Mahmood, A.; Shah, A. N.; Hashem, M.; Alamri, S.; Batool, M.; Rasheed, A.; Thabit, M. A.; Alhaithloul, H. A. S.; Qari, S. H. Trehalose: a promising osmo-protectant against salinity stress—physiological and molecular mechanisms and future prospective. Molecular Biology Reports 2022, 49((12)), 11255–11271. [Google Scholar] [CrossRef]
- Shahzad, A. N.; Qureshi, M. K.; Ullah, S.; Latif, M.; Ahmad, S.; Bukhari, S. A. H. Exogenous trehalose improves cotton growth by modulating antioxidant defense under salinity-induced osmotic stress. Pakistan Journal of Agricultural Research 2020, 33((2)), 270–279. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




