Submitted:
26 September 2024
Posted:
29 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Ethics Statement and Animal Sample Collection
2.2. Fatty Acid Content Analysis
2.3. Metabolites Extraction of Liquid
2.4. LC-MS/MS Metabolites Detection
2.5. Microbial DNA Extraction and Sequencing Method
2.6. Longissimus Dorsi Muscle Metabolites Analysis
2.7. Analysis of Intestinal Microorganisms
2.8. Integrative Analysis
2.9. Statistical Analyses
3. Results
3.1. Fatty Acid Profile in Longissimus Dorsi Muscle of SBP and LWLDP
3.2. Intestine Metabolome Analysis
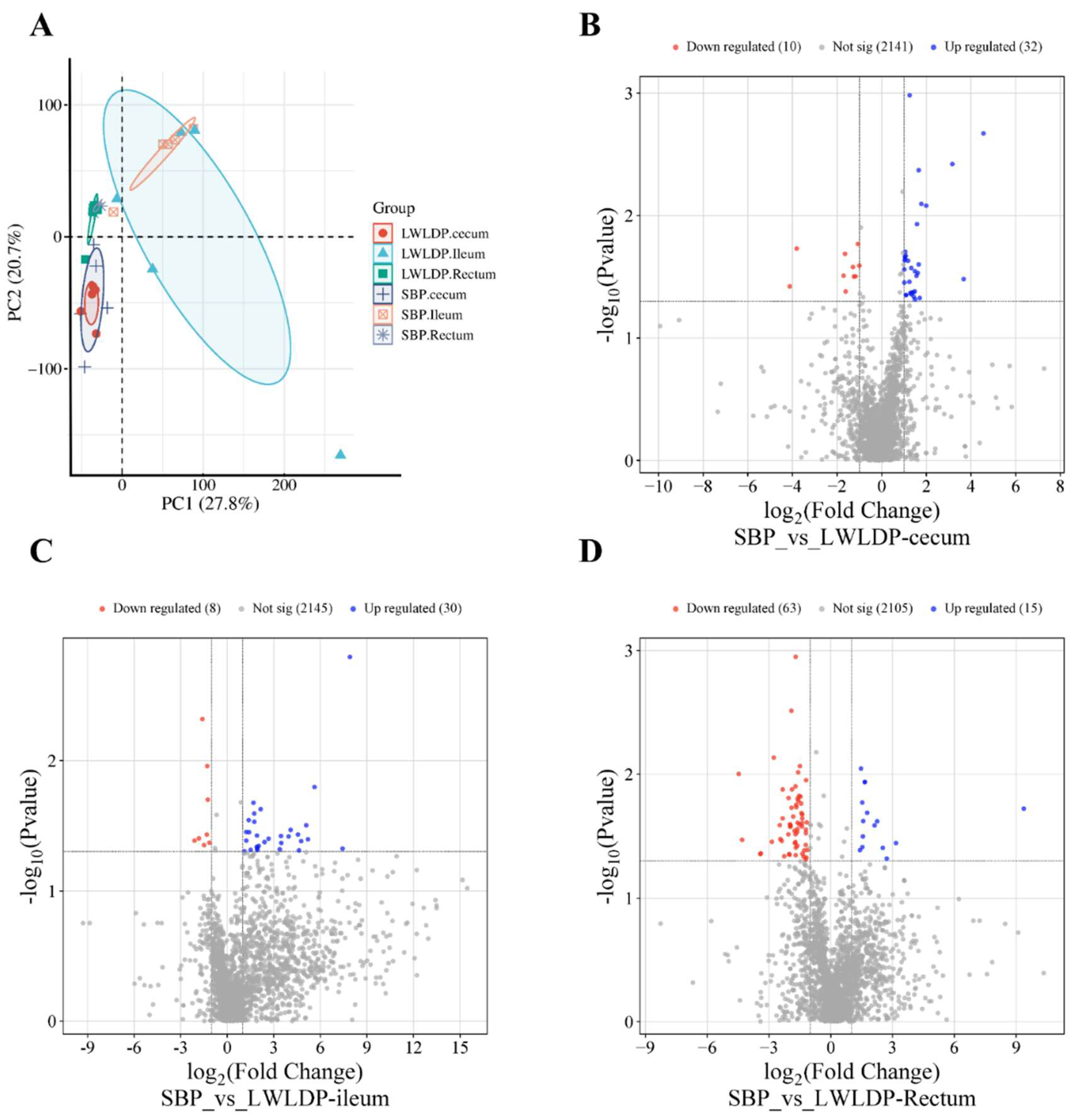
3.2.1. Differential Metabolites Analysis of Cecum, Ileum and Rectum between SBP and LWLDP Pigs Breeds
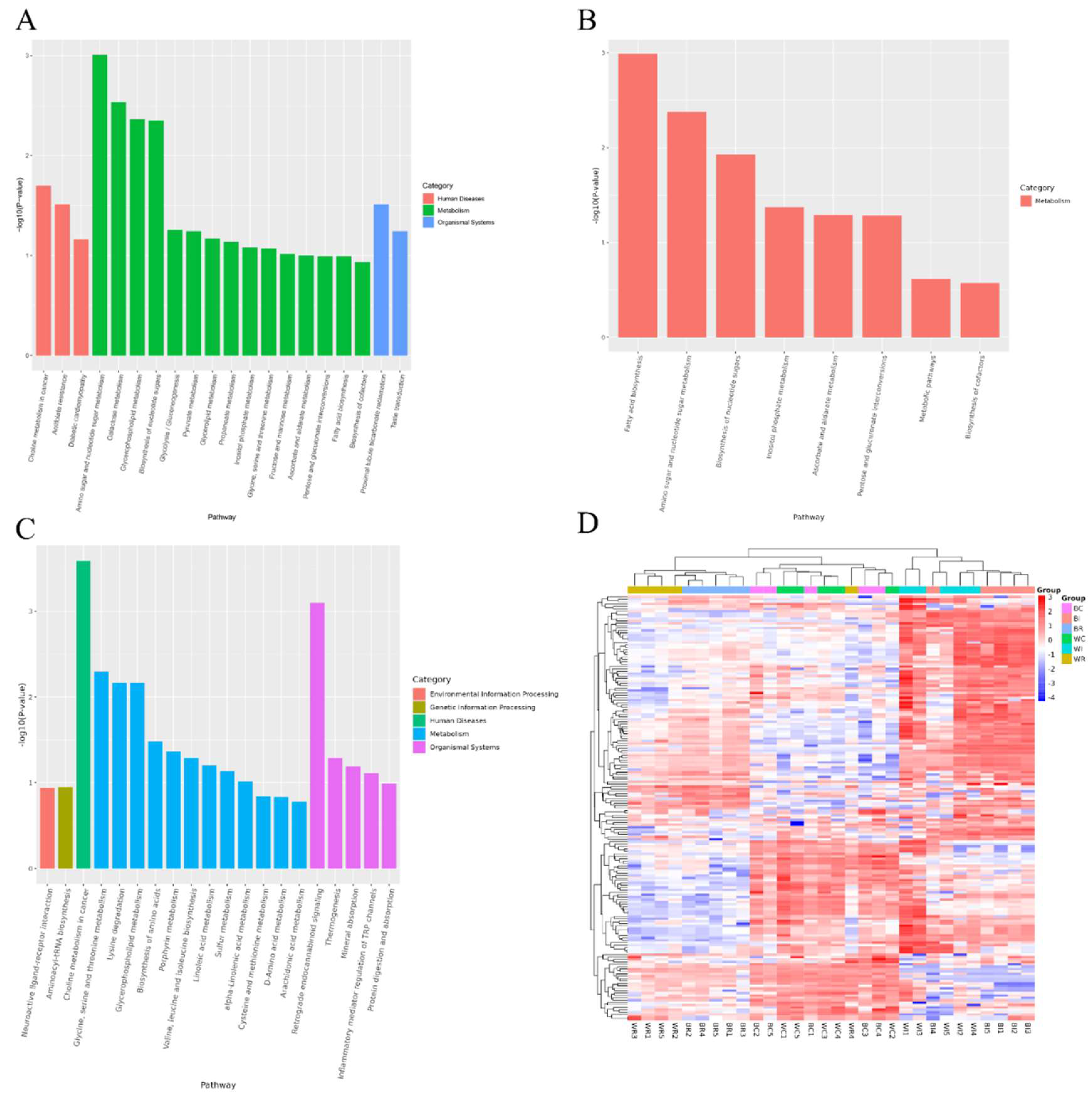
3.2.2. Functional Analysis of Ileum Cecum and Rectum Metabolites of SBP and LWLDP Pig Breeds
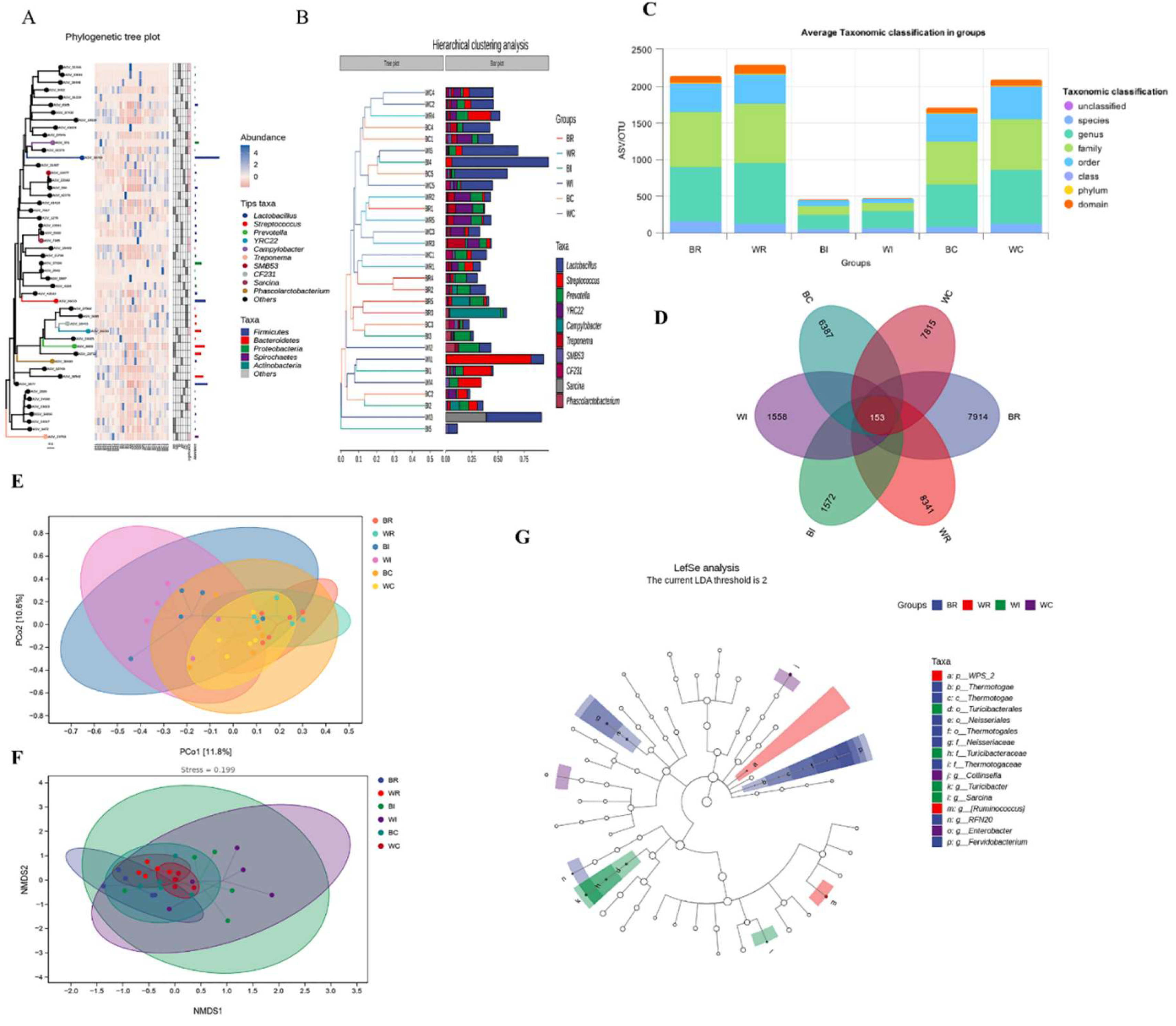
3.3. Intestine Microbiome Analysis
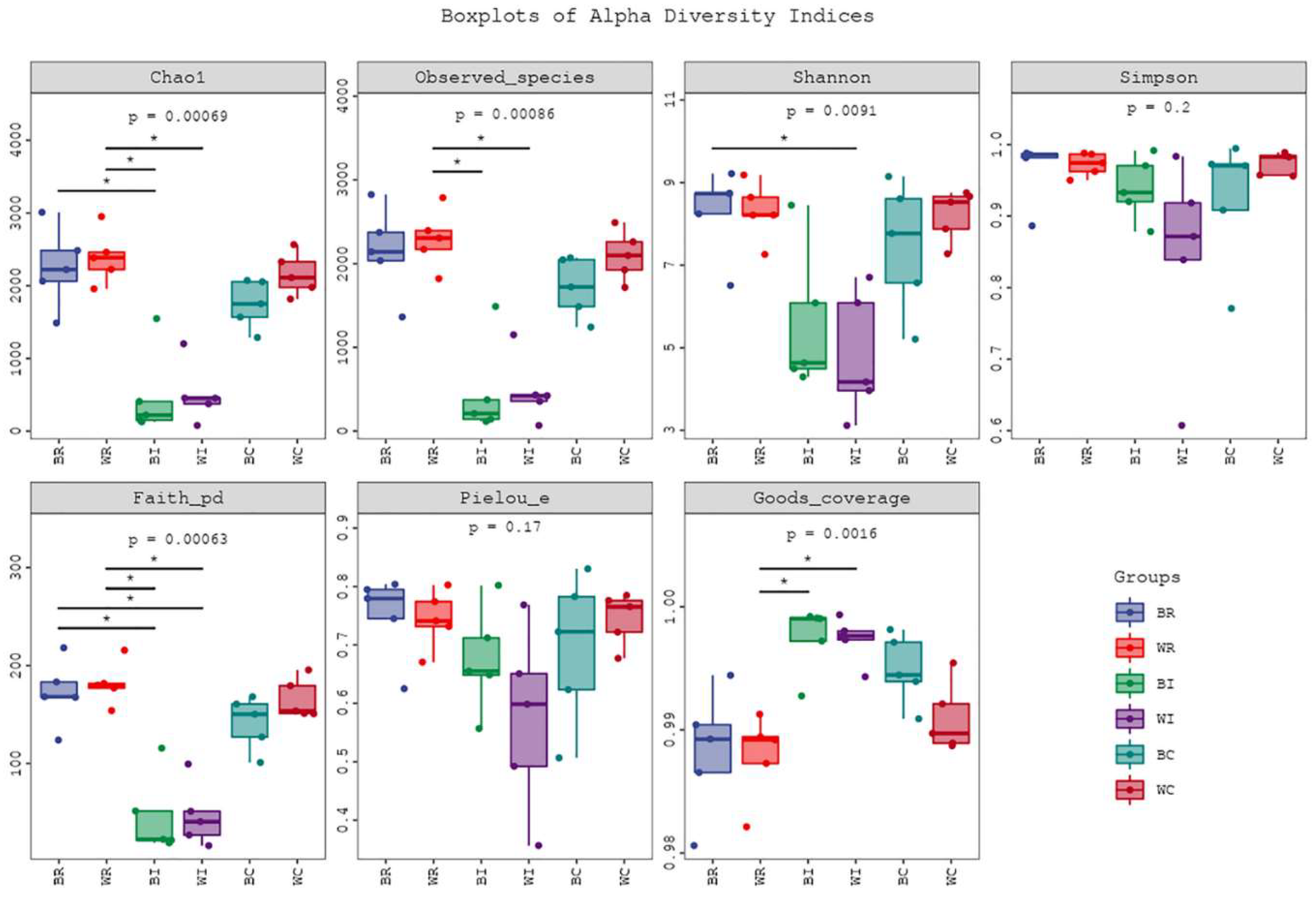
3.3.1. Taxonomy and Diversity of Intestine Microbes in Cecum, Ileum and Rectum of SBP and LWLDP Pig Breeds
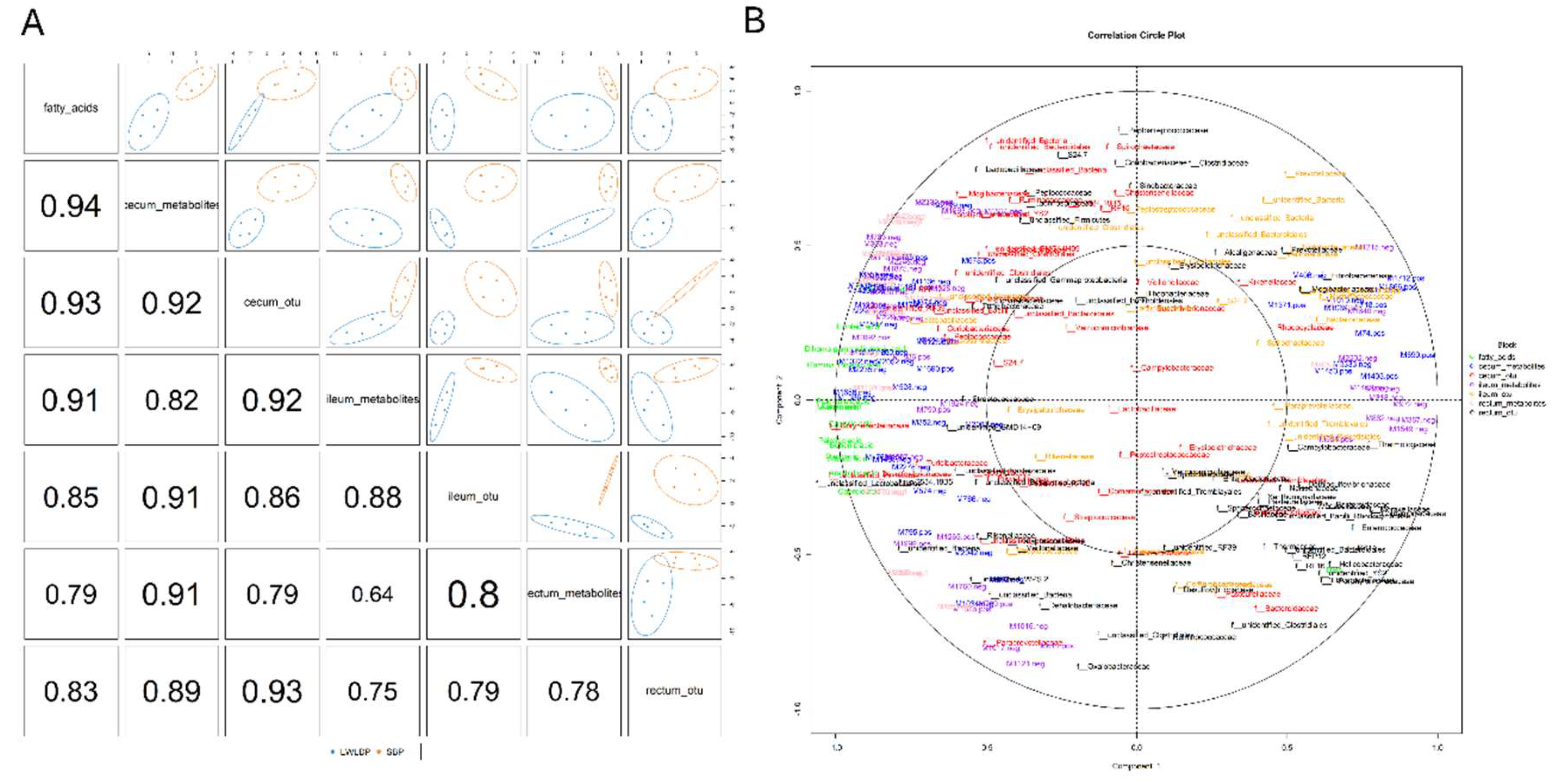
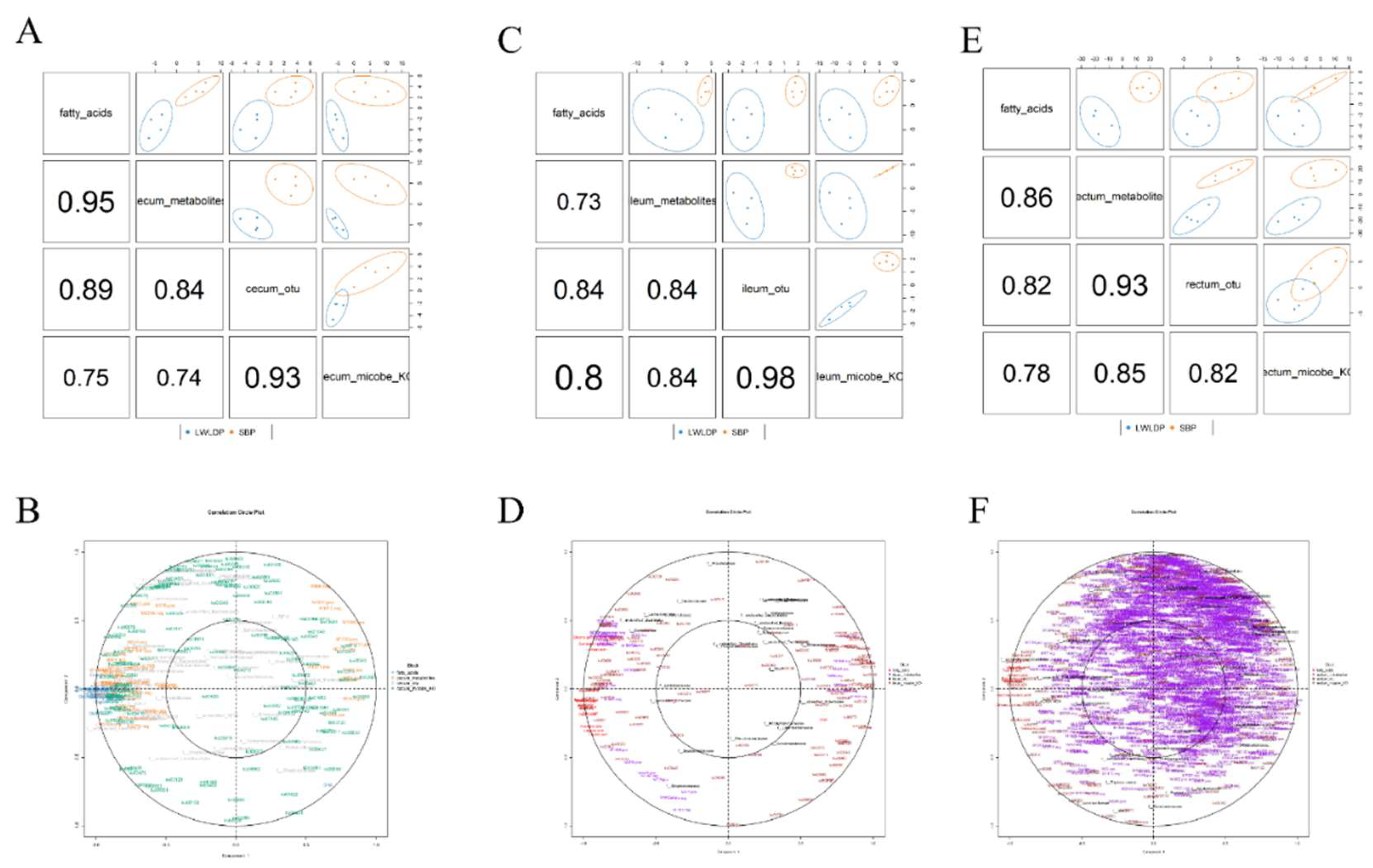
4.4. Integrative Analysis of Intestine Microbes, Metabolites and Fatty Acids
4.4.1. Combine Mix Omics Analysis
3.4.2. MixOmics Analysis of Different Regions of Intestine
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barbut, S. Automation and meat quality-global challenges. Meat science 2014, 96, 335–345. [Google Scholar] [CrossRef]
- Wu, Y.; Zhao, J.; Xu, C.; Ma, N.; He, T.; Zhao, J.; Ma, X.; Thacker, P.A. Progress towards pig nutrition in the last 27 years. Journal of the Science of Food and Agriculture 2020, 100, 5102–5110. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Gong, H.; Cui, L.; Zhang, W.; Ma, J.; Chen, C.; Ai, H.; Xiao, S.; Huang, L. Genetic correlation of fatty acid composition with growth, carcass, fat deposition and meat quality traits based on GWAS data in six pig populations. Meat Science 2019, 150, 47–55. [Google Scholar] [CrossRef]
- Wood, J.; Richardson, R.; Nute, G.; Fisher, A.; Campo, M.; Kasapidou, E.; Sheard, P.; Enser, M. Effects of fatty acids on meat quality: a review. Meat science 2004, 66, 21–32. [Google Scholar] [CrossRef]
- Kimura, I.; Ozawa, K.; Inoue, D.; Imamura, T.; Kimura, K.; Maeda, T.; Terasawa, K.; Kashihara, D.; Hirano, K.; Tani, T. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nature communications 2013, 4, 1829. [Google Scholar] [CrossRef]
- Parséus, A.; Sommer, N.; Sommer, F.; Caesar, R.; Molinaro, A.; Ståhlman, M.; Greiner, T.U.; Perkins, R.; Bäckhed, F. Microbiota-induced obesity requires farnesoid X receptor. Gut 2017, 66, 429–437. [Google Scholar] [CrossRef]
- Yin, J.; Li, Y.; Han, H.; Ma, J.; Liu, G.; Wu, X.; Huang, X.; Fang, R.; Baba, K.; Bin, P. Administration of exogenous melatonin improves the diurnal rhythms of the gut microbiota in mice fed a high-fat diet. Msystems 2020, 5. [Google Scholar] [CrossRef]
- Icaza-Chávez, M. Gut microbiota in health and disease. Revista de Gastroenterología de México (English Edition) 2013, 78, 240–248. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Reddy, D.N. Role of the normal gut microbiota. World journal of gastroenterology: WJG 2015, 21, 8787. [Google Scholar] [CrossRef]
- Yadav, S.; Jha, R. Strategies to modulate the intestinal microbiota and their effects on nutrient utilization, performance, and health of poultry. Journal of animal science and biotechnology 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Balfour Sartor, R.; Muehlbauer, M. Microbial host interactions in IBD: implications for pathogenesis and therapy. Current gastroenterology reports 2007, 9, 497–507. [Google Scholar] [CrossRef]
- Morowitz, M.J.; Carlisle, E.M.; Alverdy, J.C. Contributions of intestinal bacteria to nutrition and metabolism in the critically ill. Surgical Clinics 2011, 91, 771–785. [Google Scholar] [CrossRef]
- Xie, G.; Zhang, S.; Zheng, X.; Jia, W. Metabolomics approaches for characterizing metabolic interactions between host and its commensal microbes. Electrophoresis 2013, 34, 2787–2798. [Google Scholar] [CrossRef]
- Demain, A.L.; Fang, A. The natural functions of secondary metabolites. History of modern biotechnology I 2000, 1–39. [Google Scholar] [CrossRef]
- Roessner, U.; Dias, D.A. Metabolomics tools for natural product discovery; Springer: 2013.
- Zhao, W.; Wang, Y.; Liu, S.; Huang, J.; Zhai, Z.; He, C.; Ding, J.; Wang, J.; Wang, H.; Fan, W. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PloS one 2015, 10, e0117441. [Google Scholar] [CrossRef]
- Yang, H.; Huang, X.; Fang, S.; Xin, W.; Huang, L.; Chen, C. Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness. Scientific reports 2016, 6, 27427. [Google Scholar] [CrossRef]
- Kelly, J.; Daly, K.; Moran, A.W.; Ryan, S.; Bravo, D.; Shirazi-Beechey, S.P. Composition and diversity of mucosa-associated microbiota along the entire length of the pig gastrointestinal tract; dietary influences. Environmental microbiology 2017, 19, 1425–1438. [Google Scholar] [CrossRef]
- Xiao, Y.; Kong, F.; Xiang, Y.; Zhou, W.; Wang, J.; Yang, H.; Zhang, G.; Zhao, J. Comparative biogeography of the gut microbiome between Jinhua and Landrace pigs. Scientific reports 2018, 8, 5985. [Google Scholar] [CrossRef]
- Yang, H.; Wu, J.; Huang, X.; Zhou, Y.; Zhang, Y.; Liu, M.; Liu, Q.; Ke, S.; He, M.; Fu, H. ABO genotype alters the gut microbiota by regulating GalNAc levels in pigs. Nature 2022, 606, 358–367. [Google Scholar] [CrossRef]
- Sebastià, C.; Folch, J.M.; Ballester, M.; Estellé, J.; Passols, M.; Muñoz, M.; García-Casco, J.M.; Fernández, A.I.; Castelló, A.; Sánchez, A. Interrelation between gut microbiota, SCFA, and fatty acid composition in pigs. Msystems 2024, 9, e01049-01023. [Google Scholar] [CrossRef]
- Hwang, Y.-H.; Lee, E.-Y.; Lim, H.-T.; Joo, S.-T. Multi-Omics Approaches to Improve Meat Quality and Taste Characteristics. Food Science of Animal Resources 2023, 43, 1067. [Google Scholar] [CrossRef]
- Wu, J.; Yang, D.; Gong, H.; Qi, Y.; Sun, H.; Liu, Y.; Liu, Y.; Qiu, X. Multiple omics analysis reveals that high fiber diets promote gluconeogenesis and inhibit glycolysis in muscle. BMC genomics 2020, 21, 1–13. [Google Scholar] [CrossRef]
- Muroya, S. An insight into farm animal skeletal muscle metabolism based on a metabolomics approach. Meat science 2023, 195, 108995. [Google Scholar] [CrossRef]
- Yin, J.; Li, Y.; Tian, Y.; Zhou, F.; Ma, J.; Xia, S.; Yang, T.; Ma, L.; Zeng, Q.; Liu, G. Obese Ningxiang pig-derived microbiota rewires carnitine metabolism to promote muscle fatty acid deposition in lean DLY pigs. The Innovation 2023, 4. [Google Scholar] [CrossRef]
- Liu LiGang, L.L.; Zhang ShuMin, Z.S.; Li ZhaoHua, L.Z.; Li Na, L.N.; Jin Xin, J.X.; Yu YongSheng, Y.Y.; Zhao XiaoDong, Z.X. Comparative study on growing-finishing performance and meat quality of different hybrid combinations in pigs. 2009.
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS One 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Hall, M.; Beiko, R.G. 16S rRNA gene analysis with QIIME2. Microbiome analysis: methods and protocols 2018, 113–129. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC bioinformatics 2011, 12, 1–7. [Google Scholar] [CrossRef]
- Gaude, E.; Chignola, F.; Spiliotopoulos, D.; Spitaleri, A.; Ghitti, M.; Garcìa-Manteiga, J.M.; Mari, S.; Musco, G. muma, an R package for metabolomics univariate and multivariate statistical analysis. Current Metabolomics 2013, 1, 180–189. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K. The MetaCyc database of metabolic pathways and enzymes. Nucleic acids research 2018, 46, D633–D639. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS computational biology 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Wood, J.D.; Enser, M. Factors influencing fatty acids in meat and the role of antioxidants in improving meat quality. British journal of Nutrition 1997, 78, S49–S60. [Google Scholar] [CrossRef]
- Hoa, V.B.; Cho, S.-H.; Seong, P.-N.; Kang, S.-M.; Kim, Y.-S.; Moon, S.-S.; Choi, Y.-M.; Kim, J.-H.; Seol, K.-H. Quality characteristics, fatty acid profiles, flavor compounds and eating quality of cull sow meat in comparison with commercial pork. Asian-Australasian journal of animal sciences 2020, 33, 640. [Google Scholar] [CrossRef]
- Cheng, L.; Li, X.; Tian, Y.; Wang, Q.; Li, X.; An, F.; Luo, Z.; Shang, P.; Liu, Z.; Huang, Q. Mechanisms of cooking methods on flavor formation of Tibetan pork. Food Chemistry: X 2023, 19, 100873. [Google Scholar] [CrossRef]
- Wang, L.; Huang, Y.; Wang, Y.; Shan, T. Effects of polyunsaturated fatty acids supplementation on the meat quality of pigs: a meta-analysis. Frontiers in nutrition 2021, 8, 746765. [Google Scholar] [CrossRef]
- Oppedisano, F.; Macrì, R.; Gliozzi, M.; Musolino, V.; Carresi, C.; Maiuolo, J.; Bosco, F.; Nucera, S.; Caterina Zito, M.; Guarnieri, L. The anti-inflammatory and antioxidant properties of n-3 PUFAs: Their role in cardiovascular protection. Biomedicines 2020, 8, 306. [Google Scholar] [CrossRef]
- Qi, R.; Pfeifer, B.A.; Zhang, G. Engineering heterologous production of salicylate glucoside and glycosylated variants. Frontiers in microbiology 2018, 9, 2241. [Google Scholar] [CrossRef]
- Wang, L.; Xu, H.; Yang, H.; Zhou, J.; Zhao, L.; Zhang, F. Glucose metabolism and glycosylation link the gut microbiota to autoimmune diseases. Frontiers in Immunology 2022, 13, 952398. [Google Scholar] [CrossRef]
- Takihara, H.; Okuda, S. Glycan-related genes in human gut microbiota exhibit differential distribution and diversity in carbohydrate degradation and glycan synthesis. Frontiers in Molecular Biosciences 2023, 10, 1137303. [Google Scholar] [CrossRef]
- Andersen, S.; Møller, M.S.; Poulsen, J.-C.N.; Pichler, M.J.; Svensson, B.; Lo Leggio, L.; Goh, Y.J.; Abou Hachem, M. An 1, 4-α-glucosyltransferase defines a new maltodextrin catabolism scheme in Lactobacillus acidophilus. Applied and Environmental Microbiology 2020, 86, e00661-00620. [Google Scholar] [CrossRef]
- Lacombe, R.S.; Chouinard-Watkins, R.; Bazinet, R.P. Brain docosahexaenoic acid uptake and metabolism. Molecular Aspects of Medicine 2018, 64, 109–134. [Google Scholar] [CrossRef]
- Sinclair, A.J. Docosahexaenoic acid and the brain-what is its role? Asia Pacific journal of clinical nutrition 2019, 28, 675–688. [Google Scholar]
- Kuratko, C.N.; Barrett, E.C.; Nelson, E.B.; Norman Jr, S. The relationship of docosahexaenoic acid (DHA) with learning and behavior in healthy children: a review. Nutrients 2013, 5, 2777–2810. [Google Scholar] [CrossRef]
- EFSA Panel on Dietetic Products, N.; Allergies. Scientific Opinion on the substantiation of health claims related to EPA, DHA, DPA and maintenance of normal blood pressure (ID 502), maintenance of normal HDL-cholesterol concentrations (ID 515), maintenance of normal (fasting) blood concentrations of triglycerides (ID 517), maintenance of normal LDL-cholesterol concentrations (ID 528, 698) and maintenance of joints (ID 503, 505, 507, 511, 518, 524, 526, 535, 537) pursuant to Article 13 (1) of Regulation (EC) No 1924/2006. EFSA Journal 2009, 7, 1263. [Google Scholar] [CrossRef]
- Holub, B.J. Docosahexaenoic acid (DHA) and cardiovascular disease risk factors. Prostaglandins, Leukotrienes and Essential Fatty Acids 2009, 81, 199–204. [Google Scholar] [CrossRef]
- Wahid, S.T.; Kim, I.H. Effect of DHA supplementation on broilers' growth performance, meat quality and blood profile. Journal of Animal Physiology and Animal Nutrition 2023, 107, 703–711. [Google Scholar] [CrossRef]
- Scollan, N.D.; Price, E.M.; Morgan, S.A.; Huws, S.A.; Shingfield, K.J. Can we improve the nutritional quality of meat? Proceedings of the Nutrition Society 2017, 76, 603–618. [Google Scholar] [CrossRef]
- Carta, G.; Murru, E.; Banni, S.; Manca, C. Palmitic acid: physiological role, metabolism and nutritional implications. Frontiers in physiology 2017, 8, 902. [Google Scholar] [CrossRef]
- Beeler, J.A.; Frazier, C.R.; Zhuang, X. Putting desire on a budget: dopamine and energy expenditure, reconciling reward and resources. Frontiers in integrative neuroscience 2012, 6, 49. [Google Scholar] [CrossRef]
- Mao, X.; Yang, Q.; Chen, D.; Yu, B.; He, J. Benzoic acid used as food and feed additives can regulate gut functions. BioMed research international 2019, 2019, 5721585. [Google Scholar] [CrossRef]
- Wankhade, U.D.; Zhong, Y.; Kang, P.; Alfaro, M.; Chintapalli, S.V.; Piccolo, B.D.; Mercer, K.E.; Andres, A.; Thakali, K.M.; Shankar, K. Maternal high-fat diet programs offspring liver steatosis in a sexually dimorphic manner in association with changes in gut microbial ecology in mice. Scientific reports 2018, 8, 16502. [Google Scholar] [CrossRef]
- Usuda, H.; Okamoto, T.; Wada, K. Leaky gut: effect of dietary fiber and fats on microbiome and intestinal barrier. International journal of molecular sciences 2021, 22, 7613. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Lombard, V.; Henrissat, B. Complex carbohydrate utilization by the healthy human microbiome. PloS one 2012, 7, e28742. [Google Scholar] [CrossRef]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nature reviews Gastroenterology & hepatology 2019, 16, 461–478. [Google Scholar] [CrossRef]
- Salonen, A.; Lahti, L.; Salojärvi, J.; Holtrop, G.; Korpela, K.; Duncan, S.H.; Date, P.; Farquharson, F.; Johnstone, A.M.; Lobley, G.E. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. The ISME journal 2014, 8, 2218–2230. [Google Scholar] [CrossRef]
- Baothman, O.A.; Zamzami, M.A.; Taher, I.; Abubaker, J.; Abu-Farha, M. The role of gut microbiota in the development of obesity and diabetes. Lipids in health and disease 2016, 15, 1–8. [Google Scholar] [CrossRef]
- Silva, Y.; Bernardi, A.; Frozza, R. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol (Lausanne) 11: 25. 2020. ttps://doi.org/10.3389/fendo.2020.00025. [CrossRef]
- Metzler-Zebeli, B.U.; Klinsoda, J.; Vötterl, J.; Sharma, S.; Koger, S.; Sener-Aydemir, A. Short-, medium-, and long-chain fatty acid profiles and signaling is responsive to dietary phytase and lactic acid treatment of cereals along the gastrointestinal tract of growing pigs. Journal of animal science 2021, 99, skab117. [Google Scholar] [CrossRef]
- Zhang, L.S.; Davies, S.S. Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions. Genome medicine 2016, 8, 1–18. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: metabolism of nutrients and other food components. European journal of nutrition 2018, 57, 1–24. [Google Scholar] [CrossRef]
- Reed, K.J.; Kunz, I.G.; Scare, J.A.; Nielsen, M.K.; Turk, P.J.; Coleman, R.J.; Coleman, S.J. The pelvic flexure separates distinct microbial communities in the equine hindgut. Scientific Reports 2021, 11, 4332. [Google Scholar] [CrossRef]

| Fatty acids | SBP | LWLDP | |
|---|---|---|---|
| Mean ± std | Mean ± std | Significant level | |
| Butyric acid (C4:0) | 0.11±0.055 | 0.10±0.02 | ns |
| Caproic acid (C6:0) | 0.012±0.003 | 0.02±0.004 | ** |
| Caprylic acid (C8:0) | 0.042±0.009 | 0.08±0.02 | ** |
| Capric acid (C10:0) | 0.484±0.176 | 0.94±0.28 | ** |
| Undecanoic acid (C11:0) | 0.016±0.003 | 0.02±0.01 | ns |
| Lauric acid (C12:0) | 0.407±0.22 | 0.74±0.22 | * |
| Tridecanoic acid (C13:0) | 0.004±0.002 | 0.007±0.002 | * |
| Myristic acid (C14:0) | 6.26±3.23 | 10.96±3.05 | * |
| Myristoleic acid (C14:1n5) | 0.13±0.064 | 0.23±0.1 | ns |
| Pentadecanoic acid (C15:0) | 0.122±0.06 | 0.19±0.04 | ns |
| Palmitic acid (C16:0) | 67.40±15.28 | 101.1±20.20 | ** |
| Palmitoleic acid (C16:1n7) | 13.7±4.47 | 23.59±7.12 | * |
| Margaric acid (C17:0) | 0.74±0.41 | 1.119±0.21 | ns |
| Heptadecenoic acid (C17:1n7) | 0.62±0.33 | 0.99±0.24 | ns |
| Stearic acid (C18:0) | 40.34±10.09 | 57.9±11.72 | * |
| Elaidic acid (C18:1n9t) | 0.736±0.56 | 1.03±0.26 | ns |
| Oleic acid (C18:1n9c) | 94.60±17.27 | 132.2±26.25 | * |
| Linolelaidic acid (C18:2n6t) | 0.029±0.01 | 0.03±0.005 | ns |
| Linoleic acid (C18:2n6c) | 28.70±8.9 | 39.87±4.38 | * |
| Arachidic acid (C20:0) | 1.08±0.30 | 1.8±0.51 | * |
| γ-linolenic acid (C18:3n6) | 0.16±0.049 | 0.27±0.04 | ** |
| Gadoleic acid (C20:1) | 4.63±2.20 | 7.0±2.12 | ns |
| α-linolenic acid (C18:3n3) | 1.37±0.74 | 1.84±0.27 | ns |
| Heneicosanoic acid (C21:0) | 0.0084±0.003 | 0.014±0.004 | ns |
| Eicosadienoic acid (C20:2) | 1.95±1.05 | 2.9±0.52 | ns |
| Behenic acid (C22:0) | 0.057±0.007 | 0.10±0.016 | *** |
| Di-homo-γ-linolenic-acid (C20:3n6) | 0.63±0.16 | 0.96±0.15 | ** |
| Eicosatrienoic acid (C20:3n3) | 0.22±0.16 | 0.39±0.09 | ns |
| Arachidonic acid (C20:4n6) | 3.85±0.64 | 4.72±0.75 | ns |
| Docosadienoic acid (C22:2n6) | 0.15±0.10 | 0.076±0.03 | ns |
| Lignoceric acid (C24:0) | 0.029±0.01 | 0.04±0.008 | ns |
| Eicosapentaenoic acid (C20:5n3) | 0.071±0.03 | 0.079±0.003 | ns |
| DHA (C22:6n3) | 0.86±0.36 | 0.42±0.06 | * |
| n3 PUFA | 0.64±0.25 | 0.68±0.091 | ns |
| n6 PUFA | 1.20±0.17 | 1.50±0.22 | * |
| n3/n6 PUFA | 0.92±0.17 | 1.09±0.11 | ns |
| SFA | 117.1±29.5 | 175.32±35.69 | * |
| UFA | 152.5±35.4 | 216.78±38.83 | * |
| MUFA | 116.42±25.4 | 168.08±35.95 | * |
| PUFA | 36.11±10.1 | 48.69±5.05 | * |
| T. FA | 269.6964.6 | 392.10±73.78 | * |
| Metabolite ID | Metabolite Name | log2FC value | P-Value |
|---|---|---|---|
| M1069.pos | Di(3,7-dimethyl-1-octyl) phthalate | -0.83192 | 0.046301 |
| M1371.pos | Hecogenin | -0.98083 | 0.04355 |
| M1403.pos | (3S,4S,4aR,6R,11bS,11cS)-11c-Ethenyl-2-methyl-1,2,3,4,4a,5,6,11c-octahydro-6,4-(epoxymethano)-3,11b-methanopyrido[4,3-c] carbazole | -0.93327 | 0.012531 |
| M1453.pos | 3-Benzylpiperidine | -1.61786 | 0.041685 |
| M1615.neg | (2,2,3,3-Tetrafluoropropoxy) acetic acid | -1.00123 | 0.025629 |
| M1712.pos | (R)-4-((8S,9S,10R,13R,14S,17R)-10,13-dimethyl-3-oxo-2,3,8,9,10,11,12,13,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17-yl) pentanoic acid | -1.2868 | 0.026356 |
| M1753.pos | (R)-4-((7R,8S,9S,10R,13R,14S,17R)-7-hydroxy-10,13-dimethyl-3-oxo-2,3,6,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl) pentanoic acid | -1.06664 | 0.017033 |
| M1866.pos | N-Oleoyltaurine | -1.17102 | 0.031286 |
| M2333.neg | 10-Formyldihydrofolate | -4.12055 | 0.037883 |
| M406.neg | Inosine 5'-monophosphate (IMP) | -3.81249 | 0.018608 |
| M599.pos | Gabapentin | -1.24879 | 0.031443 |
| M718.pos | (R)-Aminocarnitine | -1.70991 | 0.030991 |
| M74.pos | PC(P-16:0/0:0) | -1.64491 | 0.02053 |
| M1022.neg | 5-KETO-GLUCONIC ACID | 1.580694 | 0.01174 |
| M1022.neg | 5-KETO-GLUCONIC ACID | 1.580694 | 0.01174 |
| M1034.neg | trans-3'-Hydroxycotinine O-. beta. -D-glucuronide | 1.655328 | 0.004261 |
| M1131.pos | N-Benzyladenine | 1.702923 | 0.047175 |
| M1135.neg | (2E)-4-Hydroxybut-2-enoic acid | 1.181196 | 0.023398 |
| M1136.neg | N-Acetylgalactosamine_6-sulfate | 3.671372 | 0.033051 |
| M122.neg | Glucose | 1.030966 | 0.023176 |
| M1386.neg | Halofenozide | 1.074294 | 0.021257 |
| M1423.pos | 2-Chloro-5,6,7,8-tetrahydroquinoxaline | 1.056675 | 0.022562 |
| M1485.neg | 1,3-Dihydroxy-7,8,9,10-tetrahydro-6H-benzo[c]chromen-6-one | 0.825711 | 0.030275 |
| M162.neg | sn-Glycerol 3-phosphate | 1.086599 | 0.044527 |
| M1690.pos | Dehydrogriseofulvin | 1.613374 | 0.029414 |
| M1740.pos | ZON | 1.655373 | 0.025052 |
| M1741.neg | 3-O-Feruloylquinic acid | 1.011234 | 0.027509 |
| M1769.neg | 5-Methoxypsoralen | 0.914868 | 0.0294 |
| M1843.neg | (1S,4R)-4-[[(2S,3aS,4S)-2-Butan-2-yl-4-hydroxy-1-oxo-3,3a-dihydro-2H-imidazo[1,2-a] indol-4-yl]methyl]-1-methyl-2,4-dihydro-1H-pyrazino[2,1-b]quinazoline-3,6-dione | 1.776069 | 0.00802 |
| Metabolite ID | Metabolite Name | log2FC value | P-Value |
|---|---|---|---|
| M1215.neg | [2.3-dihydroxypropoxy] [2-[docosa-4.7.10.13.16.19-hexaenoyloxy]-3-[octadec-9-enoyloxy] propoxy]phosphinic acid | -1.59791 | 0.004795 |
| M1462.neg | 2-Benzimidazolinone, 1-benzyl- | -0.75022 | 0.049191 |
| M1549.neg | (2-aminoethoxy) [2-[docosa-4.7.10.13.16.19-hexaenoyloxy]-3-[octadeca-1.9-dien-1-yloxy] propoxy]phosphinic acid | -1.31198 | 0.037041 |
| M1840.neg | 1-Palmitoyl-2-docosahexaenoyl-sn-glycero-3-phospho-(1'-rac-glycerol) | -1.82212 | 0.039609 |
| M2232.neg | Nicotinurate | -0.79274 | 0.047029 |
| M318.neg | Capric acid | -1.28503 | 0.011012 |
| M367.neg | cis-9-Palmitoleic acid | -0.68697 | 0.026125 |
| M524.pos | PC(P-18:0/18:1(9Z)) | -2.10508 | 0.041248 |
| M882.neg | 2-Hydroxy-2',3'-dichlorobiphenyl | -1.48729 | 0.044641 |
| M905.neg | (24E)-12,15-Dihydroxy-3-(pentopyranosyloxy)-9,19-cyclolanost-24-en-26-oic acid | -1.14334 | 0.042769 |
| M972.neg | Ethyldodecanoate | -1.24549 | 0.019967 |
| M1016.neg | [6]-Gingerdiol_3,5-diacetate | 4.088465 | 0.034114 |
| M1017.neg | Canrenone | 1.118609 | 0.049823 |
| M1087.neg | 4-Hydroxy-3-[(E)-7-hydroxy-3,7-dimethyl-4-oxooct-5-enyl]-5-[(E)-4-hydroxy-3-methylbut-2-enyl] benzoic acid | 2.15706 | 0.023635 |
| M1103.neg | 3-[(Ethylanilino)methyl] benzenesulfonic acid | 3.486875 | 0.042944 |
| M1121.neg | 8-Geranyl-7-hydroxycoumarin | 1.227386 | 0.03537 |
| M1147.neg | 5-Ethyl-N-phenyl-2-pyridinecarbothioamide | 1.908612 | 0.045952 |
| M1265.pos | PA (12:0/0:0) | 3.468987 | 0.037976 |
| M1347.pos | Progabide | 5.204754 | 0.040273 |
| M1375.pos | (2-Biphenyl) dicyclohexylphosphine | 4.559945 | 0.036992 |
| M1524.neg | Mangostine | 2.405721 | 0.042286 |
| M1635.pos | 16-Phenoxytetranorprostaglandin F2. alpha. cyclopropyl methyl amide | 7.433494 | 0.047492 |
| M1636.pos | Polyvidone | 3.963228 | 0.03826 |
| M1760.neg | Antibiotic FR 901512 | 1.689833 | 0.021109 |
| M1875.neg | N-(2-Methylphenyl) benzenesulfonamide | 1.743374 | 0.02551 |
| Metabolite ID | Metabolite Name | log2FC value | P-Value |
|---|---|---|---|
| M1057.neg | 3-Heptanone, 1,7-bis(3,4-dihydroxyphenyl)-6-methoxy- | -1.18067 | 0.047272 |
| M1061.pos | LysoPC(0:0/18:0) | -1.2026 | 0.028103 |
| M1111.pos | Flavidulol_C | -1.6351 | 0.029404 |
| M1142.pos | 1-O-Hexadecyl-2-O-(2E-butenoyl)-sn-glyceryl-3-phosphocholine | -1.61897 | 0.027758 |
| M1151.pos | LPC (18:1) | -1.47418 | 0.015168 |
| M1215.neg | [2.3-dihydroxypropoxy] [2-[docosa-4.7.10.13.16.19-hexaenoyloxy]-3-[octadec-9-enoyloxy] propoxy]phosphinic acid | -1.28737 | 0.041294 |
| M1219.pos | Bortezomib__ | -1.31754 | 0.046516 |
| M1241.pos | Americine | -3.42564 | 0.044253 |
| M1259.pos | PC (22:4(7Z,10Z,13Z,16Z)/P-18:1(9Z)) | -2.4722 | 0.025806 |
| M129.pos | LPS (18:1) | -1.9801 | 0.025347 |
| M1333.pos | 2-(3-Hydroxyphenyl)-1H-isoindole-1,3(2H)-dione | -1.6313 | 0.037885 |
| M134.pos | 1-Myristoyl-sn-glycero-3-phosphocholine (LPC (14:0/0:0)) | -1.82494 | 0.021964 |
| M135.pos | LPC (20:0) | -1.52303 | 0.025362 |
| M1453.pos | 3-Benzylpiperidine | -0.99566 | 0.039404 |
| M1522.pos | PC (18:0/18:3(6Z,9Z,12Z)) | -1.60944 | 0.01577 |
| M1091.pos | Edaravone | 0.650092 | 0.0352 |
| M1134.pos | Val-Asn | 1.534942 | 0.038822 |
| M1305.neg | Ikarugamycin | 9.372605 | 0.018966 |
| M1414.neg | 7-Oxopimara-8(14),15-dien-20-oic acid | 3.165438 | 0.036013 |
| M1479.neg | Phe(Benzoyl)-Leu-Arg | 1.469218 | 0.009001 |
| M151.neg | 4-Hydroxybenzaldehyde | 1.529151 | 0.016922 |
| M1550.pos | Sterebin_D | 1.560733 | 0.031825 |
| M1739.neg | 5-(2-Furyl)-1,3,4-oxadiazole-2(3H)-thione | 1.419276 | 0.041087 |
| M1920.neg | Erythronolactone | 0.783752 | 0.02743 |
| M1944.neg | 3-[(3-Carboxypropanoyl) amino] benzoic acid | 1.577807 | 0.023923 |
| M1979.neg | 3-(Dodecylsulfonyl)propanoic acid | 2.128976 | 0.025944 |
| M2353.neg | 16-Oxopalmitate | 2.25613 | 0.024024 |
| M299.neg | Benzoic acid | 1.6551 | 0.011532 |
| M387.neg | p-Toluquinone | 1.654802 | 0.011544 |
| M442.neg | Pseudolaric Acid B | 2.718804 | 0.048124 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




