Submitted:
02 February 2024
Posted:
06 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
Pig Samples
Metabolome Analyses
16S rRNA Microbiome Characterization
Functional Predictions
Biostatistical Analyses
3. Results
3.1. Differentially Abundant Metabolites between Infected and Control Pigs
3.2. Particular Changes Associated to Infection
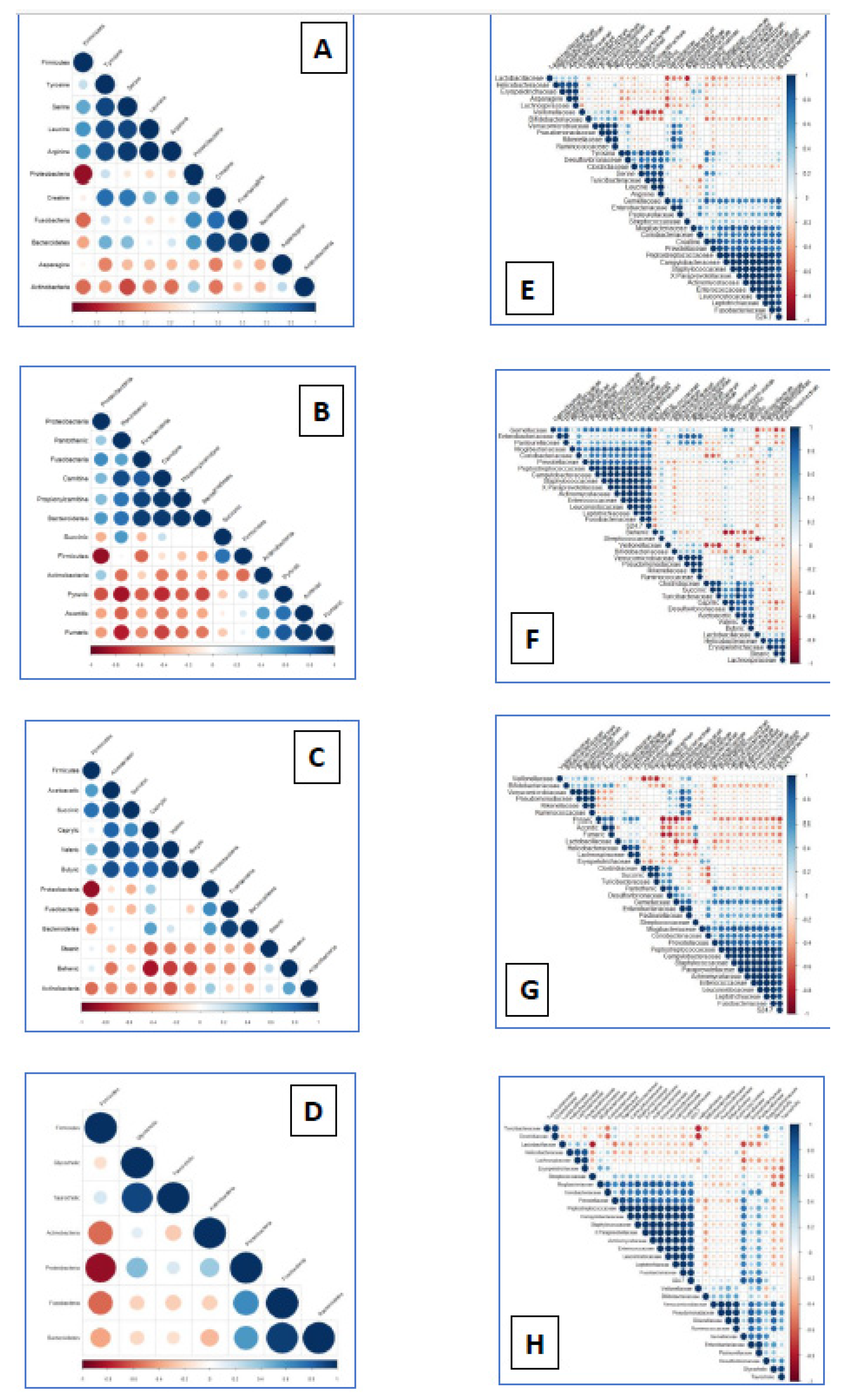
3.3. Significant Associations between Metabolome and Microbiome Changes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- EFSA; ECDC. The European Union One Health 2019 Zoonoses Report; 2021.
- Correia-Gomes, C.; Leonard, F.; Graham, D. Description of control programmes for Salmonella in pigs in Europe. Progress to date? Journal of Food Safety 2021, 41, e12916. [Google Scholar] [CrossRef]
- Collado-Romero, M.; Aguilar, C.; Arce, C.; Lucena, C.; Codrea, M.C.; Morera, L.; Bendixen, E.; Moreno, A.; Garrido, J.J. Quantitative proteomics and bioinformatic analysis provide new insight into the dynamic response of porcine intestine to Salmonella Typhimurium. Front Cell Infect Microbiol 2015, 5, 64. [Google Scholar] [CrossRef] [PubMed]
- Bellido-Carreras, N.; Argüello, H.; Zaldívar-López, S.; Jiménez-Marín, Á.; Martins, R.P.; Arce, C.; Morera, L.; Carvajal, A.; Garrido, J.J. Salmonella Typhimurium Infection Along the Porcine Gastrointestinal Tract and Associated Lymphoid Tissues. Vet Pathol 2019, 300985819843682. [Google Scholar] [CrossRef]
- Rivera-Chávez, F.; Bäumler, A.J. The Pyromaniac Inside You: Salmonella Metabolism in the Host Gut. Annu Rev Microbiol 2015, 69, 31–48. [Google Scholar] [CrossRef] [PubMed]
- Argüello, H.; Estellé, J.; Zaldívar-López, S.; Jiménez-Marín, Á.; Carvajal, A.; López-Bascón, M.A.; Crispie, F.; O’Sullivan, O.; Cotter, P.D.; Priego-Capote, F.; et al. Early Salmonella Typhimurium infection in pigs disrupts Microbiome composition and functionality principally at the ileum mucosa. Sci Rep 2018, 8, 7788. [Google Scholar] [CrossRef] [PubMed]
- Bearson, S.M.; Allen, H.K.; Bearson, B.L.; Looft, T.; Brunelle, B.W.; Kich, J.D.; Tuggle, C.K.; Bayles, D.O.; Alt, D.; Levine, U.Y.; et al. Profiling the gastrointestinal microbiota in response to Salmonella: low versus high Salmonella shedding in the natural porcine host. Infect Genet Evol 2013, 16, 330–340. [Google Scholar] [CrossRef]
- Argüello, H.; Estellé, J.; Leonard, F.C.; Crispie, F.; Cotter, P.D.; O’Sullivan, O.; Lynch, H.; Walia, K.; Duffy, G.; Lawlor, P.G.; et al. Influence of the Intestinal Microbiota on Colonization Resistance to. mSystems 2019, 4. [Google Scholar] [CrossRef]
- Borewicz, K.A.; Kim, H.B.; Singer, R.S.; Gebhart, C.J.; Sreevatsan, S.; Johnson, T.; Isaacson, R.E. Changes in the Porcine Intestinal Microbiome in Response to Infection with Salmonella enterica and Lawsonia intracellularis. PLoS One 2015, 10, e0139106. [Google Scholar] [CrossRef]
- Patel, C.; Leone, R.; Horton, M.; Powell, J. Targeting metabolism to regulate immune responses in autoimmunity and cancer. Nat Rev Drug Discov 2019, 18. [Google Scholar] [CrossRef]
- López-Bascón, M.A.; Calderón-Santiago, M.; Argüello, H.; Morera, L.; Garrido, J.J.; Priego-Capote, F. Comprehensive analysis of pig feces metabolome by chromatographic techniques coupled to mass spectrometry in high resolution mode: Influence of sample preparation on the identification coverage. Talanta 2019, 199, 303–309. [Google Scholar] [CrossRef]
- Arguello, H.; Estelle, J.; Zaldivar-Lopez, S.; Jimenez-Marin, A.; Carvajal, A.; Lopez-Bascon, M.A.; Crispie, F.; O’Sullivan, O.; Cotter, P.D.; Priego-Capote, F.; et al. Early Salmonella Typhimurium infection in pigs disrupts Microbiome composition and functionality principally at the ileum mucosa. Sci Rep 2018, 8, 7788. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Yamada, T.; Letunic, I.; Okuda, S.; Kanehisa, M.; Bork, P. iPath2.0: interactive pathway explorer. Nucleic Acids Res 2011, 39, W412–415. [Google Scholar] [CrossRef] [PubMed]
- Antunes, L.; Arena, E.; Menende, z.A.; Han, J.; Ferreira, R.; Buckner, M.; Lolic, P.; Madilao, L.; Bohlmann, J.; Borchers, C.; et al. Impact of salmonella infection on host hormone metabolism revealed by metabolomics. Infect Immun 2011, 79. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Uribe, J.; Collado-Romero, M.; Zaldivar-Lopez, S.; Arce, C.; Bautista, R.; Carvajal, A.; Cirera, S.; Claros, M.G.; Garrido, J.J. Transcriptional analysis of porcine intestinal mucosa infected with Salmonella Typhimurium revealed a massive inflammatory response and disruption of bile acid absorption in ileum. Vet Res 2016, 47, 11. [Google Scholar] [CrossRef]
- Vernocchi, P.; Del Chierico, F.; Putignani, L. Gut Microbiota Profiling: Metabolomics Based Approach to Unravel Compounds Affecting Human Health. Front Microbiol 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Wylensek, D.; Hitch, T.; Riedel, T.; Afrizal, A.; Kumar, N.; Wortmann, E.; Liu, T.; Devendran, S.; Lesker, T.; Hernández, S.; et al. A collection of bacterial isolates from the pig intestine reveals functional and taxonomic diversity. Nat Commun 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.; Kelly, B.; Logan, A.; Costa, A.; Varma, M.; Bryant, C.; Tourlomousis, P.; Däbritz, J.; Gottlieb, E.; Latorre, I.; et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 2016, 167. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Hu, S.; Du, X.; Wen, Q.; Zhong, X.; Zhou, X.; Zhou, C.; Xiong, W.; Gao, Y.; Zhang, S.; et al. Vitamin B5 Reduces Bacterial Growth via Regulating Innate Immunity and Adaptive Immunity in Mice Infected with Mycobacterium tuberculosis. Front Immunol 2018, 9. [Google Scholar] [CrossRef]
- Collado-Romero, M.; Arce, C.; Ramirez-Boo, M.; Carvajal, A.; Garrido, J.J. Quantitative analysis of the immune response upon Salmonella typhimurium infection along the porcine intestinal gut. Vet Res 2010, 41, 23. [Google Scholar] [CrossRef]
- Meena, M.; Prasad, V.; Zehra, A.; Gupta, V.; Upadhyay, R. Mannitol metabolism during pathogenic fungal-host interactions under stressed conditions. Front Microbiol 2015, 6. [Google Scholar] [CrossRef] [PubMed]
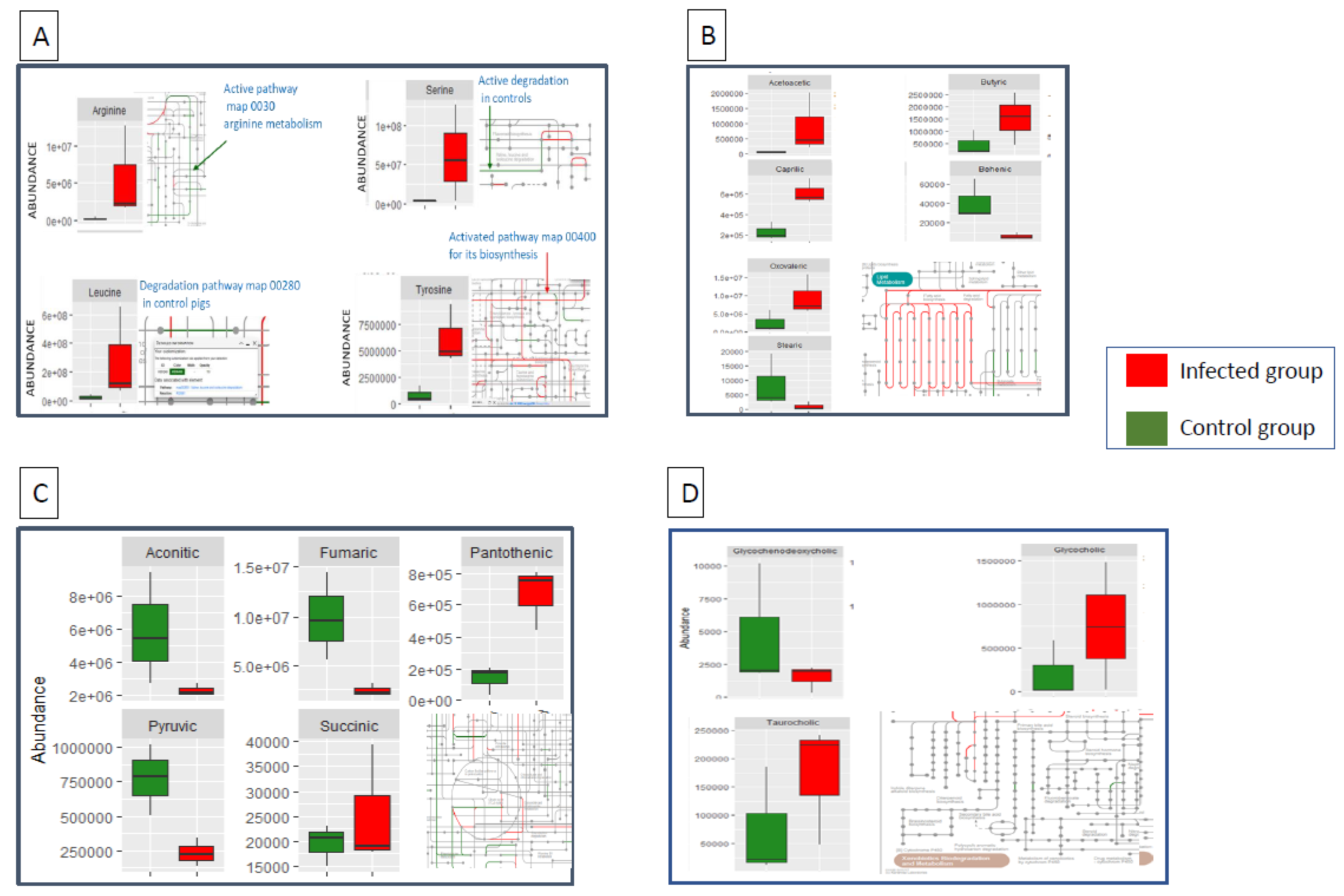
| Metabolite | Group | Abundance | P-value | |
|---|---|---|---|---|
| Mean Control | Mean Infected | |||
| Asparagine | Amino acids | 2,49E+05 | 3,10E+04 | <0.05 |
| Leucine | Amino acids | 2,23E+05 | 2,81E+06 | <0.01 |
| Phenylalanine | Amino acids | 1,06E+04 | 4,47E+03 | <0.01 |
| Serine | Amino acids | 3,25E+04 | 6,17E+05 | <0.01 |
| Tyrosine | Amino acids | 7,81E+03 | 6,20E+04 | <0.01 |
| Arginine | Amino acids | 2,15E+03 | 5,50E+04 | <0.01 |
| Creatine | Amino acids | 1,23E+04 | 6,84E+04 | <0.01 |
| Decenoylcarnitine | Amino acids | 6,72E+03 | 2,38E+03 | <0.01 |
| GCDCA | Bile acids | 4,68E+03 | 1,53E+03 | <0.05 |
| Glycocholic acid | Bile acids | 2,06E+05 | 7,45E+05 | <0.01 |
| Taurocholic acid | Bile acids | 7,30E+04 | 1,71E+05 | <0.05 |
| Tocopherol acetate | Bile acids | 1,29E+05 | 5,82E+04 | <0.05 |
| Fumaric acid | Carboxilic acids | 9,86E+04 | 2,40E+04 | <0.01 |
| Pantothenic acid | Carboxilic acids | 1,37E+03 | 6,67E+03 | <0.01 |
| Aconitic acid | Carboxylic acids | 5,87E+04 | 2,25E+04 | <0.05 |
| Pyruvic acid | Carboxylic acids | 7,72E+03 | 2,31E+03 | <0.01 |
| Succinic_acid | Carboxylic acids | 1,95E+04 | 2,54E+04 | <0.01 |
| Acetylcarnitine | Carnitines | 1,36E+04 | 5,58E+04 | <0.01 |
| Carnitine | Carnitines | 2,07E+04 | 4,85E+04 | <0.01 |
| Propionylcarnitine | Carnitines | 1,16E+04 | 4,70E+04 | <0.05 |
| Acetoacetic acid | Fatty acids | 6,44E+04 | 8,95E+05 | <0.01 |
| Butyric acid | Fatty acids | 4,56E+05 | 1,54E+06 | <0.01 |
| Oxovaleric acid | Fatty acids | 2,51E+06 | 9,47E+06 | <0.01 |
| Methylvaleric acid | Fatty acids | 6,57E+03 | 1,03E+04 | <0.05 |
| Caprylic acid | Fatty acids | 2,30E+05 | 6,13E+05 | <0.05 |
| Linoleic acid | Fatty acids | 4,85E+04 | 3,55E+04 | <0.01 |
| Stearic acid | Fatty acids | 8,60E+03 | 1,11E+03 | <0.01 |
| Behenic acid | Fatty acids | 4,16E+04 | 6,12E+03 | <0.01 |
| HODE_1 | Fatty acids | 5,19E+04 | 2,97E+03 | <0.01 |
| HODE_2 | Fatty acids | 3,65E+03 | 1,20E+04 | <0.01 |
| Hypoxanthine | Others | 3,77E+04 | 2,06E+05 | <0.01 |
| Choline | Others | 1,66E+06 | 3,66E+06 | <0.01 |
| Quinoline | Others | 5,71E+04 | 2,50E+03 | <0.01 |
| Sphinganine | Others | 1,00E+06 | 5,08E+05 | <0.01 |
| Sphingosine | Others | 3,02E+04 | 1,74E+05 | <0.01 |
| Stearoylethanolamide | Others | 6,05E+03 | 3,45E+03 | <0.01 |
| Glycericacid | Sugar derivates | 4,38E+04 | 7,66E+04 | <0.05 |
| Glycerol | Sugar derivates | 1,32E+04 | 3,28E+03 | <0.01 |
| Mannitol | Sugar derivates | 1,02E+05 | 9,71E+05 | <0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





