Submitted:
06 September 2024
Posted:
09 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Biological Material, Experimental Design and Stress Application
- ❖
- Root dehydration assay
- ❖
- Mechanical injury and virus inoculation assays
- MI+CABMV(60min) vs. control: MI_CABMV60’
- MI+CABMV(16h) vs. control: MI_CABMV16h
- MI+CPSMV(60min) vs. control: MI_CPSMV60’
- MI+CPSMV(16h) vs. control: MI_CPSMV16h
2.2. RNA Extraction and cDNA Synthesis
2.3. RNA-Seq Libraries Assembly and Differential Expression Analysis
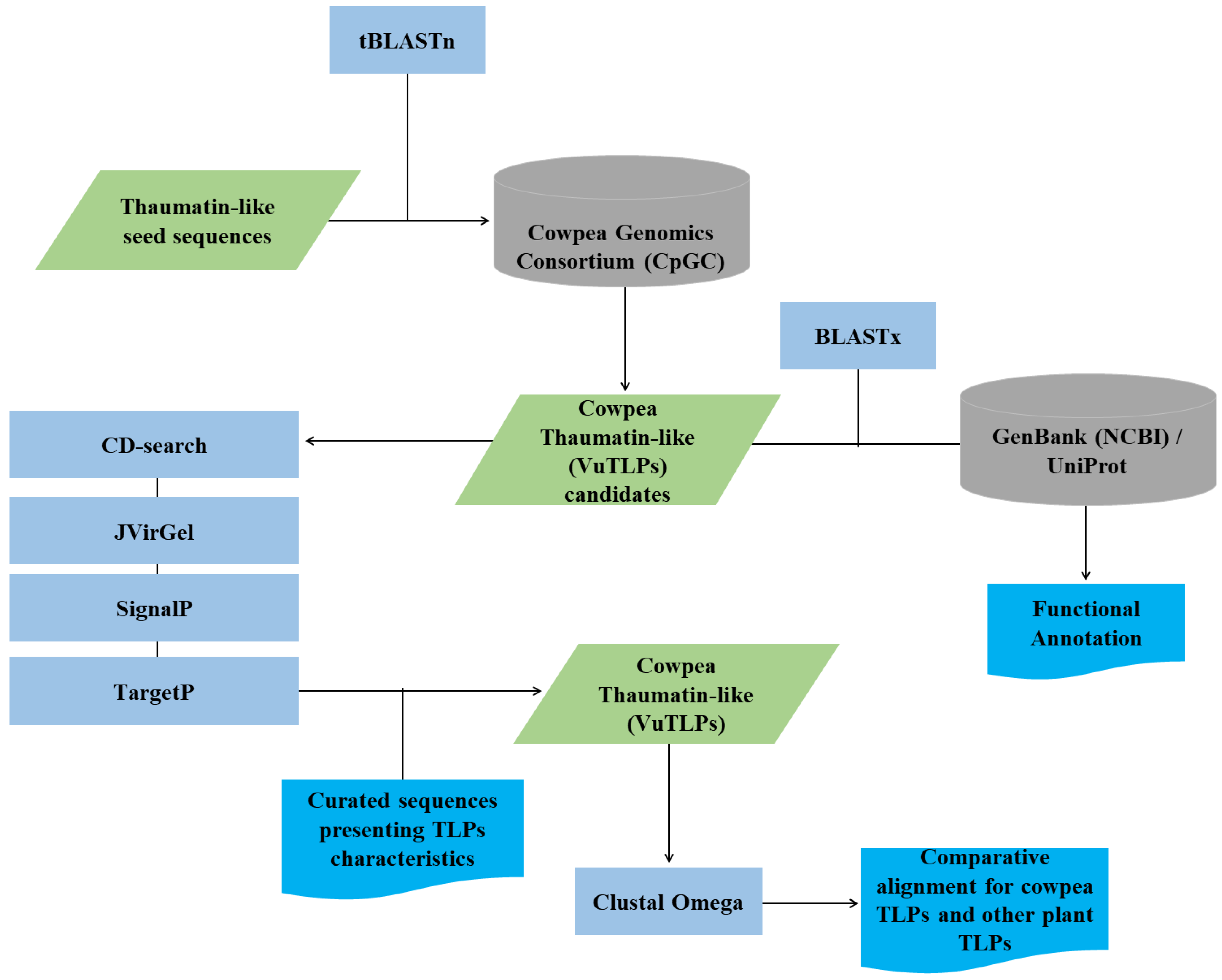
2.4. Cowpea TLPs (VuTLPs) in Silico Characterization
- Determination of the putative isoelectric point and molecular weight using ProtParam (https://web.expasy.org/protparam/; (Gasteiger et al. 2005); Figure 1);
- Prediction of signal peptide for each cowpea TLP candidate with SignalP 4.1 Server (Petersen et al. 2011; http://www.cbs.dtu.dk/services/ SignalP/);
- Prediction of the subcellular localization with TargetP 1.1 Server (Emanuelsson et al. 2007; http://www.cbs.dtu.dk/services/TargetP).
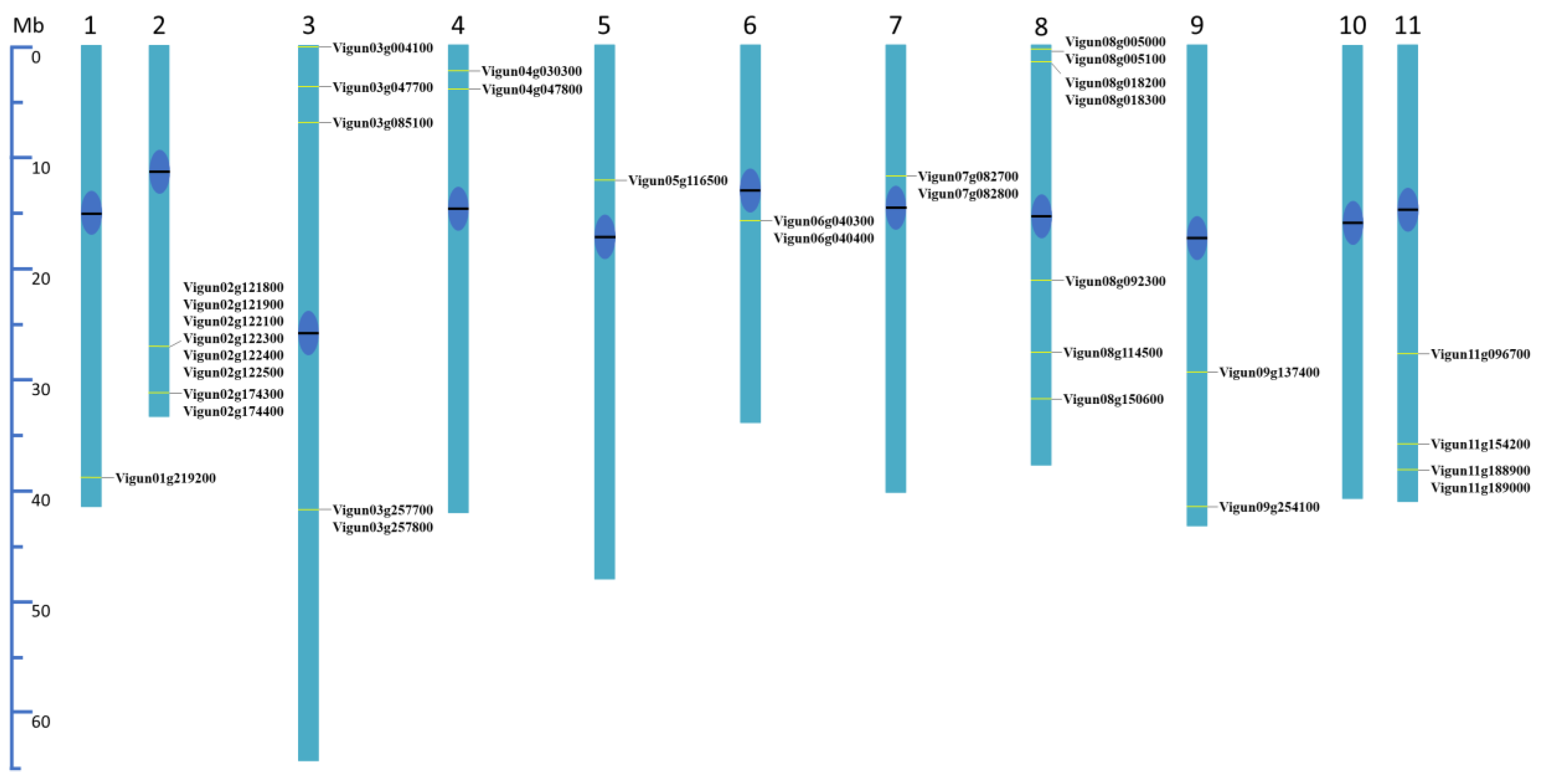
2.5. Distribution of VuTLP-Candidates in V. unguiculata Genome
2.6. NJ Analysis and VuTLPs Gene Features
2.7. Analysis of VuTLPs Duplication
- Whole genome /segmental (i.e., collinear genes in collinear blocks);
- Tandem (adjacent loci in a genome region);
- Proximal (gene pairs in nearby chromosomal region but not adjacent);
- Dispersed (other modes than segmental, tandem and proximal) duplications.
- a)
- All genes were initially classified as ‘singletons’ (i.e., without duplicates in the genome) and assigned gene ranks according to their order of appearance along chromosomes;
- b)
- BLASTp results were evaluated and the genes with BLASTp hits to other genes were re-labeled as ‘dispersed duplicates’;
- c)
- In any BLASTp hit, the two genes were re-labeled as ‘proximal duplicates’ if they had a difference of gene rank < 20 (configurable);
- d)
- In any BLASTp hit, the two genes were re-labeled as ‘tandem duplicates’ if they had a difference of gene rank = 1;
- e)
- MCScanX was executed. The anchor genes in collinear blocks were re-labeled as ‘WGD/segmental’.
- f)
- So, if a gene appeared in multiple BLASTp hits, it was assigned a unique class according to the order of priority: whole genome /segmental > tandem > proximal > dispersed.
2.8. Ratio of Synonymous (Ks) and Non-Synonymous (Ka) Substitutions per Site for Tandem Duplicated Genes
- if its value is lower than ‘1’, the duplicated gene pairs may have evolved from purifying selection (also called as negative selection; conserves the amino acid sequence);
- If Ka/Ks equal ‘1’, means neutral selection (that had no constraint for sequence divergence);
- while Ka/Ks greater than ‘1’ means positive selection (that led to different peptides).
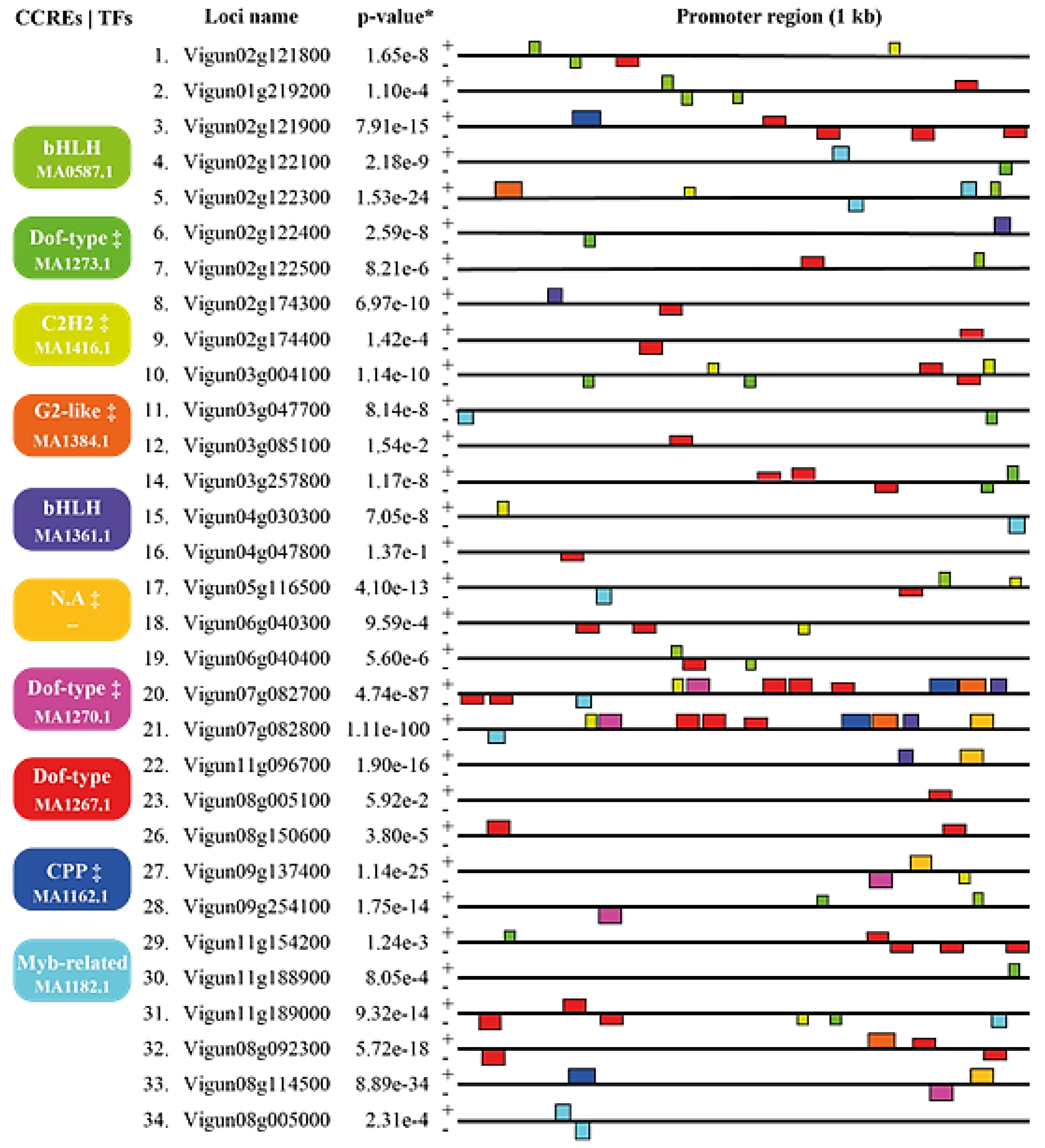
2.9. Promoter Analysis
3. Results
3.1. In Silico Identification and Characterization of VuTLPs
3.2. Mapping of VuTLPs in V. unguiculata Genome and Analysis of Gene Duplication
3.3. Ratio of Synonymous and Non-Synonymous Substitutions for Tandem Duplicated Genes
3.4. NJ Analysis and Exon-Intron Organization
3.5. Promoter VuTLPs
3.6. TLP Content and Expression in Cowpea Transcriptomes under Different Stress Types
3.6.1. Mechanical Injury (MI) and Virus Inoculation Assays
3.6.2. Root Dehydration Assay
3.6.3. MI_CABMV vs. MI_CPSMV vs. Root Dehydration Assays
3.7. RNA-Seq Data Validation by qPCR
3.7.1. MI_CABMV and MI_CPSMV Assays
3.7.2. Root Dehydration
4. Discussion
5. Conclusion
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Feller, K. MacHemer, E.L. Braun, E. Grotewold, Evolutionary and comparative analysis of MYB and bHLH plant transcription factors, Plant J. 66 (2011) 94–116. [CrossRef]
- Kohler, C. Rinaldi, S. Duplessis, M. Baucher, D. Geelen, F. Duchaussoy, B.C. Meyers, W. Boerjan, F. Martin, Genome-wide identification of NBS resistance genes in Populus trichocarpa, Plant Mol. Biol. 66 (2008) 619–636.
- A.H. Paterson, M. Freeling, H. Tang, X. Wang, Insights from the comparison of plant genome sequences. Annu. Rev. Plant. Biol. 61 (2010) 349–372. [CrossRef]
- A.M. Benko-Iseppon, S.L. Galdino, T. Calsa, E.A. Kido, A. Tossi, L.C. Belarmino, S. Crovella, Overview on plant antimicrobial peptides., Curr. Protein Pept. Sci. 11 (2010) 181–188.
- Hu, J. Jin, A.Y. Guo, H. Zhang, J. Luo, G. Gao, GSDS 2.0: an upgraded gene feature visualization server, Bioinformatics 31 (2015) 1296–1297. [CrossRef]
- Petre, I. Major, N. Rouhier, S. Duplessis, Genome-wide analysis of eukaryote thaumatin-like proteins (TLPs) with an emphasis on poplar., BMC Plant Biol. 11 (2011) 33. [CrossRef]
- Ruperti, L. Cattivelli, S. Pagni, A. Ramina, Ethylene-responsive genes are differentially regulated during abscission, organ senescence and wounding in peach (Prunus persica), J. Exp. Bot. 53 (2002) 429–437.
- Liu, X. He, W. Li, C. Chen, F. Ge, Molecular cloning of a thaumatin-like protein gene from Pyrus pyrifolia and overexpression of this gene in tobacco increased resistance to pathogenic fungi, Plant Cell. Tissue Organ Cult. 111 (2012) 29–39. [CrossRef]
- Fierens, S. Rombouts, K. Gebruers, H. Goesaert, K. Brijs, J. Beaugrand, G. Volckaert, S. Van Campenhout, P. Proost, C.M. Courtin, J.A. Delcour, TLXI, a novel type of xylanase inhibitor from wheat (Triticum aestivum) belonging to the thaumatin family, Biochem. J. 403 (2007) 583–591.
- Gasteiger, C. Hoogland, A. Gattiker, S. Duvaud, M.R. Wilkins, R.D. Appel, A. Bairoch; Protein Identification and Analysis Tools on the ExPASy Server;(In) John M. Walker (ed): The Proteomics Protocols Handbook, Humana Press (2005). pp. 571-607.
- E.A. Kido, J.R.C. Ferreira-Neto, R.L.O. Silva, L.C. Belarmino, N.M. Soares-Cavalcanti, V. Pandolfi, M.D. Silva, A.L. Nepomuceno, A.M. Benko-Iseppon, Expression dynamics and genome distribution of osmoprotectants in soybean: identifying important components to face abiotic stress, BMC Bioinformatics 14 (2013) S7.
- Sievers, A. Wilm, D. Dineen, T.J. Gibson, K. Karplus, W. Li, R. Lopez, H. McWilliam, M. Remmert, J. Söding, J.D. Thompson, D.G. Higgins, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega, Mol. Syst. Biol. 7 (2011).
- Song, R.M. Goodman, Cloning and identification of the promoter of the tobacco Sar8.2b gene, a gene involved in systemic acquired resistance, Gene. 290 (2002) 115–124.
- Breiteneder, C. Radauer, A classification of plant food allergens, J. Allergy Clin. Immunol. 113 (2004) 821–830.
- Ellegren, Comparative genomics and the study of evolution by natural selection, Mol. Ecol. 17 (2008) 4586–4596. [CrossRef]
- H. Li, W. Huang, Z.W. Liu, Y.X. Wang, J. Zhuang, Transcriptome-based analysis of Dof family transcription factors and their responses to abiotic stress in tea plant (Camellia sinensis), Int. J. Genomics. 2016 (2016). [CrossRef]
- H. Tachi, K. Fukuda-Yamada, T. Kojima, M. Shiraiwa, H. Takahara, Molecular characterization of a novel soybean gene encoding a neutral PR-5 protein induced by high-salt stress, Plant Physiol. Biochem. 47 (2009) 73–79.
- H. van der Wel, K. Loeve, Isolation and characterization of thaumatin I and II, the sweet-tasting proteins from Thaumatococcus daniellii Benth, Eur. J. Biochem. 31 (1972) 221–225.
- H. Wang, S. Zhao, Y. Gao, J. Yang, Characterization of dof transcription factors and their responses to osmotic stress in poplar (Populus trichocarpa), PLoS One. 12 (2017) 1–19. [CrossRef]
- Cao, Y. Lv, Z. Hou, X. Li, L. Ding, Expansion and evolution of thaumatin-like protein (TLP) gene family in six plants, Plant Growth Regul. 79 (2016) 299–307. [CrossRef]
- Grenier, C. Potvin, J. Trudel, A. Asselin, Some thaumatin-like proteins hydrolyse polymeric β-1,3-glucans, Plant J. 19 (1999) 473–480.
- J. Madroñero, S.P. Rodrigues, T.F.S. Antunes, P.M.V. Abreu, J.A. Ventura, A.A.R. Fernandes, P.M.B. Fernandes, Transcriptome analysis provides insights into the delayed sticky disease symptoms in Carica papaya, Plant Cell Rep. 37 (2018) 967–980.
- J. Trudel, J. Grenier, C. Potvin, a Asselin, Several thaumatin-like proteins bind to beta-1,3-glucans., Plant Physiol. 118 (1998) 1431–1438.
- J. Xiao, H. Cheng, X. Li, J. Xiao, C. Xu, S. Wang, Rice WRKY13 Regulates Cross Talk between Abiotic and Biotic Stress Signaling Pathways by Selective Binding to Different cis-Elements, Plant Physiol. 163 (2013) 1868–1882. [CrossRef]
- J.C. Misas-Villamil, R.A.L. van der Hoorn, Enzyme-inhibitor interactions at the plant-pathogen interface, Curr. Opin. Plant Biol. 11 (2008) 380–388. [CrossRef]
- J.C. Oliveros VENNY: An interactive tool for comparing lists with Venn diagrams. Available: http://bioinfogp.cnb.csic.es/tools/venny/ index.html. (2007) Accessed 17 December 2018.
- J.J. Burdon, P.H. Thrall, Coevolution of plants and their pathogens in natural habitats, Science 324 (2009) 755–756. [CrossRef]
- J.J. Liu, R. Sturrock, A.K.M. Ekramoddoullah, The superfamily of thaumatin-like proteins: its origin, evolution, and expression towards biological function, Plant. Cell Rep. 29 (2010) 419–436. [CrossRef]
- Hiroyuki, R. Terauchi, Regulation of expression of rice thaumatin-like protein: inducibility by elicitor requires promoter W-box elements, Plant Cell Rep. 27 (2008) 1521–1528.
- K. Mao, Q. Dong, C. Li, C. Liu, F. Ma, Genome Wide Identification and Characterization of Apple bHLH Transcription Factors and Expression Analysis in Response to Drought and Salt Stress, Front. Plant Sci. 8 (2017). [CrossRef]
- K. Min, S.C. Ha, P.M. Hasegawa, R.A. Bressan, D.J. Yun, K.K. Kim, Crystal structure of osmotin, a plant antifungal protein, Proteins 54 (2004) 170–173.
- K. Sugimoto, MYB-Related Transcription Factor NtMYB2 Induced by Wounding and Elicitors is a Regulator of the Tobacco Retrotransposon Tto1 and Defense-Related Genes, Plant Cell Online. 12 (2000) 2511–2528.
- L.L.B. Amorim, J.R.C. Ferreira-Neto, J.P. Bezerra-Neto, V. Pandolfi, F.T. Araújo, M.K. Silva Matos, M.G. Santos, E.A. Kido, A.M. Benko-Iseppon, Cowpea and abiotic stresses: Identification of reference genes for transcriptional profiling by qPCR, Plant Methods. 14 (2018).
- L.S. Melcher, M.B. Sela-Buurlage, S.A. Vloemans, C.P. Woloshuk, J.S. van Roekel, J. Pen, P.J. van den Elzen, B.J. Cornelissen, Extracellular targeting of the vacuolar tobacco proteins AP24, chitinase and b-1,3-glucanase in transgenic plants, Plant Mol. Biol. 21 (1993) 583–593.
- Hayashi, S. Shiro, H. Kanamori, S. Mori-Hosokawa, H. Sasaki-Yamagata, T. Sayama, M. Nishioka, M. Takahashi, M. Ishimoto, Y. Katayose, A. Kaga, K. Harada, H. Kouchi, Y. Saeki, Y. Umehara, A thaumatin-like protein, Rj4, controls nodule symbiotic specificity in soybean, Plant Cell Physiol. 55 (2014) 1679–1689.
- Kappagantu, J.M. Bullock, M.E. Nelson, K.C. Eastwell, Hop stunt viroid : Effect on Host (Humulus lupulus) Transcriptome and Its Interactions With Hop Powdery Mildew (Podospheara macularis), Mol. Plant-Microbe Interact. 30 (2017) 842–851.
- M. Lynch, J.S. Conery, The evolutionary fate and consequences of duplicate genes, Science 290 (2000) 1151–1155. [CrossRef]
- M.A. Batalia, A.F. Monzingo, S. Ernst, W. Roberts, J.D. Robertus, The crystal structure of the antifungal protein zeamatin, a member of the thaumatin-like, PR-5 protein family, Nat. Struct. Biol. 3 (1996) 19–23. [CrossRef]
- M.B. Eisen, P.T. Spellman, P.O. Brown, D. Botstein. Cluster analysis and display of genome-wide expression patterns, Proc Natl Acad Sci USA. 95 (1998) 14863–14868. [CrossRef]
- M.D. Robinson, D.J. McCarthy, G.K. Smyth, edgeR: A Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics. 26 (2009) 139–140. [CrossRef]
- M.J. Kim, B.K. Ham, H.R. Kim, I.J. Lee, Y.J. Kim, K.H. Ryu, Y.I. Park, K.H. Paek, In vitro and in planta interaction evidence between Nicotiana tabacum thaumatin-like protein 1 (TLP1) and Cucumber mosaic virus proteins, Plant Mol. Biol. 59 (2005) 981–994.
- M.W. Pfaffl, G.W. Horgan, L. Dempfle, REST 2009 Software User Guide: For gene expression analysis using real-time Sample & Assay Technologies QIAGEN Sample and Assay Technologies, (2009) 1–28.
- N.K. Singh, D.E. Nelson, D. Kuhn, P.M. Hasegawa, R.A. Bressan, Molecular cloning of osmotin and regulation of its expression by ABA and adaptation to low water potential, Plant. Physiol. 90 (1989) 1096–1101. [CrossRef]
- N.K. Singh, K.R.R. Kumar, D. Kumar, P. Shukla, P.B. Kirti, Characterization of a pathogen induced thaumatin-like protein gene AdTLP from Arachis diogoi, a wild peanut, PLoS One. 8 (2013) 1–18. [CrossRef]
- Emanuelsson, S. Brunak, G. von Heijne, H. Nielsen, Locating proteins in the cell using TargetP, SignalP and related tools, Nat. Protoc. 2 (2007) 953–971.
- Pandey, V. Ramegowda, M. Senthil-Kumar, Shared and unique responses of plants to multiple individual stresses and stress combinations: physiological and molecular mechanisms, Front. Plant Sci. 6 (2015) 1–14.
- Wang, L. Su, H. Gao, X. Jiang, X. Wu, Y. Li, Q. Zhang, Y. Wang, F. Ren, Genome-Wide Characterization of bHLH Genes in Grape and Analysis of their Potential Relevance to Abiotic Stress Tolerance and Secondary Metabolite Biosynthesis, Front. Plant Sci. 9 (2018) 1–15.
- Ghosh, C. Chakrabarti, Crystal structure analysis of NP24-I: A thaumatin-like protein, Planta. 228 (2008) 883–890. [CrossRef]
- Velazhahan, S.K. Datta, S. Muthukrishnan, Pathogenesis-related proteins in plants, in: S.K. Datta, S. Muthukrishnan (Eds.), CRC press Boca Raton, Florida, United States, 1999, pp. 107-129.
- R.C. Misra, Sandeep, M. Kamthan, S. Kumar, S. Ghosh, A thaumatin-like protein of Ocimum basilicum confers tolerance to fungal pathogen and abiotic stress in transgenic Arabidopsis., Sci. Rep. 6 (2016) 25340. [CrossRef]
- R.D.M. Page, TREEVIEW: An application to display phylogenetic trees on personal computers. Comp Appl Biosci. 12 (1996) 357–358.
- R.W. Skadsen, P. Sathish, H.F. Kaeppler, Expression of thaumatin-like permatin PR-5 genes switches from the ovary wall to the aleurone in developing barley and oat seeds, Plant Sci. 156 (2000) 11–22. [CrossRef]
- Chowdhury, A. Basu, S. Kundu, Overexpression of a New Osmotin-Like Protein Gene (SindOLP) Confers Tolerance against Biotic and Abiotic Stresses in Sesame, Front. Plant Sci. 8 (2017) 410.
- Gupta, J.A. Stamatoyannopoulos, T.L. Bailey, W.S. Noble, Quantifying similarity between motifs, Genome Biol. 8 (2007). [CrossRef]
- S. Kumar, G. Stecher, K. Tamura, MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets., Mol. Biol. Evol. 33 (2016) msw054. [CrossRef]
- S. Lonardi, M. Muñoz-amatriaín, Q. Liang, S. Shu, I. Steve, The genome of cowpea (Vigna unguiculata [ L .] Walp.), (2019) 1–49.
- S. Mellacheruvu, S. Tamirisa, D.R. Vudem, V.R. Khareedu, Pigeonpea Hybrid-Proline-Rich Protein (CcHyPRP) Confers Biotic and Abiotic Stress Tolerance in Transgenic Rice, Front. Plant Sci. 6 (2016) 1–13. [CrossRef]
- S. Singh, S. Chand, N.K. Singh, T.R. Sharma, Genome-wide distribution, organisation and functional characterization of disease resistance and defence response genes across rice species, PLoS One 10 (2015) 1–29.
- S.F. Altschul, W. Gish, W. Miller, E.W. Myers, D.J. Lipman, T. Pennsylvania, U. Park, Basic Local Alignment Search Tool, J Mol Biol. 215 (1990) 403–410. [CrossRef]
- L. Bailey, C. Elkan, Fitting a mixture model by expectation maximization to discover motifs in biopolymers, Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28-36, AAAI Press, Menlo Park, California, 1994.
- T.N. Petersen, S. Brunak, G. von Heijne, H. Nielsen, SignalP 4.0: discriminating signal peptides from transmembrane regions, Nat. Methods 8 (2011) 785–786. [CrossRef]
- W.C. Hon, M. Griffith, A. Mlynarz, Y.C. Kwok, D.S.C. Yang, Antifreeze proteins in winter rye are similar to pathogenesis-related proteins, Plant Physiol. 109 (1995) 879–889.
- W.K. Roberts, C.P. Selitrennikoff, Zeamatin, an antifungal protein from maize with membrane-permeabilizing activity, J. Gen. Microbiol. 136 (1990) 1771–1778.
- X. Wang, C. Tang, L. Deng, G. Cai, X. Liu, B. Liu, Q. Han, H. Buchenauer, G. Wei, D. Han, L. Huang, Z. Kang, Characterization of a pathogenesis-related thaumatin-like protein gene TaPR5 from wheat induced by stripe rust fungus, Physiol. Plant. 139 (2010) 27–38.
- X. Yan, H. Qiao, X. Zhang, C. Guo, M. Wang, Y. Wang, X. Wang, Analysis of the grape (Vitis vinifera L.) thaumatin-like protein (TLP) gene family and demonstration that TLP29 contributes to disease resistance, Sci. Rep. 7 (2017) 1–14. [CrossRef]
- X. Zhu, L. Qi, X. Liu, S. Cai, H. Xu, R. Huang, J. Li, X. Wei, Z. Zhang, The Wheat Ethylene Response Factor Transcription Factor Pathogen-induced ERF1 Mediates Host Responses to Both the Necrotrophic Pathogen Rhizoctonia cerealis and Freezing Stresses, Plant Physiol. 164 (2014) 1499–1514. [CrossRef]
- Y. Xu, F.L. Chang, D. Liu, M.L. Narasimhan, K.G. Raghothama, P.M. Hasegawa, R.A. Bressan, Plant defense genes are synergistically induced by ethylene and methyl jasmonate, Plant. Cell 6 (1994) 1077–1085.
- Y. Yang, H. Guan, F. Wei, Z. Li, S. Yang, J. Huang, Genome-wide identification of thaumatin-like protein family genes in Panax notoginseng and analysis of their responses to Fusarium solani infection, Genet Resour Crop Evol (2024) 71, 2267–2279. [CrossRef]
- Y. Zhang, H. Yan, X. Wei, J. Zhang, H. Wang, D. Liu, Expression analysis and functional characterization of a pathogen-induced thaumatin-like gene in wheat conferring enhanced resistance to Puccinia triticina, J Plant Interactions 12 (2017) 332–339. [CrossRef]
- Y.C. Jung, H.J. Lee, S.S. Yum, W.Y. Soh, D.Y. Cho, C.K. Auh, T.K. Lee, H.C. Soh, Y.S. Kim, S.C. Lee, Drought-inducible - but ABA-independent - thaumatin-like protein from carrot (Daucus carota L.), Plant Cell Rep. 24 (2005) 366–373.
- Z. Wang, M. Gerstein, M. Snyder, RNA-Seq: a revolutionary tool for transcriptomics, Nature Rev. Genet. 10 (2009) 57–63.
| Gene | Loci | Duplication pattern | Signal peptide | MW (kDa) | pI | REDDD motif | Subcelular location | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VuTLP1 | Vigun01g219200 | WGD or Segmental | YES | 23.27 | 7.32 | R | E | D | D | D | Extracelular |
| VuTLP2 | Vigun02g121800 | WGD or Segmental | YES | 30.75 | 4.35 | R | E | D | D | D | Extracelular |
| VuTLP3 | Vigun02g121900 | WGD or Segmental | YES | 28.48 | 4.05 | R | E | D | D | D | Extracelular |
| VuTLP4 | Vigun02g122100 | Proximal | YES | 21.57 | 5.66 | R | E | D | D | D | Extracelular |
| VuTLP5 | Vigun02g122300 | Tandem | NO | 31.03 | 7.31 | R | E | Y | E | D | Membrana Plasmática |
| VuTLP6 | Vigun02g122400 | Tandem | YES | 28.11 | 8.19 | Q | E | D | E | D | Extracelular |
| VuTLP7 | Vigun02g122500 | Tandem | YES | 21.36 | 6.86 | R | E | Y | E | D | Extracelular |
| VuTLP8 | Vigun02g174300 | WGD or Segmental | YES | 21.56 | 4.04 | R | E | D | D | D | Extracelular |
| VuTLP9 | Vigun02g174400 | Tandem | YES | 24.06 | 6.74 | R | E | D | D | D | Extracelular |
| VuTLP10 | Vigun03g004100 | WGD or Segmental | YES | 23.80 | 8.76 | R | E | D | D | D | Extracelular |
| VuTLP11 | Vigun03g047700 | WGD or Segmental | YES | 21.48 | 4.29 | R | E | D | D | D | Extracelular |
| VuTLP12 | Vigun03g085100 | Dispersed | YES | 28.55 | 6.06 | R | E | D | N | D | Extracelular |
| VuTLP13 | Vigun03g257700 | WGD or Segmental | YES | 28.72 | 4.19 | R | E | D | D | D | Extracelular |
| VuTLP14 | Vigun03g257800 | WGD or Segmental | YES | 27.88 | 5.71 | R | E | D | D | D | Extracelular |
| VuTLP15 | Vigun04g030300 | WGD or Segmental | YES | 24.89 | 8.40 | R | E | D | D | D | Extracelular |
| VuTLP16 | Vigun04g047800 | Dispersed | YES | 25.18 | 7.32 | R | E | D | D | D | Extracelular |
| VuTLP17 | Vigun05g116500 | WGD or Segmental | YES | 23.47 | 7.32 | R | E | D | D | D | Extracelular |
| VuTLP18 | Vigun06g040300 | Tandem | YES | 108.18 | 5.22 | R | E | Y | Q | - | Membrana Plasmática |
| VuTLP19 | Vigun06g040400 | Tandem | NO | 13.75 | 5.46 | R | E | D | D | - | Extracelular |
| VuTLP20 | Vigun07g082700 | Tandem | NO | 27.34 | 4.77 | R | E | D | D | D | Extracelular |
| VuTLP21 | Vigun07g082800 | Tandem | NO | 27.34 | 4.77 | R | E | D | D | D | Extracelular |
| VuTLP22 | Vigun08g005000 | WGD or Segmental | YES | 30.35 | 4.21 | R | E | D | D | D | Extracelular |
| VuTLP23 | Vigun08g005100 | Tandem | YES | 30.42 | 4.24 | R | E | D | D | D | Extracelular |
| VuTLP24 | Vigun08g018200 | Tandem | NO | 29.20 | 4.65 | R | E | D | D | D | Extracelular |
| VuTLP25 | Vigun08g018300 | Tandem | YES | 22.77 | 4.30 | R | E | D | D | D | Extracelular |
| VuTLP26 | Vigun08g092300 | Dispersed | YES | 28.05 | 8.36 | R | E | D | D | D | Extracelular |
| VuTLP27 | Vigun08g114500 | WGD or Segmental | NO | 33.67 | 4.93 | R | E | D | D | D | Extracelular |
| VuTLP28 | Vigun08g150600 | WGD or Segmental | YES | 23.48 | 7.52 | R | E | D | D | D | Extracelular |
| VuTLP29 | Vigun09g137400 | WGD or Segmental | YES | 29.89 | 7.32 | R | E | D | D | D | Extracelular |
| VuTLP30 | Vigun09g254100 | WGD or Segmental | YES | 31.53 | 4.86 | R | E | D | D | D | Extracelular |
| VuTLP31 | Vigun11g096700 | Dispersed | YES | 23.26 | 4.11 | R | E | D | D | D | Extracelular |
| VuTLP32 | Vigun11g154200 | Dispersed | YES | 24.16 | 6.80 | R | Q | G | D | D | Extracelular |
| VuTLP33 | Vigun11g188900 | WGD or Segmental | YES | 33.65 | 4.00 | R | E | D | D | D | Extracelular |
| VuTLP34 | Vigun11g189000 | WGD or Segmental | YES | 30.44 | 8.15 | R | E | D | D | D | Extracelular |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





