1. Introduction
Sunlight is the most prominent factor for plant development, which is generally dynamic over land surface and ranges from milliseconds to hours [
1,
2]. The sunlight quantity was affected by the instantaneous amount of light, as well as the cumulative amount of light delivered each day. Changes in light density or duration for crops are usually caused by the variation of the solar irradiance and shading from overlapping leaves of surrounding plants [
3]. In recent years, solar irradiance has decreased by 28%-49% because of heavy aerosol and haze pollution induced by the development of industry and vehicle exhaust [
4]. Except for environmental factors, plants also have to face the insufficient light caused by global climate change. So light insufficient has been an ongoing global phenomenon nowadays and will strongly be impacting on crop production [
5]. So it’s worthy to research on the effect of light supplementary on plant breeding.
Rapeseed (
Brassica napus L.) is one of the most important source for protein and oil produced throughout the world [
6]. Southwest of China is a representative rapeseed production region, but rainy and cloudy conditions often occur in Sichuan and Chongqing these two districts during the whole developmental stage of rapeseed (Octorber to next March), that will cause low light intensity and has severely negative impacts on rapeseed quality and yield. Yunnan province is the district with much more light intensity and duration, so the seed oil content or yield of rapeseed is usually higher than other districts. Previous study found that the electron transport rate, photochemical efficiency and the net photosynthetic rate have effect on photosynthetic production, and also less sunlight leads to decrease in the chlorophyll a/b ratio and key enzyme activities in photosynthesis (e.g. phosphoenolpyruvate carboxylase) [
7,
8,
9]. So measuring the photosynthetic indicators will help us understanding crop effective light utilization especially in greenhouse farming.
The seed oil content of rapeseed is generally between 35% and 50%, which is mainly decide by genetic effects and genotype × environment interactions (GE). Transcriptome sequencing was widely used to investigate the formation mechanism of seed oil content in the recent years. Rapeseed seeds from four varieties were analysis based on transcriptome sequencing, revealing that the genes involved in lipid synthesis are conserved and species-specific [
10]. The three different parts of oil palm fruits and seeds were analysis by transcriptomes to found that EgWRI1-1 and EgWRI1-2 transcription factors were heavily transcribed in the process of oil accumulation in pericarp and endosperm [
11]. The expression of lipid metabolites and oil synthesis-related genes were compared in high and low oil content varieties, and the results showed that there were significant differences in different parts of seeds [
12]. So some key regulatory genes related to oil synthesis can be identified by transcriptome analysis of differentially expressed genes (DEGs) in the seeds of rapeseed, providing a theoretical basis for future high-oil rapeseed breeding.
However, to date, there were few studies that focused on the effect of light quatity on the plant that during their entire life cycle. Therefore, a light quatity control experiment (rapeseed grown under natural condition and light supplement by LED) was conducted in Chongqing, China in this study. The main objectives of this study were to investigate the DEGs and phenotypic of leaf growth and seed oil content regulated by light quatity in rapeseed. This finding can provide information of light efficiently utilizing for the rapeseed breeding regions affected by solar irradiance.
2. Method
2.1. Plant Materials and Experimental Design
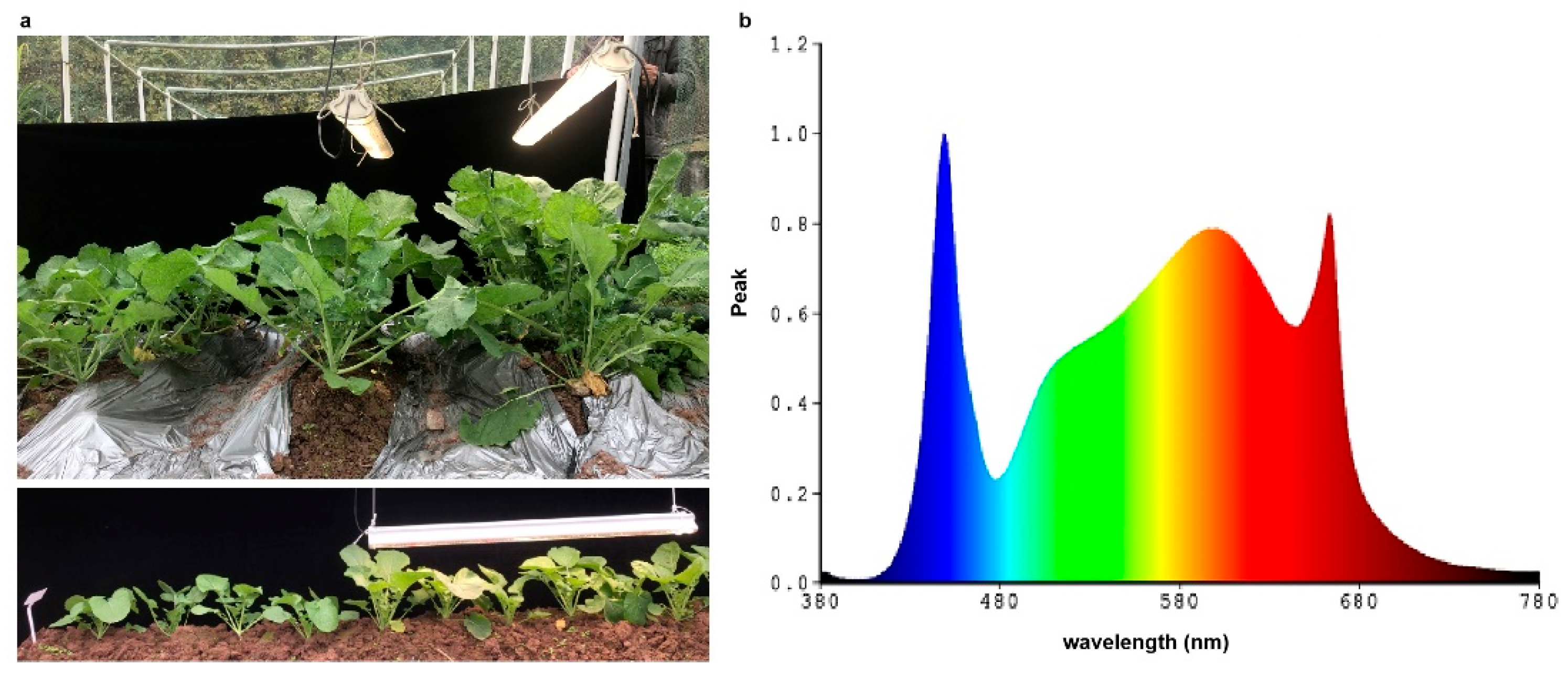
The Brassica napus variety, Qingyou 3, was used in this experiment as it is broadly adaptable in China. The variety Qingyou 3 also have high seed oil content and yield. Field experimentswere conducted in Beibei district (29◦48′N, 106◦24′E), Chongqing, China in 2021. Seedlings were raised in a seedbed for 30 days, then uniform healthy plants were transplanted in each pot; the pots contain 50.0 × 20.0 cm apart. There were 30 pots for each treatment, with a total of 60 pots. Half a month after transplanting, compare to natural conditions as control and light supplement treatment by LEDs was applied to the test groups. Light supplement conditions were provided by LEDs (KS-Z-D60, China) (
Figure 1a). The light supplement time is from 7:00 am to 19:00 pm. Spectrometer (HR-550) was used to measure the luminance. The wavelength of LEDs is shown in
Figure 1b.
After 30 days of light treatment, the young leaves from representative individual plants were collected and then mixed as leaf samples. And leaves were frozen in liquid nitrogen immediately and stored at -80 °C until for transcriptome sequencing and hormone (ABA and IAA) analysis. Free IAA and ABA measurements were analyzed by liquid chromatography-tandem mass spectrometry as described previously [
13]. Seed of 30 and 40 days after pollination (DAP) were also collected for RNA extraction, followed by transcriptome sequencing. So 6 samples with 3 biological replicates each were collected to transcriptome sequencing.
At harvest, 10 representative individual plants from each treatment were selected. Seed oil content and protein content were determined using a near-infrared rapid quality analyser (NIR System 6500, FOSS, Sweden) according to the method of Dimov et al. (2012).
2.2. Microscopic Observations
Three middle segments of leaves (1 cm
2) were sampled to do paraffin section and fixed in FAA at 4℃ under vacuum as described previously [
14]. Leaves section from different light condition were imaged, leaves thickness contains spongy tissue and palisade tissue in each segment of the leaves were measured.
2.3. Photosynthetic Performance Test
The rapeseed leaves were used for net photosynthetic efficiency, stomatal conductance and intercellular CO2 concentration tests using an Li-6400 (Li-Cor, Lincoln, NE) on 30 days after treatment and before the flowering stage. Three fully expanded leaves of each plant was measured on a sunny day at 10: 00 am, and the mean value was used for data analysis.
2.4. Differential Expression Analysis and Functional Annotation
http://www.genoscope.cns.fr/brassicanapus/data/ was used as the reference genome. Total RNA was extracted from rapeseed leaves and seeds at 30 and 40 DPA. RNA from all these samples were conducted transcriptom sequencing by Majorbio Technologies Co., Ltd. (Shanghai, China). Per kilobase of transcript per million mapped reads (RPKM) were read as gene expression levels. Differentially expressed genes (DEGs) were defined with an false discovery rate (FDR)<0.01 and |log2(fold change)|≥1. Two samples were compared using the “LEDs vs Control” method, if the expression level of a DEG in sample “LEDs” was higher than that in “Control”, the DEG was upregulated; otherwise, it was downregulated. To further characterize the function of DEGs, they were mapped to Gene Ontology (GO) classifications using Blast2GO (Conesa and Götz, 2008) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment was analysis by KOBAS (Xie et al., 2011).The data were analyzed on the online platform of Majorbio Cloud Platform.
2.5. qRT-PCR Analysis and Statistical Analysis
The primers were designed using The primers were designed using PRIMER-BLAST of NCBI’s primer designing tool (Table S1). The Bio-Rod quantitative PCR instrument was adopted to perform real-time fluorescence quantitative PCR. The following thermocycle conditions was used: 95 ◦C for 5 min, 38 cycles at 95 ◦C for 15 s, 56 ◦C for 15 s, and 72 ◦C for 15 s, respectively. The B. napus actin gene (AF1l1812) was selected as a reference gene in the study. All data were obtained from three biological repetitions and experiments were repeated three times. Student’s t-tests demonstrated significant differences (*, P < 0.05; **, P < 0.01) between CK and treatment.
3. Results
3.1. Light Supplement Have Effect on Leaves Development and the Seed Oil Composition of B. napus Plants
The value of natural light intensity on the land was usually 4,006.7 lux, while the light intensity was 8,093.3 lux under the LEDs, the irradiation in field trial was significantly increased by the LEDs (
Figure 1a). The wavelength of LEDs is shown in
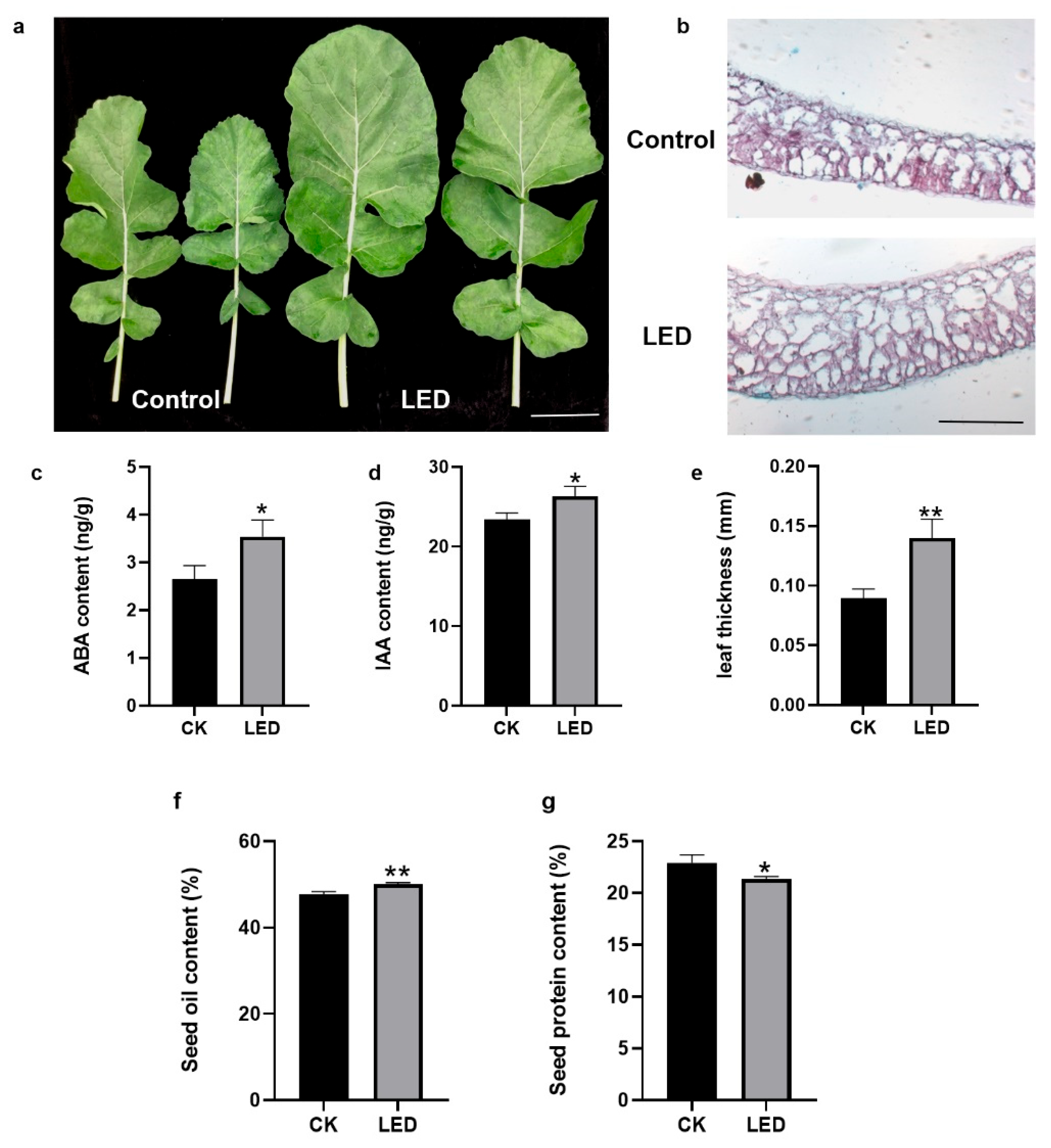
Figure 1b. To explore the change of leaf growth induced by light intensity supplement, the leaf size, leaf anatomical structure, hormone concentration and photosynthetic efficiency in rapeseed leaves were investigated. Obviously the size of leaves increased under LEDs than corresponding leaves on control condition (
Figure 2a), which consistent with the results reported by Deng et al. (2018). The thickness of leaves under LEDs increased by 50% compared to the control which was observed by cross-section microscopic images (
Figure 2b,c). The net photosynthetic efficiency, stomatal conductance and intercellular CO
2 concentration were also significantly increased in the plants under LEDs (
Table 1). And the ABA and IAA concentration significantly increased in the plants under LEDs (
Figure 2d,e). The result indicates the intensity of light that reaches the leaves have influences on the process of leaves growth. We also measured mature seed oil content and protein content by NIR. After light supplement, the seed oil content is up to 50%, significantly more than control, although the protein content has decreased slightly (
Figure 2f,g).
3.2. Overview of Transcriptome Analysis and DEGs
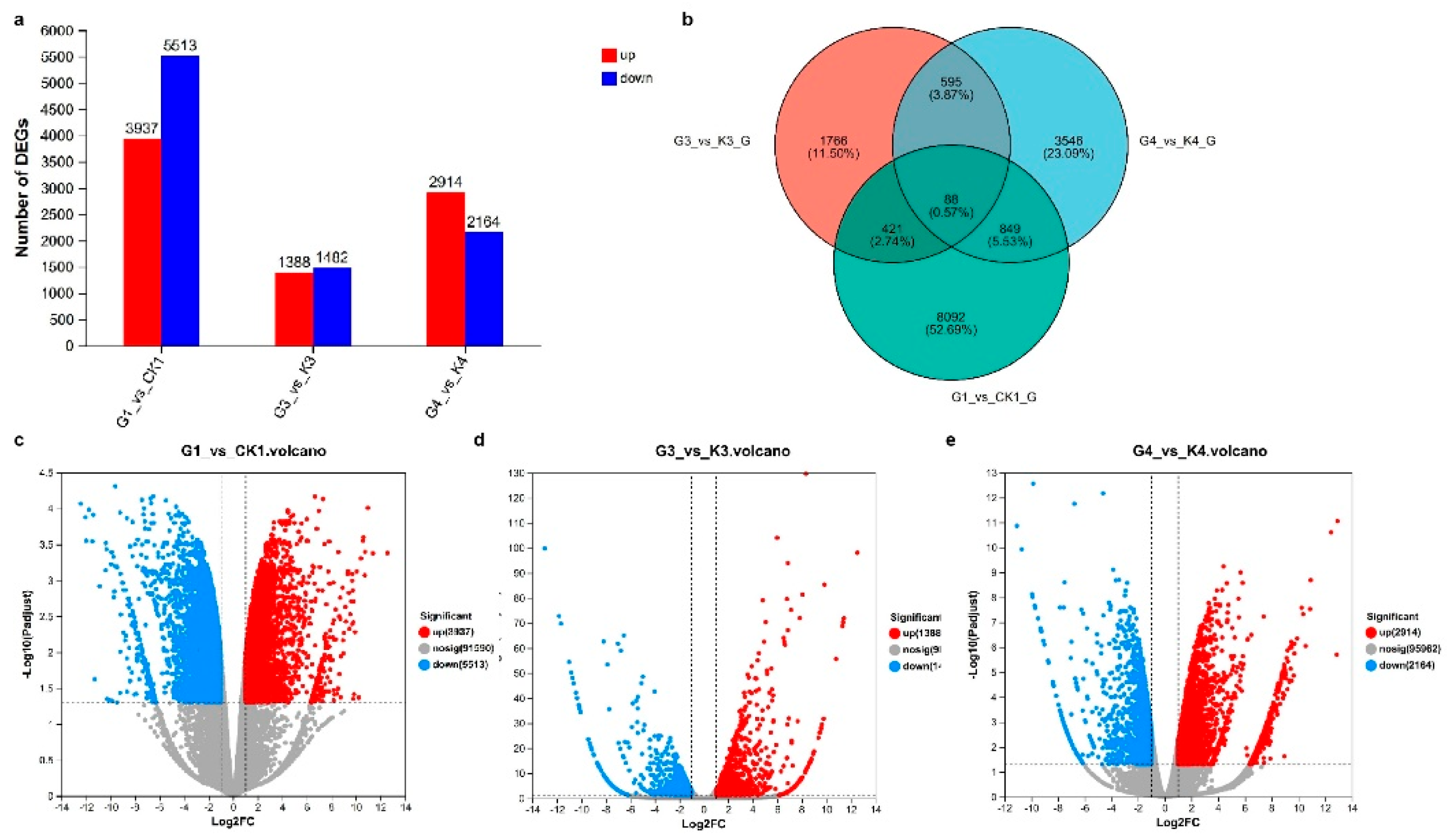
Total eighteen samples from six groups including K1 (leaves under control), G1 (leaves under LED), K30 (30 DPA seed of control), G30 (30 DPA seed under LED), K40 (40 DPA seed of control) and G40 (40 DPA seed under LED) were subjected to high throughput Illumina sequencing to obtain an overview of
B. napus leaves and seed under light quantity change related gene expression profiles. The differentially expressed gene analysis was performed through pair-wise comparison in three comparison groups (G1 vs. K1, G3 vs. K3, G4 vs. K4) that generated a total of 17398 differentially expressed genes among which 9815 (5513, 1388 and 2914, respectively) were up regulated and 7583 (3937, 1482 and 2164, respectively) were down regulated (
Figure 3a). The expression level up and down regulated genes in different comparison groups was visualized through volcano plots (>2-fold change, False discovery rate < 0.05) (
Figure 3c–e). The color differences indicate up regulation (red color) and down regulation (green color). To visualize the common DEGs in three comparison groups, a Venn diagram was showed that 88 common DEGs exist in all comparison groups, whereas 683 DEGs were common expressed in G3 vs. K3 and G4 vs. K4 (
Figure 3b).
3.3. Enrichment Characteristics of KEGG and GO of DEGs
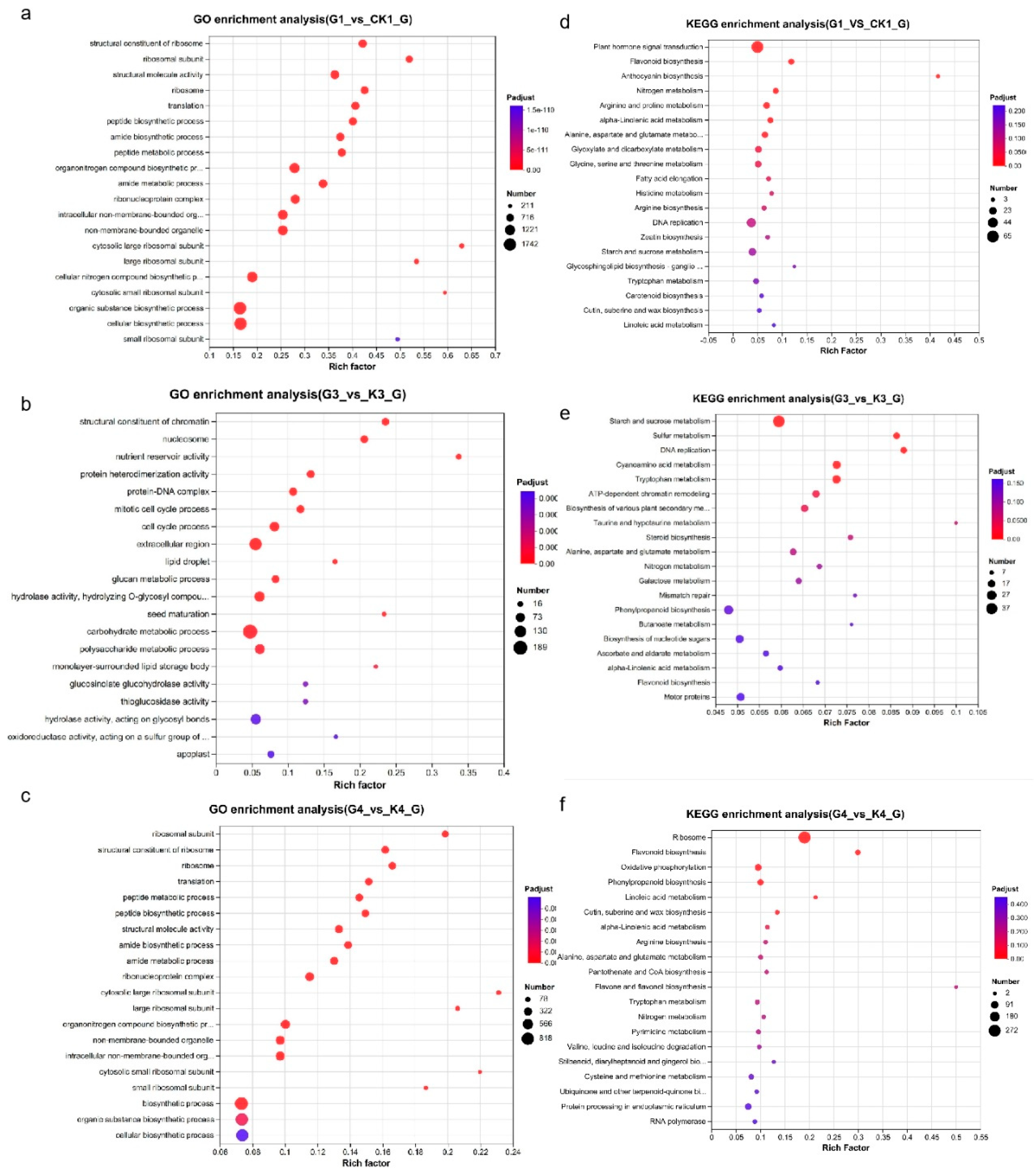
For GO term, the DEGs in G1 vs. K1 are mainly assigned to “cellular biosynthetic process”, “organic substance biosynthetic process”, “organonitrogen compound biosynthetic process” (
Figure 4c). The GO terms of “biosynthetic process”, “organic substance biosynthetic process”, “cellular biosynthetic process” were commonly enriched in G4 vs. K4 (
Figure 4b). And the the most enriched GO categories in G3 vs. K3 were “carbohydrate metabolic process”, “extracellular region”, “polysaccharide metabolic process”, “cell cycle process” and so on (
Figure 4a). The KEGG enrichment analysis of G1 vs. K1 showed that the most significant DEGs were enriched in Plant hormone signal transduction and Phenylpropanoid biosynthesis, and so on (
Figure 4f). After treatment under LEDs for 30DPA seeds, the DEGs were mainly enriched in Starch and sucrose metabolism and so on (
Figure 4d). After treatment under LEDs for 40DPA seeds, Phenylpropanoid biosynthesis were the most enriched in G4 vs. K4 except ribosome (
Figure 4e).
3.4. Light Supplement Induced DEGs in Leaves
In terms of hormonal regulation pathways, DEGs of leaves after treatment under LEDs were found in the expressions of Auxin efflux carrier, Aux/IAA and SAUR, which are essential genes can respond to auxin induction in a very short period of time, and these genes are called early auxin response genes [
15]. The expression of Auxin efflux carrier (BnaA09g09160D), Aux/IAA (BnaC06g40600D) and SAUR (BnaA09g56290D) exhibited a significant 8.6-fold 9.4-fold, and 6.6-fold up-regulation, respectively. And the expression of PYL6 (BnaA03g19030D) and PYL6 (BnaA07g38130D) were up-regulation by 4.0 and 4.6 times, respectively. Nevertheless while, the expression levels of Abscisic acid 8’-hydroxylase (ABAH), which are crucial genes in the ABA metabolism pathway, exhibited up-regulation. AS expected, we identified five up-regulation DEGs coding for Chlorophyll A-B binding protein processes. It has excellent blue-green light capture ability and strong light protection ability, which is the energy basis for the rapid growth and reproduction of diatom cells [
16]. GRAS gene family is a kind of transcription factor widely distributed in plants, which plays an important role in plant growth and development, biological and abiotic stress, light signal, hormone signal response and other processes. Some GRAS transcription factor members were up-regulated, which may play a vital role in leaves growth and development. We also screened other transcription factors (TFs) with differential expression profiles belonging to important families involved in plant development pathways, such as MYB, MADS, TCP were identified with increasing trend of expression after LEDs. Furthermore, the key genes in Cell division control protein exhibited distinct expression patterns between control and LEDs’ treatments (
Figure 4 and Supplementary Figure S1).
3.5. Candidate Gene Partaking in Seed Oil Content Synthesis
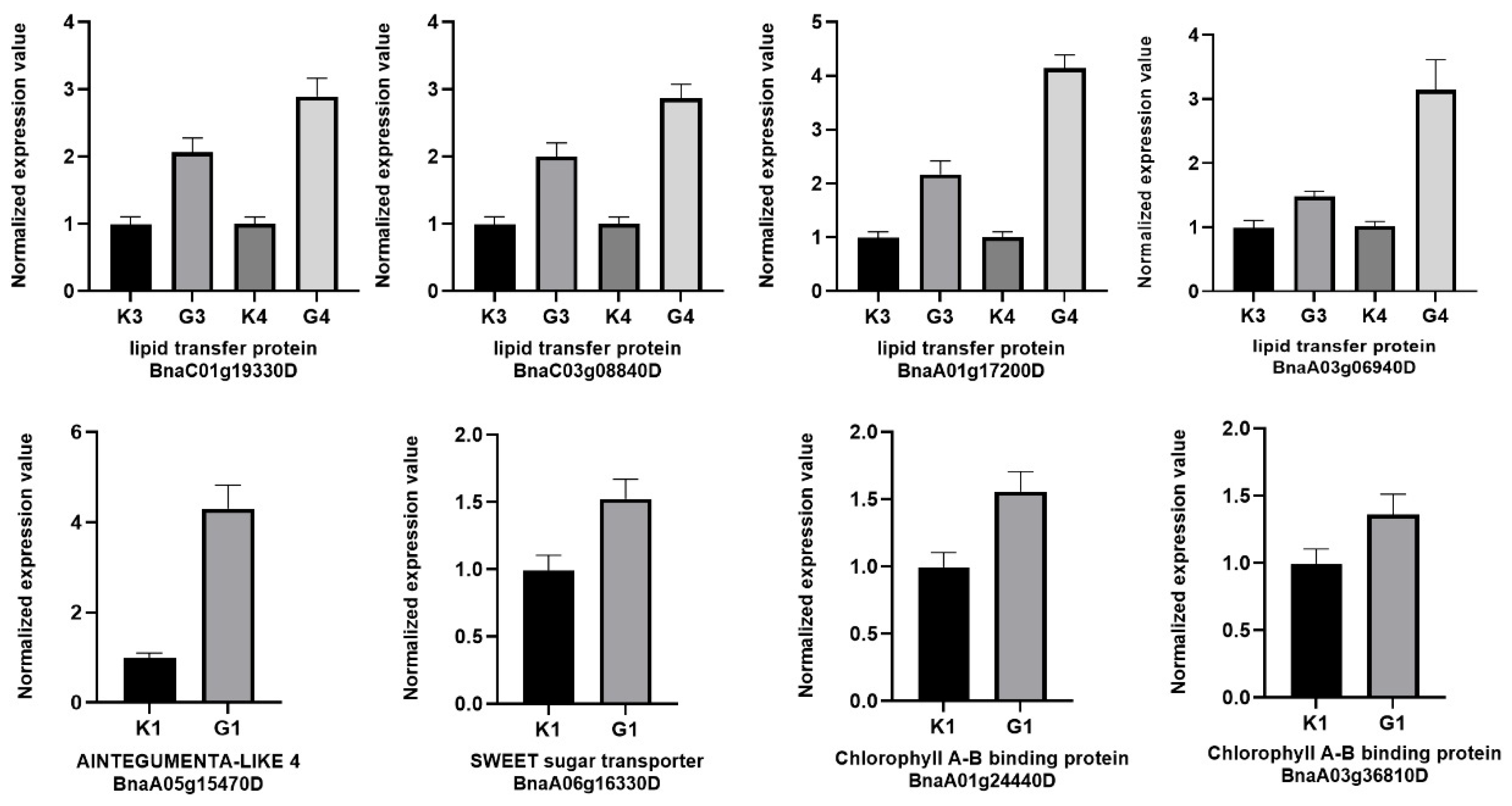
Notable changes were observed in the genes involved in seed storage protein, plant lipid transfer protein, Late embryogenesis abundant protein (LEA), and SWEET sugar transporter in seed profile. In terms of plant lipid transfer protein, the expression of BnaC01g19330D exhibited a significant 2-fold up-regulation under LEDs of 30DPA seeds and 1.75 -fold up-regulation under LEDs of 40DPA seeds. the expression levels of BnaA01g17200D exhibited a 3.9-fold and 1.5-fold increase, respectively. BnaC03g08840D exhibited a remarkable 3.4-fold up-regulation under LEDs of 40DPA seeds, nevertheless, the expression of exhibited 1.5-fold up-regulation under LEDs of 30DPA seeds. Furthermore, the expression levels of BnaAnng06900D increased by 2.9-fold of 40DPA seeds and 1.4-fold of 30DPA seeds, respectively. For LEAs, LEA-3 and LEA-25 were most significantly up-regulated under LEDs of 30DPA seeds but down-regulated under LEDs of 40DPA seeds. On the contract, SWEET sugar transporter were most significantly down-regulated under LEDs of 30DPA seeds but up-regulated under LEDs of 40DPA seeds. A large proportion of TFs are up regulated in G4 vs. K4, indicating that most gene expressions are stimulated under LEDs at late stage of seed development.
3.6. qRT-PCR Analysis
The real-time quantitative PCR analysis was performed to validate the expression of 8 randomly selected genes in sample under LEDs and in their relative controls. The qRT-PCR results were found to be consistent with the RNA-seq data (
Figure 5).
4. Discussion
Light-emitting diodes (LEDs) was often used for manipulating precisely in indoors farming, which are under controlled conditions such as light spectral composition, location, and timing [
17]. Among the visible light, the red-orange light (600~700 nm) and blue-violet light (400~500 nm) are major absorbed element, and only a small amount of green light (500~600 nm) can be absorbed by green plants [
18]. The combined spectrum of red and blue light can improve the dry matter quality, deciduous number and seed yield of wheat (Monostori et al., 2018), and also increase the dry matter quality of lettuce [
19]. So utilization of LEDs forms a vital part of efforts in current indoor farming.
The result implied that these hormones may boost the mechanisms of growth and developmental processes in the plants under LEDs, as the YABBY genes play important roles in leaf lamina development in Arabidopsis thaliana [
20], snapdragon (
Antirrhinum majus) [
21], and rice (
Oryza sativa) [
22]. And OsIAA1 exhibits different expression patterns under light and dark conditions, indicating that certain Aux/IAA genes are also involved in the light signaling pathway [
23]. The result implied that these hormones may boost the mechanisms of growth and developmental processes in the plants under LEDs. Photosynthesis, photosystem Ⅱ assembly are one of enriched biological pathways by KEGG and among the differentially expressed genes related to the photosynthetic system, the expression of the BnaCnng55860D, BnaA09g26570D, BnaC02g44590D, BnaA07g07570D, BnaC08g38660D and BnaAnng00160D gene was significantly increased (Supplementary Table S2). So the present study conclude that these genes serve as positive regulators which indirectly promote leaves development.
DEGs were highly clustered in flavonoid and Phenylpropanoid biosynthesis process, suggesting that these genes may perform its function through these pathways. The “flavonoid” was identified as the one of the most significantly enriched pathway in KEGG analysis (Rich Factor = 0.23 and Q value < 0.001), because during the development of seed maturation, oil storage change may also relate to the color change of seed coat. Furthermore, the key genes in bidirectional sugar transporter SWEET (i.e., SWEET1) exhibited distinct expression patterns between control and light treatments. In our study, a group of Xyloglucan endotransglucosylase/hydrolase protein (XTH) were constantly up-regulated in leaf tissues, as XTH22 were highly expressed in response to light supplement rather than in control condition, the function is related to the elongation of cell walls [
24]. The results showed that the leaves development and seed oil content increased by the increasing of radiation. The current study displayed a dissection of underlying mechanism of light-regulated growth in
B. napus by light-mediated transcriptional regulation network.
Taken together, these results indicated that light supplement increased leaf size by photosynthesis efficiency, cell proliferation and hormone content. And the result means LEDs is useful to increase biomass in the plant production. The results was same as the analysis of the model plant species
Arabidopsis thaliana that the plant response to shade is because of regulating the plant response to changes in light intensity and quality by integrating light and auxin signals [
25].
5. Conclusions
The results of the study comparing the B. rassica napus under natural condition and light supplement by LEDs, which explain the reason of the seed oil content is higher in some district that contain sufficient solar resource. This result is is a significant contribution to the light utilization in crop breeding, with the help of high-throughput technology. This study specifically highlights the fact that the seed oil content is not controlled by a single gene, but rather, involves the coordinated expression of multiple genes. The identification of these genes in this study opens new avenues for the light utilization in crop breeding and has implications for the development of more high photosynthetic efficiency cultivars through transgenic technology. In conclusion, this study emphasizes the importance of understanding of the complex processes involved in plant light utilization.
Author Contributions
The author Xingying Yan conducted a series of studies and wrote a paper related to this paper.Wenqin Bai conducted the field trial. Taocui Huang provide the seed of Qingyou 3#.
Funding
This work was financially supported by Chongqing Academy of Agricultural Sciences.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- Kaiserli E, Páldi K, O’Donnell L, Batalov O, Pedmale Ullas V, Nusinow Dmitri A, Kay Steve A, Chory J: Integration of Light and Photoperiodic Signaling in Transcriptional Nuclear Foci. Developmental Cell 2015, 35:311-321. [CrossRef]
- Huang J, Zhao X, Chory J: The Arabidopsis Transcriptome Responds Specifically and Dynamically to High Light Stress. Cell Reports 2019, 29:4186-4199.e4183. [CrossRef]
- Way DA, Pearcy RW: Sunflecks in trees and forests: from photosynthetic physiology to global change biology. Tree Physiology 2012, 32:1066-1081. [CrossRef]
- Niemeier U, Schmidt H, Alterskjær K, Kristjánsson JE: Solar irradiance reduction via climate engineering: Impact of different techniques on the energy balance and the hydrological cycle. Journal of Geophysical Research: Atmospheres 2013, 118:11,905-911,917. [CrossRef]
- Wild M, Gilgen H, Roesch A, Ohmura A, Long CN, Dutton EG, Forgan B, Kallis A, Russak V, Tsvetkov A: From Dimming to Brightening: Decadal Changes in Solar Radiation at Earth’s Surface. Science 2005, 308:847-850. [CrossRef]
- Wittkop B, Snowdon RJ, Friedt W: Status and perspectives of breeding for enhanced yield and quality of oilseed crops for Europe. Euphytica 2009, 170:131-140. [CrossRef]
- Dai Y, Shen Z, Liu Y, Wang L, Hannaway D, Lu H: Effects of shade treatments on the photosynthetic capacity, chlorophyll fluorescence, and chlorophyll content of Tetrastigma hemsleyanum Diels et Gilg. Environmental and Experimental Botany 2009, 65:177-182.
- Wang L, Deng F, Ren W-J: Shading tolerance in rice is related to better light harvesting and use efficiency and grain filling rate during grain filling period. Field Crops Research 2015, 180:54-62. [CrossRef]
- Deng F, Wang L, Pu S-L, Mei X-F, Li S-X, Li Q-P, Ren W-J: Shading stress increases chalkiness by postponing caryopsis development and disturbing starch characteristics of rice grains. Agricultural and Forest Meteorology 2018, 263:49-58. [CrossRef]
- Troncoso-Ponce MA, Kilaru A, Cao X, Durrett TP, Fan J, Jensen JK, Thrower N, Pauly M, Wilkerson CG, Ohlrogge JB: Comparative deep transcriptional profiling of four developing oilseeds. The Plant Journal 2011, 68:1014 - 1027. [CrossRef]
- Dussert S, Guerin C, Andersson M, Joët T, Tranbarger TJ, Pizot M, Sarah G, Omore A, Durand-Gasselin T, Morcillo F: Comparative Transcriptome Analysis of Three Oil Palm Fruit and Seed Tissues That Differ in Oil Content and Fatty Acid Composition Plant Physiology 2013, 162:1337-1358. [CrossRef]
- Lu S, Sturtevant D, Aziz M, Jin C, Li Q, Chapman KD, Guo L: Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oil Brassica napus L. seeds. The Plant journal : for cell and molecular biology 2018, 94:915-932. [CrossRef]
- Zeng J, Yan X, Bai W, Zhang M, Chen Y, Li X, Hou L, Zhao J, Ding X, Liu R, et al: Carpel-specific down-regulation of GhCKXs in cotton significantly enhances seed and fiber yield. J Exp Bot 2022, 73:6758-6772. [CrossRef]
- Zeng J, Zhang M, Hou L, Bai W, Yan X, Hou N, Wang H, Huang J, Zhao J, Pei Y: Cytokinin inhibits cotton fiber initiation by disrupting PIN3a-mediated asymmetric accumulation of auxin in the ovule epidermis. J Exp Bot 2019, 70:3139-3151. [CrossRef]
- Bao D, Chang S, Li X, Qi Y: Advances in the study of auxin early response genes: Aux/IAA, GH3, and SAUR. The Crop Journal 2024. [CrossRef]
- Büchel C: Evolution and function of light harvesting proteins. Journal of Plant Physiology 2015, 172:62-75. [CrossRef]
- Vialet-Chabrand S, Matthews JSA, Simkin AJ, Raines CA, Lawson T: Importance of Fluctuations in Light on Plant Photosynthetic Acclimation. Plant Physiology 2017, 173:2163-2179. [CrossRef]
- Vaštakaitė-Kairienė V, Samuolienė G, Šveikauskas V, Laužikė K, Jurkonienė S: The Influence of End-of-Day Blue Light on the Growth, Photosynthetic, and Metabolic Parameters of Lettuce at Different Development Stages. In Plants, vol. 11; 2022. [CrossRef]
- Zhang T, Shi Y, Piao F, Sun Z: Effects of different LED sources on the growth and nitrogen metabolism of lettuce. Plant Cell, Tissue and Organ Culture (PCTOC) 2018, 134:231 - 240. [CrossRef]
- Sarojam R, Sappl PG, Goldshmidt A, Efroni I, Floyd SK, Eshed Y, Bowman JL: Differentiating Arabidopsis Shoots from Leaves by Combined YABBY Activities The Plant Cell 2010, 22:2113-2130. [CrossRef]
- Stahle MI, Kuehlich J, Staron L, von Arnim AG, Golz JF: YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. The Plant cell 2009, 21:3105-3118. [CrossRef]
- Ohmori Y, Toriba T, Nakamura H, Ichikawa H, Hirano H-Y: Temporal and spatial regulation of DROOPING LEAF gene expression that promotes midrib formation in rice. The Plant journal : for cell and molecular biology 2011, 65:77-86. [CrossRef]
- Ku S-J, Park J-Y, Ha S-B, Kim J: Overexpression of IAA1 with domain II mutation impairs cell elongation and cell division in inflorescences and leaves of Arabidopsis. Journal of Plant Physiology 2009, 166:548-553. [CrossRef]
- He J-X, Fujioka S, Li T-C, Kang SG, Seto H, Takatsuto S, Yoshida S, Jang J-C: Sterols Regulate Development and Gene Expression in Arabidopsis. Plant Physiology 2003, 131:1258-1269. [CrossRef]
- Xie X, Cheng H, Hou C, Ren M: Integration of Light and Auxin Signaling in Shade Plants: From Mechanisms to Opportunities in Urban Agriculture. In International Journal of Molecular Sciences, vol. 23; 2022. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).