Submitted:
31 July 2024
Posted:
31 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zeng, Z.Q.; Zhuang, W.Y. The Genera Rugonectria and Thelonectria (Hypocreales, Nectriaceae) in China. MycoKeys 55: 101-120 2019, 55, 101–120. [Google Scholar] [CrossRef] [PubMed]
- Rossman, A.Y.; Seifert, K.A.; Samuels, G.J.; Minnis, A.M.; Schroers, H.J.; Lombard, L.; Crous, P.W.; Põldmaa, K.; Cannon, P.F.; Summerbell, R.C.; et al. Genera in Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales) Proposed for Acceptance or Rejection. IMA Fungus 2013, 4, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Booth, C. Studies of Pyrenomycetes Nectria (Part 1). Mycol Pap 1959, 73, 1–115. [Google Scholar]
- Rogerson, C.T. The Hypocrealean Fungi (Ascomycetes, Hypocreales). Mycologia 1970, 62, 865–910. [Google Scholar] [CrossRef] [PubMed]
- Samuels, G. A Revision of the Fungi Formerly Classified as Nectria Subgenus Hyponectria. Mem N Y Bot Gard 1976, 26, 1–126. [Google Scholar]
- Seifert, K. A Monograph of Stilbella and Some Allied Hyphomycetes. Stud Mycol 1985, 27, 1–235. [Google Scholar]
- GJ Samuels, D.B. Variation in Nectria Radicicola and Its Anamorph Cylindrocarpon Destructans. Mycol Res 1990, 94, 433–442. [Google Scholar] [CrossRef]
- AY Rossman, G.S.C.R.R.L. Genera of Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales, Ascomycetes). Stud Mycol 1999, 42, 1–248. [Google Scholar]
- E Lieckfeldt, K.S. An Evaluation of the Use of ITS Sequences in the Taxonomy of the Hypocreales. Stud Mycol 2000, 45, 35–44. [Google Scholar]
- Rossman, A. Towards Monophyletic Genera in the Holomorphic Hypocreales. Stud Mycol 2000, 45, 27–34. [Google Scholar]
- Schroers, H.-J. A Monograph of Bionectria (Ascomycota, Hypocreales, Bionectriaceae) and Its Clonostachys Anamorphs. Stud Mycol 2001, 46, 1–214. [Google Scholar]
- Hirooka, Y.; Kobayashi, T. Taxonomic Studies of Nectrioid Fungi in Japan. I: The Genus Neonectria. Mycoscience 2007 48:1 2007, 48, 53–62. [Google Scholar] [CrossRef]
- Luo, J.; Zhuang, W.Y. Three New Species of Neonectria (Nectriaceae, Hypocreales) with Notes on Their Phylogenetic Positions. Mycologia 2010, 102, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Goh, J.; Oh, Y.; Park, Y.-H.; Mun, H.Y.; Park, S.; Cheon, W. The Korean Journal of Mycology Isolation and Characterization of Previously Undescribed Seventeen Fungal Species Belonging to the Order Hypocreales in Korea. The Korean Journal of Mycology 2022, 50, 1–29. [Google Scholar] [CrossRef]
- Zeng, Z.Q.; Zhuang, W.Y. New Species of Nectriaceae (Hypocreales) from China. Journal of Fungi 2022, Vol. 8, Page 1075 2022, 8, 1075. [Google Scholar] [CrossRef] [PubMed]
- Habtewold, J.Z.; Helgason, B.L.; Yanni, S.F.; Janzen, H.H.; Ellert, B.H.; Gregorich, E.G. Litter Composition Has Stronger Influence on the Structure of Soil Fungal than Bacterial Communities. Eur J Soil Biol 2020, 98, 103190. [Google Scholar] [CrossRef]
- Lombard, L.; van der Merwe, N.A.; Groenewald, J.Z.; Crous, P.W. Generic Concepts in Nectriaceae. Stud Mycol 2015, 80, 189–245. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 Fungal Pathogens in Molecular Plant Pathology. Mol Plant Pathol 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Xu, H.; Liu, L.; Li, H.; Gao, Z. Draft Genome Sequence of Neonectria Sp. DH2 Isolated from Meconopsis Grandis Prain in Tibet. 3 Biotech 2020, 10, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Impacts of the International Code of Nomenclature for Algae, Fungi and Plants (Melbourne Code) on the Scientific Names of Plant Pathogenic Fungi. APS Features. APSnet Feature Articles 2013. [Google Scholar] [CrossRef]
- Bennett, J.W.; Klich, M. Mycotoxins. Clin Microbiol Rev 2003, 16, 497–516. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.R.; Hu, X.P.; Jiang, C.J.; Qi, J.; Wu, Y.C.; Li, W.; Zeng, Y.J.; Li, C.F.; Liu, S.X. Diversity and Antimicrobial Activity of Endophytic Fungi Isolated from Cephalotaxus Hainanensis Li, a Well-known Medicinal Plant in China. Lett Appl Microbiol 2015, 61, 484–490. [Google Scholar] [CrossRef] [PubMed]
- Sofian, F.F.; Suzuki, T.; Supratman, U.; Harneti, D.; Maharani, R.; Salam, S.; Abdullah, F.F.; Koseki, T.; Tanaka, K.; Kimura, K. ichi; et al. Cochlioquinone Derivatives Produced by Coculture of Endophytes, Clonostachys Rosea and Nectria Pseudotrichia. Fitoterapia 2021, 155, 105056. [Google Scholar] [CrossRef] [PubMed]
- JIANG, S.; QIAN, D.; YANG, N.; TAO, J.; DUAN, J. Biodiversity and Antimicrobial Activity of Endophytic Fungi in Angelica Sinensis. Chin Herb Med 2013, 5, 264–271. [Google Scholar] [CrossRef]
- Verma, A.; Shameem, N.; Jatav, H.S.; Sathyanarayana, E.; Parray, J.A.; Poczai, P.; Sayyed, R.Z. Fungal Endophytes to Combat Biotic and Abiotic Stresses for Climate-Smart and Sustainable Agriculture. Front Plant Sci 2022, 13, 953836. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Wei, D.Q.; Shen, M.; Zhou, Z.P. Endophytes and Their Role in Phytoremediation. Fungal Divers 2012, 54, 11–18. [Google Scholar] [CrossRef]
- Yu, Z. Whole RDNA Analysis Reveals Novel and Endophytic Fungi in Bletilla Ochracea (Orchidaceae). 2008.
- Christensen, M. A View of Fungal Ecology. Mycologia 1989, 81, 1–19. [Google Scholar] [CrossRef]
- Jumpponen, A. Dark Septate Endophytes - Are They Mycorrhizal? Mycorrhiza 2001, 11, 207–211. [Google Scholar] [CrossRef]
- Photita, W.; Lumyong, S.; Lumyong, P.; Mckenzie, E.H.C.; Hyde, K.D.; Photita, W.; Lumyong, S.; Lumyong, P.; Hyde, M.E.H.C. Fungal Diversity Are Some Endophytes of Musa Acuminata Latent Pathogens?
- Bernstein, N. Bernstein, N. Effects of Salinity on Root Growth. Plant Roots: The Hidden Half, Fourth Edition 2013, 595–612. [CrossRef]
- Gardes, M. An Orchid-Fungus Marriage: Physical Promiscuity, Conflict and Cheating. New Phytol 2002, 154, 4–7. [Google Scholar] [CrossRef]
- Promputtha, I.; Lumyong, S.; Dhanasekaran, V.; McKenzie, E.H.C.; Hyde, K.D.; Jeewon, R. A Phylogenetic Evaluation of Whether Endophytes Become Saprotrophs at Host Senescence. Microb Ecol 2007, 53, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.; Boyle, C. The Endophytic Continuum. Mycol Res 2005, 109, 661–686. [Google Scholar] [CrossRef]
- Yu, Z. Whole RDNA Analysis Reveals Novel and Endophytic Fungi in Bletilla Ochracea (Orchidaceae). 2008.
- Morrison, E.S.; Thomas, P.; Ogram, A.; Kahveci, T.; Turner, B.L.; Chanton, J.P. Characterization of Bacterial and Fungal Communities Reveals Novel Consortia in Tropical Oligotrophic Peatlands. Microb Ecol 2021, 82, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Bao, D.F.; Hyde, K.D.; Maharachchikumbura, S.S.N.; Perera, R.H.; Thiyagaraja, V.; Hongsanan, S.; Wanasinghe, D.N.; Shen, H.W.; Tian, X.G.; Yang, L.Q.; et al. Taxonomy, Phylogeny and Evolution of Freshwater Hypocreomycetidae (Sordariomycetes). Fungal Diversity 2023 121:1 2023, 121, 1–94. [Google Scholar] [CrossRef]
- Shearer, C.A.; Raja, H.A.; Miller, A.N.; Nelson, P.; Tanaka, K.; Hirayama, K.; Marvanová, L.; Hyde, K.D.; Zhang, Y. The Molecular Phylogeny of Freshwater Dothideomycetes. Stud Mycol 2009, 64, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Salgado-Salazar, C.; Rossman, A.Y.; Chaverri, P. Not as Ubiquitous as We Thought: Taxonomic Crypsis, Hidden Diversity and Cryptic Speciation in the Cosmopolitan Fungus Thelonectria Discophora (Nectriaceae, Hypocreales, Ascomycota). PLoS One 2013, 8, e76737. [Google Scholar] [CrossRef] [PubMed]
- Shearer, C.A.; Descals, E.; Kohlmeyer, B.; Kohlmeyer, J.; Marvanová, L.; Padgett, D.; Porter, D.; Raja, H.A.; Schmit, J.P.; Thorton, H.A.; et al. Fungal Biodiversity in Aquatic Habitats. Biodivers Conserv 2007, 16, 49–67. [Google Scholar] [CrossRef]
- Bärlocher, F. Research on Aquatic Hyphomycetes: Historical Background and Overview. 1992, 1–15. [CrossRef]
- Webster, J. Experiments with Spores of Aquatic Hyphomycetes: I. Sedimentation, and Impaction on Smooth Surfaces. Ann Bot 1959, 23, 595–611. [Google Scholar] [CrossRef]
- Dix, N.J.; Webster, J. Fungal Ecology. 1995. [CrossRef]
- Webster, J.; Shearer, C.A.; Spooner, B.M. Mollisia Casaresiae (Ascomycetes) the Teleomorph of Casaresia Sphagnorum, an Aquatic Fungus. Nova Hedwigia 1993, 57, 3–4. [Google Scholar]
- Vasconcelos Rissi, D.; Ijaz, M.; Baschien, C. Comparative Genome Analysis of the Freshwater Fungus Filosporella fistucella. [CrossRef]
- Brown, A.D. Compatible Solutes and Extreme Water Stress in Eukaryotic Micro-Organisms. Adv Microb Physiol 1978, 17, 181–242. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.H. Cold Adaptation in Arctic and Antarctic Fungi. New Phytologist 2001, 151, 341–353. [Google Scholar] [CrossRef]
- Mycorrhizal Fungi: Their Habitats and Nutritional Strategies. Environmental and Microbial Relationships 2007, 229–256. [CrossRef]
- Hassan, N.; Rafiq, M.; Hayat, M.; Shah, A.A.; Hasan, F. Psychrophilic and Psychrotrophic Fungi: A Comprehensive Review. Reviews in Environmental Science and Bio/Technology 2016 15:2 2016, 15, 147–172. [Google Scholar] [CrossRef]
- Weinstein, R.N.; Montiel, P.O.; Johnstone, K. Influence of Growth Temperature on Lipid and Soluble Carbohydrate Synthesis by Fungi Isolated from Fellfield Soil in the Maritime Antarctic. Mycologia 2000, 92, 222–229. [Google Scholar] [CrossRef]
- Sui, Y.; Wisniewski, M.; Droby, S.; Norelli, J.; Liu, J. Recent Advances and Current Status of the Use of Heat Treatments in Postharvest Disease Management Systems: Is It Time to Turn up the Heat? Trends Food Sci Technol 2016, 51, 34–40. [Google Scholar] [CrossRef]
- Zhgun, A.A. Fungal BGCs for Production of Secondary Metabolites: Main Types, Central Roles in Strain Improvement, and Regulation According to the Piano Principle. International Journal of Molecular Sciences 2023, Vol. 24, Page 11184 2023, 24, 11184. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Pei, R.; Zhang, Y.; Tu, Y.; He, B. Light Regulation of Secondary Metabolism in Fungi. J Biol Eng 2023, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yogabaanu, U.; Weber, J.F.F.; Convey, P.; Rizman-Idid, M.; Alias, S.A. Antimicrobial Properties and the Influence of Temperature on Secondary Metabolite Production in Cold Environment Soil Fungi. Polar Sci 2017, 14, 60–67. [Google Scholar] [CrossRef]
- Parain, E.C.; Rohr, R.P.; Gray, S.M.; Bersier, L.F. Increased Temperature Disrupts the Biodiversity–Ecosystem Functioning Relationship. 2019, 193, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Morera, A.; Martínez de Aragón, J.; Bonet, J.A.; Liang, J.; de-Miguel, S. Performance of Statistical and Machine Learning-Based Methods for Predicting Biogeographical Patterns of Fungal Productivity in Forest Ecosystems. For Ecosyst 2021, 8, 1–14. [Google Scholar] [CrossRef]
- Freire, B.; Ladra, S.; Parama, J.R. Memory-Efficient Assembly Using Flye. IEEE/ACM Trans Comput Biol Bioinform 2021, 1–1. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E. V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and Open Software for Comparing Large Genomes. Genome Biol 2004, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lachance, M.A.; Lee, D.K.; Hsiang, T. Delineating Yeast Species with Genome Average Nucleotide Identity: A Calibration of ANI with Haplontic, Heterothallic Metschnikowia Species. Antonie van Leeuwenhoek, International Journal of General and Molecular Microbiology 2020, 113, 2097–2106. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for Automated Genomic Discovery of Transposable Element Families. Proc Natl Acad Sci U S A 2020, 117, 9451–9457. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, D. RepeatMasker. https://home.liebertpub.com/bsi 2004, 1, 36–39. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. TRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res 2021, 49, 9077–9096. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, L.; Brůna, T.; Hoff, K.J.; Ebel, M.; Lomsadze, A.; Borodovsky, M.; Stanke, M. BRAKER3: Fully Automated Genome Annotation Using RNA-Seq and Protein Evidence with GeneMark-ETP, AUGUSTUS, and TSEBRA. Genome Res 2024. [Google Scholar] [CrossRef] [PubMed]
- Brůna, T.; Lomsadze, A.; Borodovsky, M. GeneMark-EP+: Eukaryotic Gene Prediction with Self-Training in the Space of Genes and Proteins. NAR Genom Bioinform 2020, 2. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. AUGUSTUS: Ab Initio Prediction of Alternative Transcripts. Nucleic Acids Res 2006, 34, W435–W439. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol 2019, 20, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol Biol Evol 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R.; Teeling, E. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nature Methods 2017 14:6 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 Predicts All Five Types of Signal Peptides Using Protein Language Models. Nature Biotechnology 2022 40:7 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; Von Heijne, G. Predicting Subcellular Localization of Proteins Based on Their N-Terminal Amino Acid Sequence. J Mol Biol 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Hallgren, J.; Tsirigos, K.D.; Damgaard Pedersen, M.; Juan, J.; Armenteros, A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM Predicts Alpha and Beta Transmembrane Proteins Using Deep Neural Networks. bioRxiv, 2022. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nature Methods 2014 12:1 2014, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Ge, Q.; Yan, Y.; Zhang, X.; Huang, L.; Yin, Y. DbCAN3: Automated Carbohydrate-Active Enzyme and Substrate Annotation. Nucleic Acids Res 2023, 51, W115–W121. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Dodds, P.N. EffectorP 3.0: Prediction of Apoplastic and Cytoplasmic Effectors in Fungi and Oomycetes. Molecular Plant-Microbe Interactions 2022, 35, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Erickson, E.; Gado, J.E.; Avilán, L.; Bratti, F.; Brizendine, R.K.; Cox, P.A.; Gill, R.; Graham, R.; Kim, D.J.; König, G.; et al. Sourcing Thermotolerant Poly(Ethylene Terephthalate) Hydrolase Scaffolds from Natural Diversity. Nature Communications 2022 13:1 2022, 13, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, M.A.; Yurkov, A.; Nowrousian, M.; Stukenbrock, E.H. Lifestyle Transitions in Basidiomycetous Fungi Are Reflected by TRNA Composition and Translation Efficiency of Metabolic Genes. bioRxiv, 2023. [Google Scholar] [CrossRef]
- Fijarczyk, A.; Hessenauer, P.; Hamelin, R.C.; Landry, C.R. Lifestyles Shape Genome Size and Gene Content in Fungal Pathogens. bioRxiv, 2022. [Google Scholar] [CrossRef]
- Raffaele, S.; Kamoun, S. Genome Evolution in Filamentous Plant Pathogens: Why Bigger Can Be Better. Nature Reviews Microbiology 2012 10:6 2012, 10, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Talhinhas, P.; Carvalho, R.; Loureiro, J. The Use of Flow Cytometry for Fungal Nuclear DNA Quantification. Cytometry Part A 2021, 99, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Ting, A.S.Y.; Chaverri, P.; Edrada-Ebel, R.A. Editorial: Endophytes and Their Biotechnological Applications. Front Bioeng Biotechnol 2021, 9, 795174. [Google Scholar] [CrossRef] [PubMed]
- Gouda, S.; Das, G.; Sen, S.K.; Shin, H.S.; Patra, J.K. Endophytes: A Treasure House of Bioactive Compounds of Medicinal Importance. Front Microbiol 2016, 7, 219261. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Plaza, C.; Ochoa-Hueso, R.; Trivedi, C.; Wang, J.; Trivedi, P.; Zhou, G.; Piñeiro, J.; Martins, C.S.C.; Singh, B.K.; et al. Litter and Soil Biodiversity Jointly Drive Ecosystem Functions. Glob Chang Biol 2023, 29, 6276–6285. [Google Scholar] [CrossRef]
- Mirabile, G.; Ferraro, V.; Mancuso, F.P.; Pecoraro, L.; Cirlincione, F. Biodiversity of Fungi in Freshwater Ecosystems of Italy. Journal of Fungi 2023, Vol. 9, Page 993 2023, 9, 993. [Google Scholar] [CrossRef]
- Bhunjun, C.S.; Phukhamsakda, C.; Hyde, K.D.; McKenzie, E.H.C.; Saxena, R.K.; Li, Q. Do All Fungi Have Ancestors with Endophytic Lifestyles? Fungal Diversity 2023 125:1 2023, 125, 73–98. [Google Scholar] [CrossRef]
- Gorbunova, V.; Seluanov, A.; Mita, P.; McKerrow, W.; Fenyö, D.; Boeke, J.D.; Linker, S.B.; Gage, F.H.; Kreiling, J.A.; Petrashen, A.P.; et al. The Role of Retrotransposable Elements in Ageing and Age-Associated Diseases. Nature 2021 596:7870 2021, 596, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Latzel, V.; Puy, J.; Thieme, M.; Bucher, E.; Götzenberger, L.; de Bello, F. Phenotypic Diversity Influenced by a Transposable Element Increases Productivity and Resistance to Competitors in Plant Populations. Journal of Ecology 2023, 111, 2376–2387. [Google Scholar] [CrossRef]
- Liu, C.; Li, B.; Dong, Y.; Lin, H. Endophyte Colonization Enhanced Cadmium Phytoremediation by Improving Endosphere and Rhizosphere Microecology Characteristics. J Hazard Mater 2022, 434, 128829. [Google Scholar] [CrossRef] [PubMed]
- Mascarin, G.M.; Jaronski, S.T. The Production and Uses of Beauveria Bassiana as a Microbial Insecticide. World Journal of Microbiology and Biotechnology 2016 32:11 2016, 32, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, C.B. de; Santana, M.F. Prediction of the Secretomes of Endophytic and Nonendophytic Fungi Reveals Similarities in Host Plant Infection and Colonization Strategies. Mycologia 2020, 112, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Oggenfuss, U.; Croll, D. Recent Transposable Element Bursts Are Associated with the Proximity to Genes in a Fungal Plant Pathogen. PLoS Pathog 2023, 19, e1011130. [Google Scholar] [CrossRef] [PubMed]
- Muszewska, A.; Steczkiewicz, K.; Stepniewska-Dziubinska, M.; Ginalski, K. Transposable Elements Contribute to Fungal Genes and Impact Fungal Lifestyle. Scientific Reports 2019 9:1 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Grandaubert, J.; Lowe, R.G.T.; Soyer, J.L.; Schoch, C.L.; Van De Wouw, A.P.; Fudal, I.; Robbertse, B.; Lapalu, N.; Links, M.G.; Ollivier, B.; et al. Transposable Element-Assisted Evolution and Adaptation to Host Plant within the Leptosphaeria Maculans-Leptosphaeria Biglobosa Species Complex of Fungal Pathogens. BMC Genomics 2014, 15, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Torres, D.E.; Thomma, B.P.H.J.; Seidl, M.F. Transposable Elements Contribute to Genome Dynamics and Gene Expression Variation in the Fungal Plant Pathogen Verticillium Dahliae. Genome Biol Evol 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Seidl, M.F.; Faino, L.; Shi-Kunne, X.; Van Den Berg, G.C.M.; Bolton, M.D.; Thomma, B.P.H.J. The Genome of the Saprophytic Fungus Verticillium Tricorpus Reveals a Complex Effector Repertoire Resembling That of Its Pathogenic Relatives. 2015, 28, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.N.; Brookes, L.M.; Edwards, H.M.; Fisher, M.C.; Jervis, P.; Kappel, D.; Sewell, T.R.; Shelton, J.M.G.; Skelly, E.; Rhodes, J.L. Cross-Disciplinary Genomics Approaches to Studying Emerging Fungal Infections. Life 2020, Vol. 10, Page 315 2020, 10, 315. [Google Scholar] [CrossRef] [PubMed]
- Bennett, E.A.; Coleman, L.E.; Tsui, C.; Pittard, W.S.; Devine, S.E. Natural Genetic Variation Caused by Transposable Elements in Humans. Genetics 2004, 168, 933–951. [Google Scholar] [CrossRef] [PubMed]
- Castanera, R.; López-Varas, L.; Borgognone, A.; LaButti, K.; Lapidus, A.; Schmutz, J.; Grimwood, J.; Pérez, G.; Pisabarro, A.G.; Grigoriev, I. V.; et al. Transposable Elements versus the Fungal Genome: Impact on Whole-Genome Architecture and Transcriptional Profiles. PLoS Genet 2016, 12, e1006108. [Google Scholar] [CrossRef] [PubMed]
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. (Pat) Repetitive DNA in Eukaryotic Genomes. Chromosome Research 2015, 23, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Gazis, R.; Kuo, A.; Riley, R.; LaButti, K.; Lipzen, A.; Lin, J.; Amirebrahimi, M.; Hesse, C.N.; Spatafora, J.W.; Henrissat, B.; et al. The Genome of Xylona Heveae Provides a Window into Fungal Endophytism. Fungal Biol 2016, 120, 26–42. [Google Scholar] [CrossRef] [PubMed]
- Vincent, D.; Rafiqi, M.; Job, D. The Multiple Facets of Plant–Fungal Interactions Revealed Through Plant and Fungal Secretomics. Front Plant Sci 2020, 10, 487828. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.I.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes Database (CAZy): An Expert Resource for Glycogenomics. Nucleic Acids Res 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, H.; Wang, C.; Xu, J.R. Correction to Comparative Analysis of Fungal Genomes Reveals Different Plant Cell Wall Degrading Capacity in Fungi [BMC Genomics 14(2013) 274]. BMC Genomics 2014, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ospina-Giraldo, M.D.; Griffith, J.G.; Laird, E.W.; Mingora, C. The CAZyome of Phytophthora Spp.: A Comprehensive Analysis of the Gene Complement Coding for Carbohydrate-Active Enzymes in Species of the Genus Phytophthora. BMC Genomics 2010, 11, 1–16. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, H.; Wang, C.; Xu, J.R. Comparative Analysis of Fungal Genomes Reveals Different Plant Cell Wall Degrading Capacity in Fungi. BMC Genomics 2013, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Czislowski, E.; Zeil-rolfe, I.; Aitken, E.A.B. Effector Profiles of Endophytic Fusarium Associated with Asymptomatic Banana (Musa Sp.) Hosts. International Journal of Molecular Sciences 2021, Vol. 22, Page 2508 2021, 22, 2508. [Google Scholar] [CrossRef] [PubMed]
- Promputtha, I.; Hyde, K.D.; McKenzie, E.H.C.; Peberdy, J.F.; Lumyong, S. Can Leaf Degrading Enzymes Provide Evidence That Endophytic Fungi Becoming Saprobes? Fungal Divers 2010, 41, 89–99. [Google Scholar] [CrossRef]
- Sarkar, S.; Dey, A.; Kumar, V.; Batiha, G.E.S.; El-Esawi, M.A.; Tomczyk, M.; Ray, P. Fungal Endophyte: An Interactive Endosymbiont With the Capability of Modulating Host Physiology in Myriad Ways. Front Plant Sci 2021, 12, 701800. [Google Scholar] [CrossRef] [PubMed]
- Pointing, S.B.; Parungao, M.M.; Hyde, K.D. Production of Wood-Decay Enzymes, Mass Loss and Lignin Solubilization in Wood by Tropical Xylariaceae. Mycol Res 2003, 107, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Pointing, S.B.; Pelling, A.L.; Smith, G.J.D.; Hyde, K.D.; Reddy, C.A. Screening of Basidiomycetes and Xylariaceous Fungi for Lignin Peroxidase and Laccase Gene-Specific Sequences. Mycol Res 2005, 109, 115–124. [Google Scholar] [CrossRef]
- Koide, K.; Osono, T.; Takeda, H. Fungal Succession and Decomposition of Camellia Japonica Leaf Litter. Ecol Res 2005, 20, 599–609. [Google Scholar] [CrossRef]
- Bucher, V.V.C.; Hyde, K.D.; Pointing, S.B.; Reddy, C.A.; Reddy, S.B. Production of Wood Decay Enzymes, Mass Loss and Lignin Solubilization in Wood by Marine Ascomycetes and Their Anamorphs.
- Purahong, W.; Hyde, K.D. Effects of Fungal Endophytes on Grass and Non-Grass Litter Decomposition Rates. Fungal Divers 2011, 47, 1–7. [Google Scholar] [CrossRef]
- Barrett, K.; Jensen, K.; Meyer, A.S.; Frisvad, J.C.; Lange, L. Fungal Secretome Profile Categorization of CAZymes by Function and Family Corresponds to Fungal Phylogeny and Taxonomy: Example Aspergillus and Penicillium. Scientific Reports 2020 10:1 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.M.; Zhang, Y. Plant Immunity: Danger Perception and Signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef] [PubMed]
- Zúñiga, E.; Romero, J.; Ollero-Lara, A.; Lovera, M.; Arquero, O.; Miarnau, X.; Torguet, L.; Trapero, A.; Luque, J. Inoculum and Infection Dynamics of Polystigma Amygdalinum in Almond Orchards in Spain. Plant Dis 2020, 104, 1239–1246. [Google Scholar] [CrossRef] [PubMed]
- Ali, G.S.; Reddy, A.S.N. PAMP-Triggered Immunity. Plant Signal Behav 2008, 3, 423–426. [Google Scholar] [CrossRef]
- Chellappan, B.V.; El-Ganainy, S.M.; Alrajeh, H.S.; Al-Sheikh, H. In Silico Characterization of the Secretome of the Fungal Pathogen Thielaviopsis Punctulata, the Causal Agent of Date Palm Black Scorch Disease. Journal of Fungi 2023, 9, 303. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu Rev Plant Biol 2009, 60, 379–407. [Google Scholar] [CrossRef] [PubMed]
- Mattoo, A.J.; Nonzom, S. Endophytic Fungi: Understanding Complex Cross-Talks. Symbiosis 2021 83:3 2021, 83, 237–264. [Google Scholar] [CrossRef]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal Effectors and Plant Susceptibility. Annu Rev Plant Biol 2015, 66, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, A.; Ghosh, S.; Sahoo, D.; Jha, G. Fungal Effectors, the Double Edge Sword of Phytopathogens. Current Genetics 2020 67:1 2020, 67, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Todd, J.N.A.; Carreón-Anguiano, K.G.; Islas-Flores, I.; Canto-Canché, B. Fungal Effectoromics: A World in Constant Evolution. International Journal of Molecular Sciences 2022, Vol. 23, Page 13433 2022, 23, 13433. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Wang, Y.; McDowell, J. Focus on Effector-Triggered Susceptibility. 2017; 31, 5. [Google Scholar] [CrossRef]
- Hammond-Kosack, K.E.; Jones, J.D.G. Plant Disease Resistance Genes. Annu Rev Plant Biol 1997, 48, 575–607. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Cheng, Y. Advances in Fungal Elicitor-Triggered Plant Immunity. International Journal of Molecular Sciences 2022, Vol. 23, Page 12003 2022, 23, 12003. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, Y.; Li, B.; Zhang, Z.; Qin, G.; Chen, T.; Tian, S. Molecular Mechanisms Underlying Multi-Level Defense Responses of Horticultural Crops to Fungal Pathogens. Hortic Res 2022, 9. [Google Scholar] [CrossRef] [PubMed]
- Petit-Houdenot, Y.; Fudal, I. Complex Interactions between Fungal Avirulence Genes and Their Corresponding Plant Resistance Genes and Consequences for Disease Resistance Management. Front Plant Sci 2017, 8, 250670. [Google Scholar] [CrossRef] [PubMed]
- Balint-Kurti, P. The Plant Hypersensitive Response: Concepts, Control and Consequences. Mol Plant Pathol 2019, 20, 1163–1178. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A. Global Warming Could Drive the Emergence of New Fungal Pathogens. Nature Microbiology 2023 8:12 2023, 8, 2217–2219. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.K.; Delgado-Baquerizo, M.; Egidi, E.; Guirado, E.; Leach, J.E.; Liu, H.; Trivedi, P. Climate Change Impacts on Plant Pathogens, Food Security and Paths Forward. Nature Reviews Microbiology 2023 21:10 2023, 21, 640–656. [Google Scholar] [CrossRef] [PubMed]
- Seidel, D.; Wurster, S.; Jenks, J.D.; Sati, H.; Gangneux, J.P.; Egger, M.; Alastruey-Izquierdo, A.; Ford, N.P.; Chowdhary, A.; Sprute, R.; et al. Impact of Climate Change and Natural Disasters on Fungal Infections. Lancet Microbe 2024, 5, e594–e605. [Google Scholar] [CrossRef]
- Stauder, C.M.; Garnas, J.R.; Morrison, E.W.; Salgado-Salazar, C.; Kasson, M.T. Characterization of Mating Type Genes in Heterothallic Neonectria Species, with Emphasis on N. Coccinea, N. Ditissima, and N. Faginata. Mycologia 2020, 112, 880–894. [Google Scholar] [CrossRef] [PubMed]
- Latorre, B.A.; Rioja, M.E.; Lillo, C.; Muñoz, M. The Effect of Temperature and Wetness Duration on Infection and a Warning System for European Canker (Nectria Galligena) of Apple in Chile. Crop Protection 2002, 21, 285–291. [Google Scholar] [CrossRef]
- Langer, G.J.; Bußkamp, J. Fungi Associated With Woody Tissues of European Beech and Their Impact on Tree Health. Front Microbiol 2021, 12, 702467. [Google Scholar] [CrossRef] [PubMed]
- Purahong, W.; Tanunchai, B.; Wahdan, S.F.M.; Buscot, F.; Schulze, E.D. Molecular Screening of Microorganisms Associated with Discolored Wood in Dead European Beech Trees Suffered from Extreme Drought Event Using next Generation Sequencing. Plants 2021, 10, 2092. [Google Scholar] [CrossRef] [PubMed]
- Morrison, E.W.; Kasson, M.T.; Heath, J.J.; Garnas, J.R. Pathogen and Endophyte Assemblages Co-Vary With Beech Bark Disease Progression, Tree Decline, and Regional Climate. Frontiers in Forests and Global Change 2021, 4, 673099. [Google Scholar] [CrossRef]
- Saremi, H.; Burgess, L.W.; Backhouse, D. Temperature Effects on the Relative Abundance of Fusarium Species in a Model Plant–Soil Ecosystem. Soil Biol Biochem 1999, 31, 941–947. [Google Scholar] [CrossRef]
- Suárez-Estrella, F.; Vargas-García, M.C.; Elorrieta, M.A.; López, M.J.; Moreno, J. Temperature Effect on Fusarium Oxysporum f.Sp. Melonis Survival during Horticultural Waste Composting. J Appl Microbiol 2003, 94, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Knapp, B.D.; Huang, K.C. The Effects of Temperature on Cellular Physiology. Annu Rev Biophys 2022, 51, 499–526. [Google Scholar] [CrossRef] [PubMed]
- Bärlocher, F.; Seena, S.; Wilson, K.P.; Dudley Williams, D. Raised Water Temperature Lowers Diversity of Hyporheic Aquatic Hyphomycetes. Freshw Biol 2008, 53, 368–379. [Google Scholar] [CrossRef]
- Dang, C.K.; Schindler, M.; Chauvet, E.; Gessner, M.O. Temperature Oscillation Coupled with Fungal Community Shifts Can Modulate Warming Effects on Litter Decomposition. Ecology 2009, 90, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Větrovský, T.; Kohout, P.; Kopecký, M.; Machac, A.; Man, M.; Bahnmann, B.D.; Brabcová, V.; Choi, J.; Meszárošová, L.; Human, Z.R.; et al. A Meta-Analysis of Global Fungal Distribution Reveals Climate-Driven Patterns. Nature Communications 2019 10:1 2019, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Author, C.; Zhao, W. Endophytic Fungi in Green Manure Crops; Friends or Foe? Mycosphere 2023, 14, 7019. [Google Scholar] [CrossRef]
- Fenoy, E.; Pradhan, A.; Pascoal, C.; Rubio-Ríos, J.; Batista, D.; Moyano-López, F.J.; Cássio, F.; Casas, J.J. Elevated Temperature May Reduce Functional but Not Taxonomic Diversity of Fungal Assemblages on Decomposing Leaf Litter in Streams. Glob Chang Biol 2022, 28, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Follstad Shah, J.J.; Kominoski, J.S.; Ardón, M.; Dodds, W.K.; Gessner, M.O.; Griffiths, N.A.; Hawkins, C.P.; Johnson, S.L.; Lecerf, A.; LeRoy, C.J.; et al. Global Synthesis of the Temperature Sensitivity of Leaf Litter Breakdown in Streams and Rivers. Glob Chang Biol 2017, 23, 3064–3075. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Sun, J.; Xie, P.; Guo, C.; Zhu, K.; Tian, K. Climate Warming Enhances Microbial Network Complexity by Increasing Bacterial Diversity and Fungal Interaction Strength in Litter Decomposition. Science of The Total Environment 2024, 908, 168444. [Google Scholar] [CrossRef] [PubMed]
- Boyero, L.; Pearson, R.G.; Gessner, M.O.; Barmuta, L.A.; Ferreira, V.; Graça, M.A.S.; Dudgeon, D.; Boulton, A.J.; Callisto, M.; Chauvet, E.; et al. A Global Experiment Suggests Climate Warming Will Not Accelerate Litter Decomposition in Streams but Might Reduce Carbon Sequestration. Ecol Lett 2011, 14, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Simões, S.; Gonçalves, A.L.; Jones, T.H.; Sousa, J.P.; Canhoto, C. Air Temperature More than Drought Duration Affects Litter Decomposition under Flow Intermittency. Science of The Total Environment 2022, 829, 154666. [Google Scholar] [CrossRef] [PubMed]
- Shearer, C.A.; Webster, J. Aquatic Hyphomycete Communities in the River Teign. IV. Twig Colonization. Mycol Res 1991, 95, 413–420. [Google Scholar] [CrossRef]
- Cobo-Díaz, J.F.; Baroncelli, R.; Le Floch, G.; Picot, A. A Novel Metabarcoding Approach to Investigate Fusarium Species Composition in Soil and Plant Samples. FEMS Microbiol Ecol 2019, 95, 84. [Google Scholar] [CrossRef] [PubMed]
- Barros, J.; Seena, S. Fungi in Freshwaters: Prioritising Aquatic Hyphomycetes in Conservation Goals. Water 2022, Vol. 14, Page 605 2022, 14, 605. [Google Scholar] [CrossRef]
- Chen, H.; Raffaele, S.; Dong, S. Silent Control: Microbial Plant Pathogens Evade Host Immunity without Coding Sequence Changes. FEMS Microbiol Rev 2021, 45, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hinsch, J.; Galuszka, P.; Tudzynski, P. Functional Characterization of the First Filamentous Fungal TRNA-Isopentenyltransferase and Its Role in the Virulence of Claviceps Purpurea. New Phytologist 2016, 211, 980–992. [Google Scholar] [CrossRef] [PubMed]
- dos Reis, M.; Savva, R.; Wernisch, L. Solving the Riddle of Codon Usage Preferences: A Test for Translational Selection. Nucleic Acids Res 2004, 32, 5036–5044. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.; Kuo, A.; Nagy, L.G.; Morin, E.; Barry, K.W.; Buscot, F.; Canbäck, B.; Choi, C.; Cichocki, N.; Clum, A.; et al. Convergent Losses of Decay Mechanisms and Rapid Turnover of Symbiosis Genes in Mycorrhizal Mutualists. Nature Genetics 2015 47:4 2015, 47, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Collemare, J.; Billard, A.; Böhnert, H.U.; Lebrun, M.H. Biosynthesis of Secondary Metabolites in the Rice Blast Fungus Magnaporthe Grisea: The Role of Hybrid PKS-NRPS in Pathogenicity. Mycol Res 2008, 112, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Liu, S.; Xu, F.; Jiang, S.; Yan, J.; He, Q.; Liu, W.; Lin, C.; Zheng, F.; Wang, X.; et al. Powdery Mildews Are Characterized by Contracted Carbohydrate Metabolism and Diverse Effectors to Adapt to Obligate Biotrophic Lifestyle. Front Microbiol 2018, 9, 423656. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, J.; Yan, J.; Guo, M.; Xu, L.; Hou, L.; Zou, Q. Comparative Genome Analysis of Plant Ascomycete Fungal Pathogens with Different Lifestyles Reveals Distinctive Virulence Strategies. BMC Genomics 2022, 23, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rokas, A.; Mead, M.E.; Steenwyk, J.L.; Raja, H.A.; Oberlies, N.H. Biosynthetic Gene Clusters and the Evolution of Fungal Chemodiversity. Nat Prod Rep 2020, 37, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Mózsik, L.; Iacovelli, R.; Bovenberg, R.A.L.; Driessen, A.J.M. Transcriptional Activation of Biosynthetic Gene Clusters in Filamentous Fungi. Front Bioeng Biotechnol 2022, 10, 901037. [Google Scholar] [CrossRef] [PubMed]
- García-Estrada, C.; Domínguez-Santos, R.; Kosalková, K.; Martín, J.F. Transcription Factors Controlling Primary and Secondary Metabolism in Filamentous Fungi: The β-Lactam Paradigm. Fermentation 2018, Vol. 4, Page 47 2018, 4, 47. [Google Scholar] [CrossRef]
- Roberts, D.W.; St. Leger, R.J. Metarhizium Spp., Cosmopolitan Insect-Pathogenic Fungi: Mycological Aspects. Adv Appl Microbiol 2004, 54, 1–70. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Translating Biosynthetic Gene Clusters into Fungal Armor and Weaponry. Nature Chemical Biology 2015 11:9 2015, 11, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Gluck-Thaler, E.; Haridas, S.; Binder, M.; Grigoriev, I. V.; Crous, P.W.; Spatafora, J.W.; Bushley, K.; Slot, J.C. The Architecture of Metabolism Maximizes Biosynthetic Diversity in the Largest Class of Fungi. Mol Biol Evol 2020, 37, 2838–2856. [Google Scholar] [CrossRef] [PubMed]
- Blacutt, A.A.; Gold, S.E.; Voss, K.A.; Gao, M.; Glenn, A.E. Fusarium Verticillioides: Advancements in Understanding the Toxicity, Virulence, and Niche Adaptations of a Model Mycotoxigenic Pathogen of Maize. Phytopathology 2018, 108, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, M.A.; Rauf, A.; Ashfaq, M.; Ahmad, F. Exploring Biological Control Strategies for Managing Fusarium Mycotoxins. Nanohybrid Fungicides: New Frontiers in Plant Pathology 2024, 257–293. [CrossRef]
- Yazid, S.N.E.; Jinap, S.; Ismail, S.I.; Magan, N.; Samsudin, N.I.P. Phytopathogenic Organisms and Mycotoxigenic Fungi: Why Do We Control One and Neglect the Other? A Biological Control Perspective in Malaysia. Compr Rev Food Sci Food Saf 2020, 19, 643–669. [Google Scholar] [CrossRef] [PubMed]
- Rossman, A. Morphological and Molecular Perspectives on Systematics of the Hypocreales. Mycologia 1996, 88, 1–19. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, X.; Cai, J.; Chen, Y.; Li, B.; Guo, Z.; Huang, G. Genomic Characteristics and Comparative Genomics Analysis of the Endophytic Fungus Sarocladium Brachiariae. BMC Genomics 2019, 20, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Yoder, O.C.; Turgeon, B.G. Fungal Genomics and Pathogenicity. Curr Opin Plant Biol 2001, 4, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Christianson, D.W. Structural Biology and Chemistry of the Terpenoid Cyclases. Chem Rev 2006, 106, 3412–3442. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.M.; Croteau, R. Cyclization Enzymes in the Biosynthesis of Monoterpenes, Sesquiterpenes, and Diterpenes. 2000, 53–95. [CrossRef]
- Crutcher, F.K.; Parich, A.; Schuhmacher, R.; Mukherjee, P.K.; Zeilinger, S.; Kenerley, C.M. A Putative Terpene Cyclase, Vir4, Is Responsible for the Biosynthesis of Volatile Terpene Compounds in the Biocontrol Fungus Trichoderma Virens. Fungal Genetics and Biology 2013, 56, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, J.; Cooper, C.M.; Purchase, R. Natural Products Desk Reference. Natural Products Desk Reference 2015, 1–222. [Google Scholar] [CrossRef]
- Schmidt-Dannert, C. Biosynthesis of Terpenoid Natural Products in Fungi. Adv Biochem Eng Biotechnol 2014, 148, 19–61. [Google Scholar] [CrossRef]
- Akiyama, K.; Matsuzaki, K.I.; Hayashi, H. Plant Sesquiterpenes Induce Hyphal Branching in Arbuscular Mycorrhizal Fungi. Nature 2005 435:7043 2005, 435, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Kai, M.; Effmert, U.; Berg, G.; Piechulla, B. Volatiles of Bacterial Antagonists Inhibit Mycelial Growth of the Plant Pathogen Rhizoctonia Solani. Arch Microbiol 2007, 187, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Foppen, F.H.; Gribanovski-Sassu, O. Lipids Produced by Epicoccum Nigrum in Submerged Culture. Biochemical Journal 1968, 106, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, T.; Minami, A.; Oikawa, H. Recent Advances in the Biosynthesis of Ribosomally Synthesized and Posttranslationally Modified Peptides of Fungal Origin. The Journal of Antibiotics 2022 76:1 2022, 76, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Vignolle, G.A.; Mach, R.L.; Mach-Aigner, A.R.; Derntl, C. Novel Approach in Whole Genome Mining and Transcriptome Analysis Reveal Conserved RiPPs in Trichoderma Spp. BMC Genomics 2020, 21, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Deicke, M.; Mohr, J.F.; Roy, S.; Herzsprung, P.; Bellenger, J.P.; Wichard, T. Metallophore Profiling of Nitrogen-Fixing Frankia Spp. to Understand Metal Management in the Rhizosphere of Actinorhizal Plants. Metallomics 2019, 11, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, S.M.; Duckworth, O.W.; Harrington, J.M.; Schenkeveld, W.D.C. Metallophores and Trace Metal Biogeochemistry. Aquat Geochem 2015, 21, 159–195. [Google Scholar] [CrossRef]
- Kuzyk, S.B.; Hughes, E.; Yurkov, V. Discovery of Siderophore and Metallophore Production in the Aerobic Anoxygenic Phototrophs. Microorganisms 2021, 9, 959. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Aty, A.S. Fungicidal Activity of Indole Derivatives against Some Plant Pathogenic Fungi. J Pestic Sci 2010, 35, 431–440. [Google Scholar] [CrossRef]
- Shen, Q.; Liu, L.; Wang, L.; Wang, Q. Indole Primes Plant Defense against Necrotrophic Fungal Pathogen Infection. PLoS One 2018, 13, e0207607. [Google Scholar] [CrossRef] [PubMed]
- Svara, J.; Weferling, N.; Hofmann, T. Phosphorus Compounds, Organic. In Ullmann’s Encyclopedia of Industrial Chemistry; 2006. [Google Scholar] [CrossRef]
- Metcalf, W.W.; Van Der Donk, W.A. Biosynthesis of Phosphonic and Phosphinic Acid Natural Products. Annu Rev Biochem 2009, 78, 65–94. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.Y.; Won, T.H.; Raffa, N.; Baccile, J.A.; Wisecaver, J.; Keller, N.P.; Rokas, A.; Schroeder, F.C. Fungal Isocyanide Synthases and Xanthocillin Biosynthesis in Aspergillus Fumigatus. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Won, T.H.; Bok, J.W.; Nadig, N.; Venkatesh, N.; Nickles, G.; Greco, C.; Lim, F.Y.; González, J.B.; Turgeon, B.G.; Keller, N.P.; et al. Copper Starvation Induces Antimicrobial Isocyanide Integrated into Two Distinct Biosynthetic Pathways in Fungi. Nature Communications 2022 13:1 2022, 13, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Massarotti, A.; Brunelli, F.; Aprile, S.; Giustiniano, M.; Tron, G.C. Medicinal Chemistry of Isocyanides. Chem Rev 2021, 121, 10742–10788. [Google Scholar] [CrossRef] [PubMed]
- Raffa, N.; Won, T.H.; Sukowaty, A.; Candor, K.; Cui, C.; Halder, S.; Dai, M.; Landero-Figueroa, J.A.; Schroeder, F.C.; Keller, N.P. Dual-Purpose Isocyanides Produced by Aspergillus Fumigatus Contribute to Cellular Copper Sufficiency and Exhibit Antimicrobial Activity. Proc Natl Acad Sci U S A 2021, 118, e2015224118. [Google Scholar] [CrossRef] [PubMed]
- Crawford, J.M.; Portmann, C.; Zhang, X.; Roeffaers, M.B.J.; Clardy, J. Small Molecule Perimeter Defense in Entomopathogenic Bacteria. Proc Natl Acad Sci U S A 2012, 109, 10821–10826. [Google Scholar] [CrossRef]
- Harris, N.C.; Sato, M.; Herman, N.A.; Twigg, F.; Cai, W.; Liu, J.; Zhu, X.; Downey, J.; Khalaf, R.; Martin, J.; et al. Biosynthesis of Isonitrile Lipopeptides by Conserved Nonribosomal Peptide Synthetase Gene Clusters in Actinobacteria. Proc Natl Acad Sci U S A 2017, 114, 7025–7030. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.Y.; Chen, J.; Tang, Y.; Zhou, J.; Guo, Y.; Chang, W. chen Current Understanding toward Isonitrile Group Biosynthesis and Mechanism. Chin J Chem 2021, 39, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Sweany, R.R.; Breunig, M.; Opoku, J.; Clay, K.; Spatafora, J.W.; Drott, M.T.; Baldwin, T.T.; Fountain, J.C. Why Do Plant-Pathogenic Fungi Produce Mycotoxins? Potential Roles for Mycotoxins in the Plant Ecosystem. Phytopathology 2022, 112, 2044–2051. [Google Scholar] [CrossRef] [PubMed]
- Drott, M.T.; Lazzaro, B.P.; Brown, D.L.; Carbone, I.; Milgroom, M.G. Balancing Selection for Aflatoxin in Aspergillus Flavus Is Maintained through Interference Competition with, and Fungivory by Insects. Proceedings of the Royal Society B: Biological Sciences 2017, 284. [Google Scholar] [CrossRef]
- Fenteany, G.; Standaert, R.F.; Reichard, G.A.; Corey, E.J.; Schreiber, S.L. A Beta-Lactone Related to Lactacystin Induces Neurite Outgrowth in a Neuroblastoma Cell Line and Inhibits Cell Cycle Progression in an Osteosarcoma Cell Line. Proceedings of the National Academy of Sciences 1994, 91, 3358–3362. [Google Scholar] [CrossRef] [PubMed]
- Meesil, W.; Muangpat, P.; Sitthisak, S.; Rattanarojpong, T.; Chantratita, N.; Machado, R.A.R.; Shi, Y.M.; Bode, H.B.; Vitta, A.; Thanwisai, A. Genome Mining Reveals Novel Biosynthetic Gene Clusters in Entomopathogenic Bacteria. Scientific Reports 2023 13:1 2023, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Tian, Y.; Li, J.; Xu, G.; Zhang, Y.; Chen, S.; Chen, Y.; Tang, X. Complete Genome Sequence of Bacillus Cereus Z4, a Biocontrol Agent against Tobacco Black Shank, Isolated from the Western Pacific Ocean. Mar Genomics 2023, 72, 101071. [Google Scholar] [CrossRef] [PubMed]
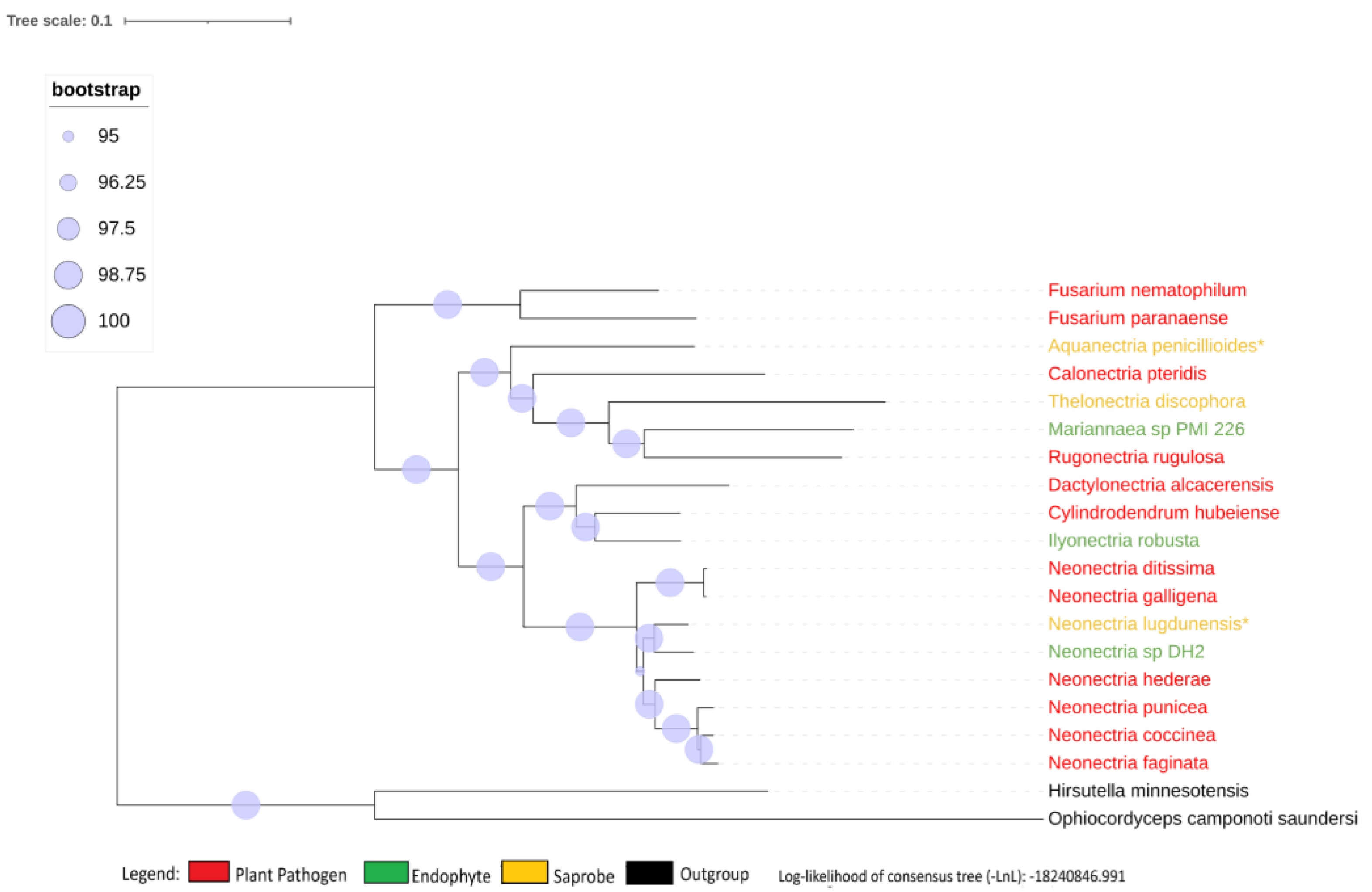
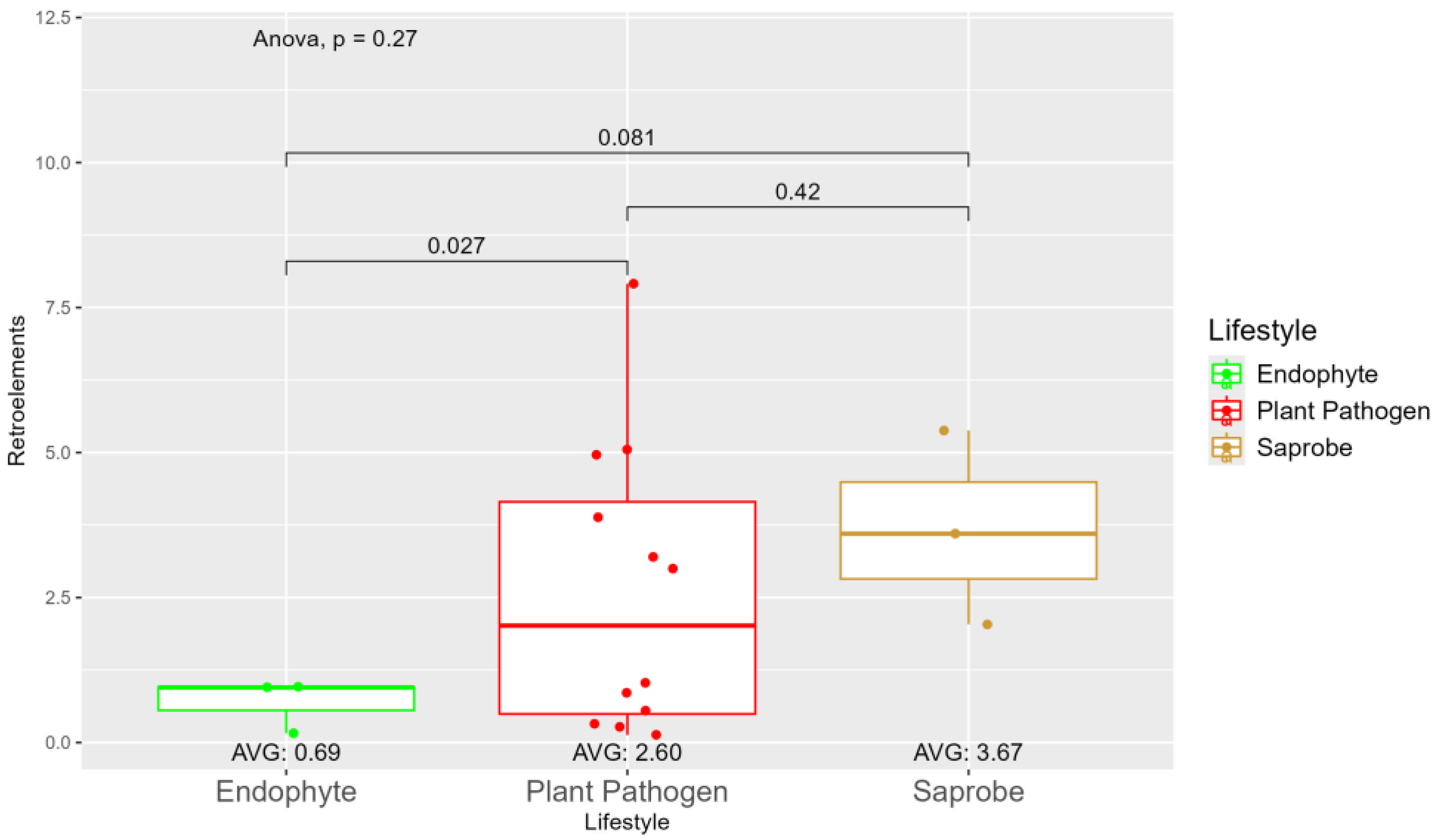
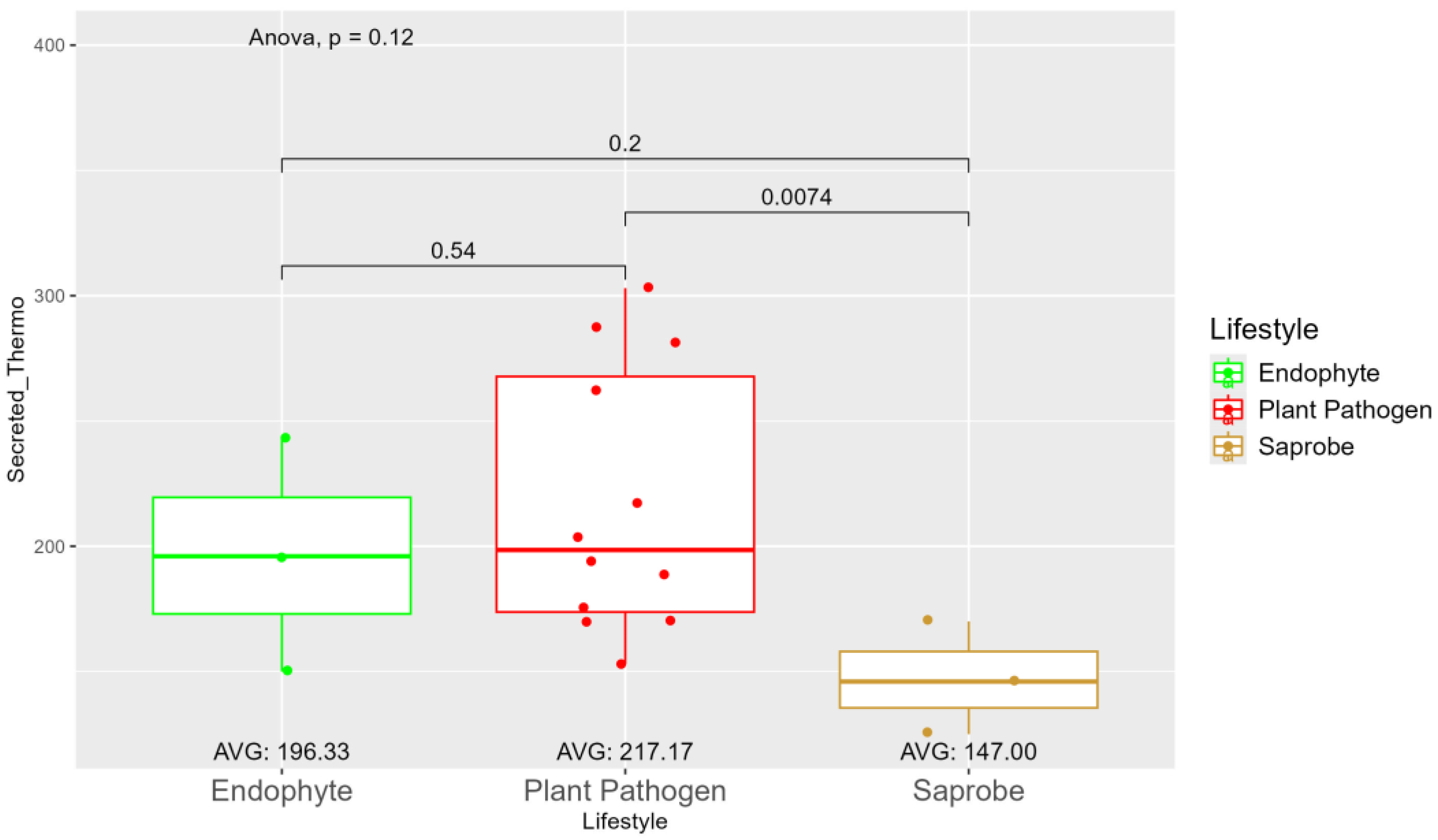
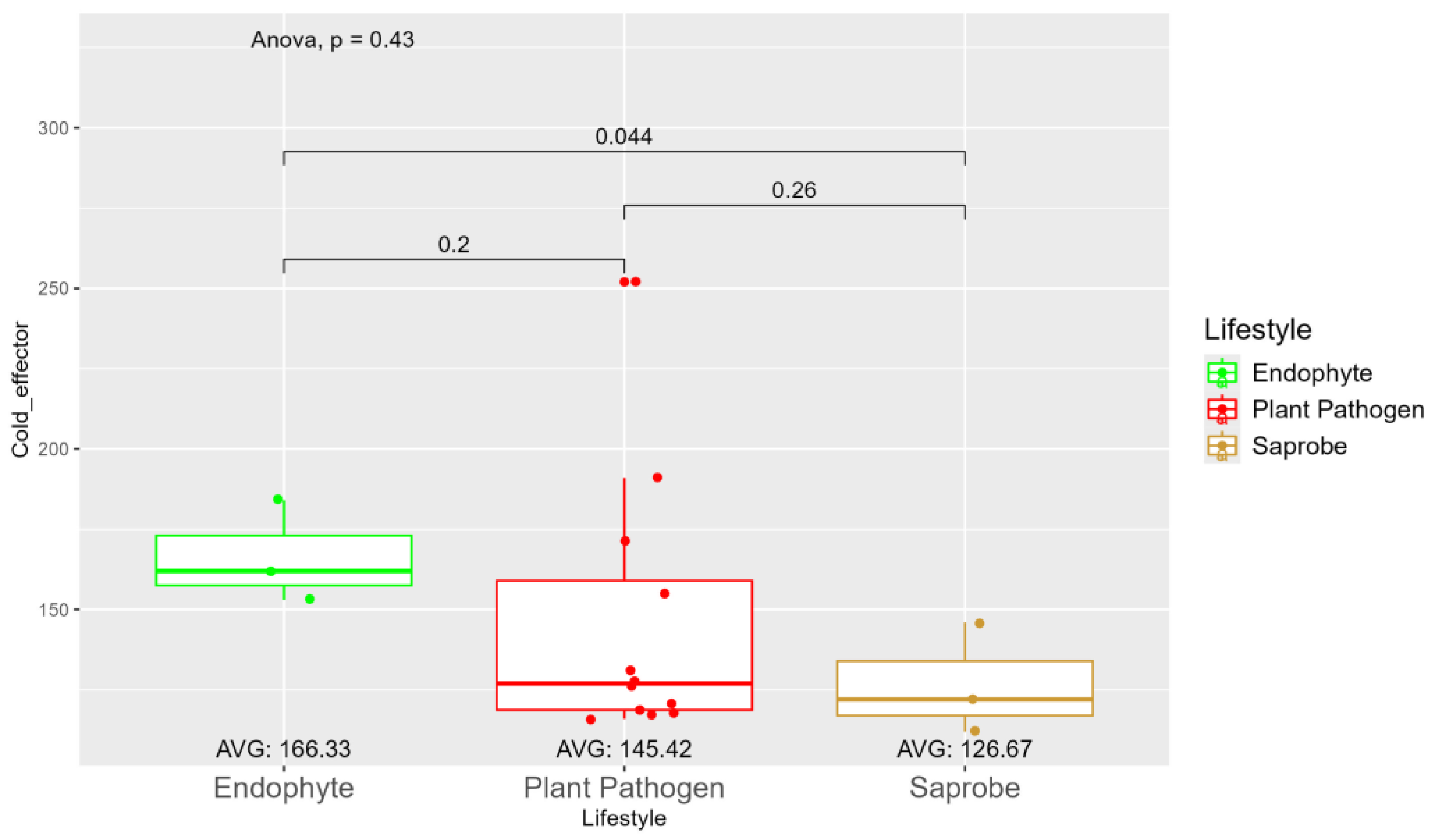
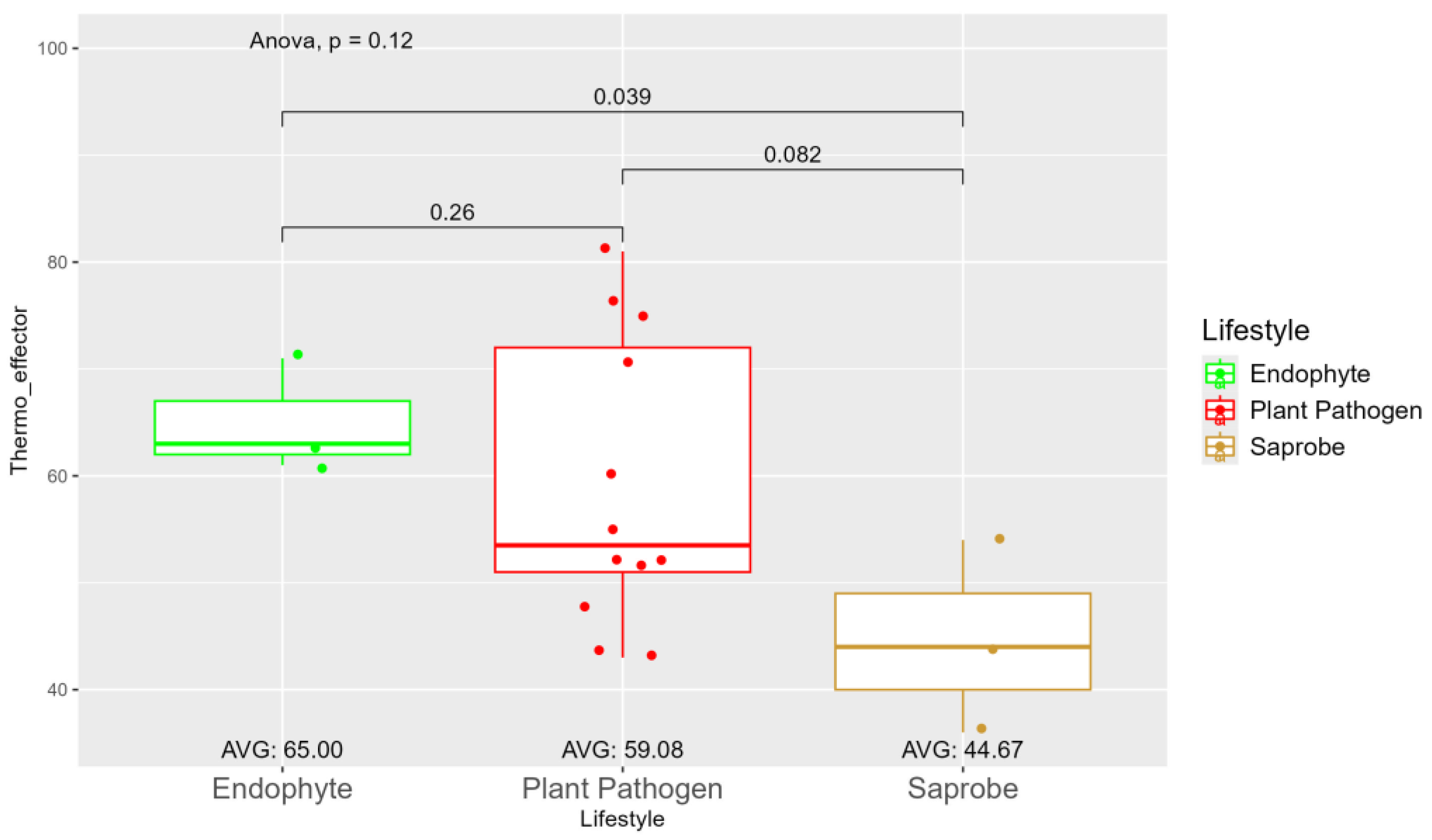
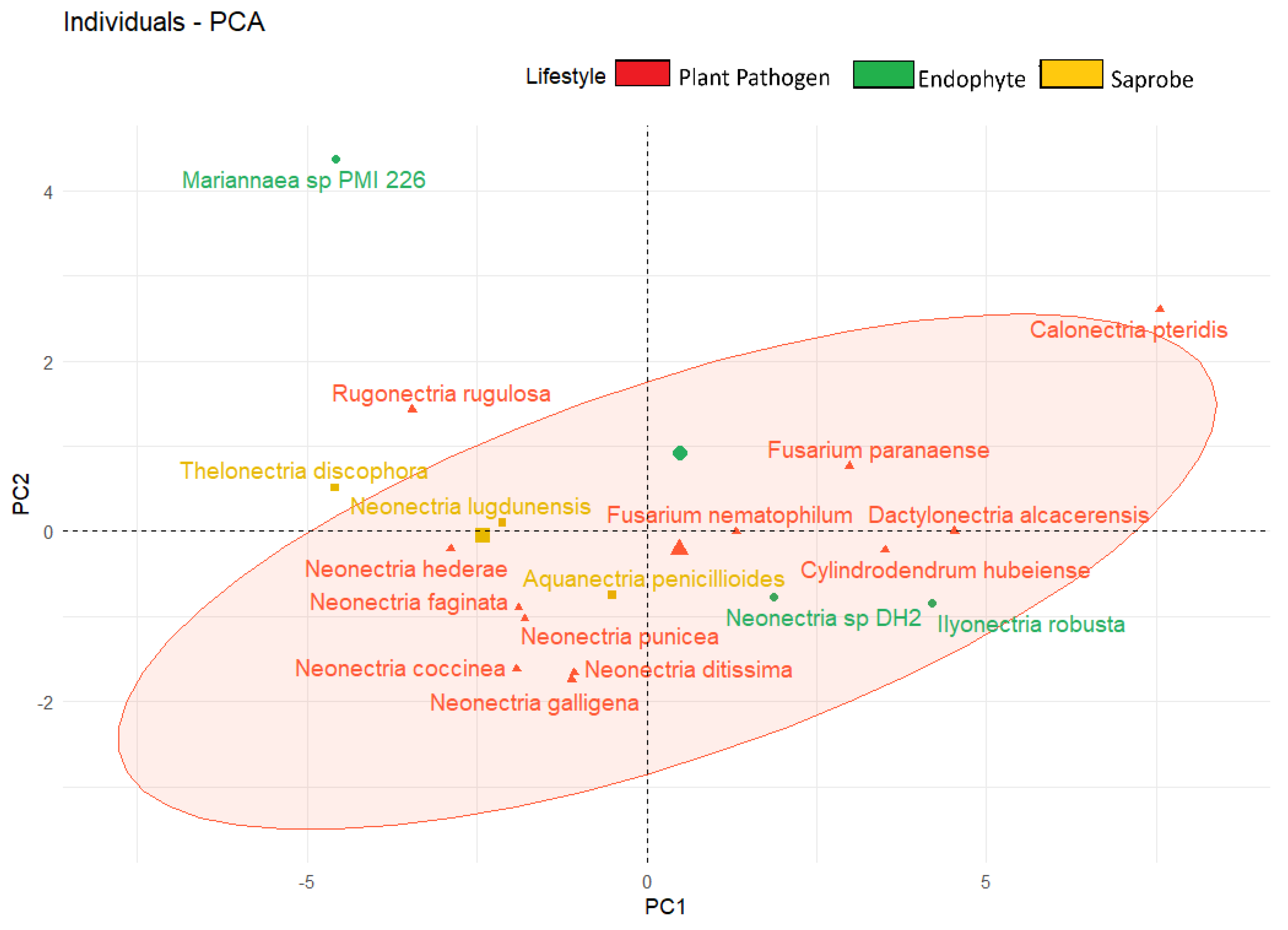
| Species | Genome Length (Mbp) | Genome Compl. (%) | GC Content (%) | N50 | Assembly Accession |
|---|---|---|---|---|---|
| Ilyonectria robusta | 59.64 | 97.5% | 51.68 | 1.2Mb | GCF_021365365.1 |
| Calonectria pteridis | 58.37 | 97.7% | 50.21 | 3.2Mb | GCA_022837005.1 |
| Dactylonectria alcacerensis | 61.76 | 97.9% | 49.86 | 4.3Mb | GCA_029931735.1 |
| Fusarium paranaense | 53.40 | 98.2% | 49.21 | 859.2Kb | GCA_027886155.1 |
| Fusarium nematophilum | 50.82 | 96.4% | 53.92 | 148.30Kb | GCA_033030565.1 |
| Cylindrodendrum hubeiense | 48.81 | 96.5% | 51.81 | 85.2Kb | GCA_014621425.1 |
| Rugonectria rugulosa | 46.95 | 97.6% | 51.43 | 56.0Kb | GCA_023509875.1 |
| Neonectria sp DH2 | 45.82 | 94.8% | 52.99 | 45.8Mb | GCA_003934905.1 |
| Aquanectria penicillioides | 53.76 | 95.0% | 47.94 | 4.93Mbp | GCA_003415625.1 |
| Neonectria ditissima | 44.95 | 97.5% | 51.83 | 1.8Mb | GCA_001305505.1 |
| Neonectria lugdunensis | 44.78 | 97.6% | 52.17 | 44.7Mb | To be added during review |
| Neonectria punicea | 41.47 | 96.8% | 52.72 | 41.4Mb | GCA_003385315.1 |
| Neonectria galligena | 41.00 | 96.7% | 53.9 | 31.3Kb | GCA_013759035.1 |
| Neonectria coccinea | 42.74 | 97.3% | 51.65 | 178.8Kb | GCA_019137265.1 |
| Neonectria faginata | 42.94 | 97.4% | 52.48 | 4.4Mb | GCA_030864175.1 |
| Neonectria hederae | 43.28 | 97.5% | 49.43 | 248.9Kb | GCA_003385265.1 |
| Thelonectria discophora | 41.60 | 97.8% | 54.16 | 41.6Mb | GCA_911649645.1 |
| Mariannaea sp PMI 226 | 42.25 | 97.7% | 48.55 | 2.9Mb | GCA_024336345.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





