Submitted:
21 July 2024
Posted:
22 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Preprocessing of RNA-Sequencing Data
2.2. Principal Component Analysis
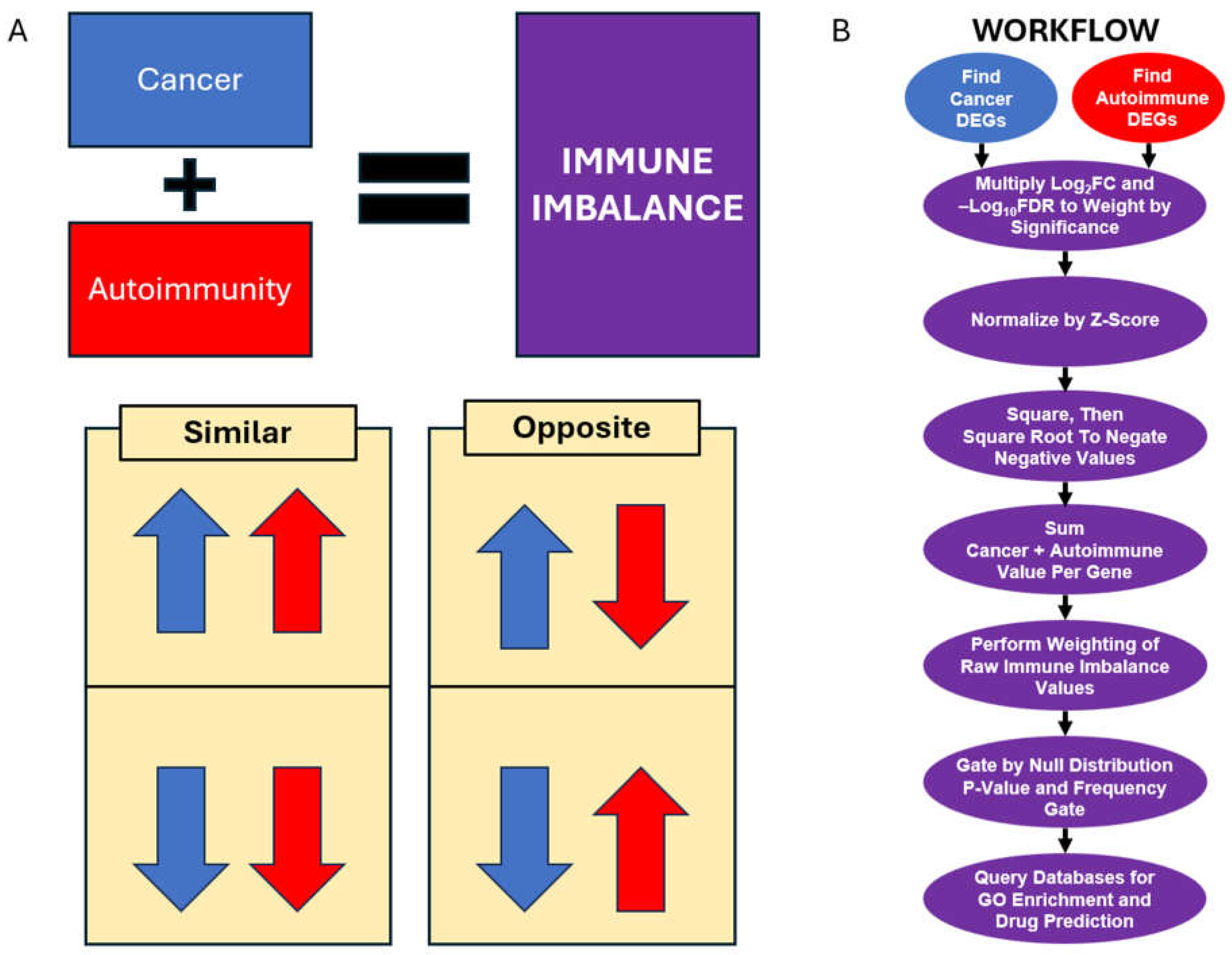
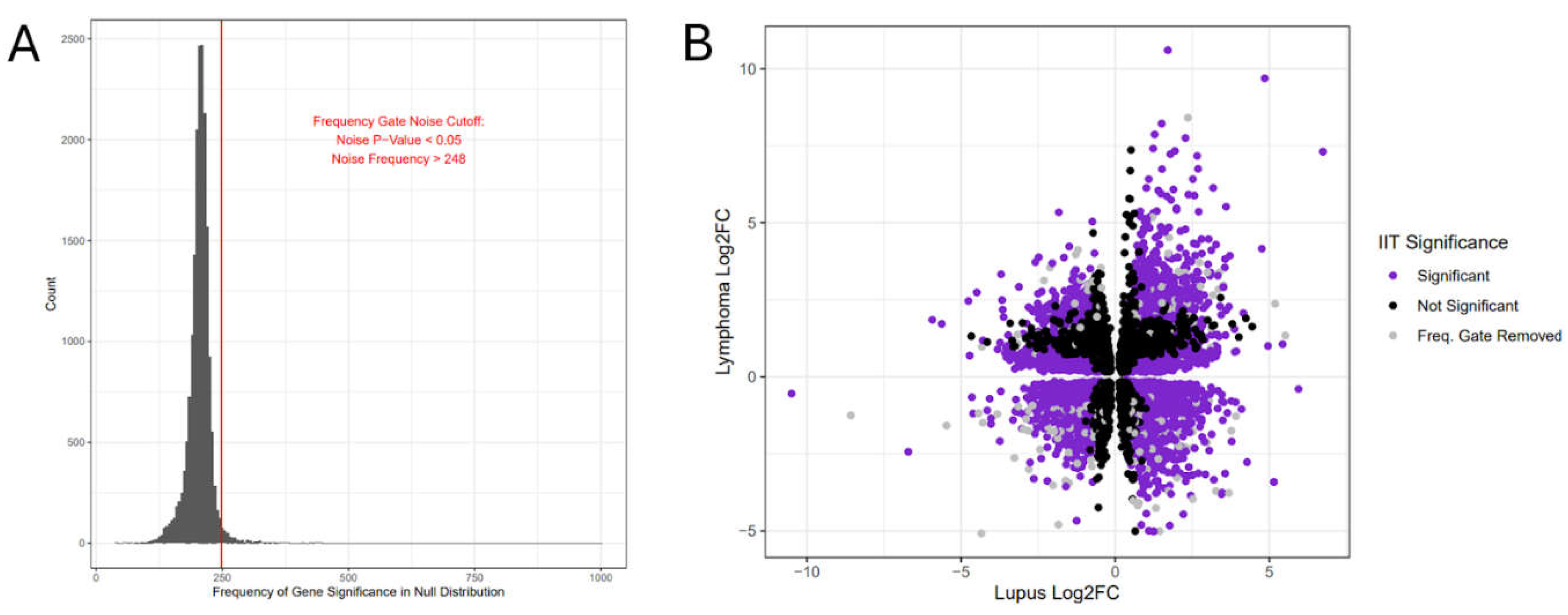
2.3. Immune Imbalance Determination
2.4. Immune Imbalance Validation
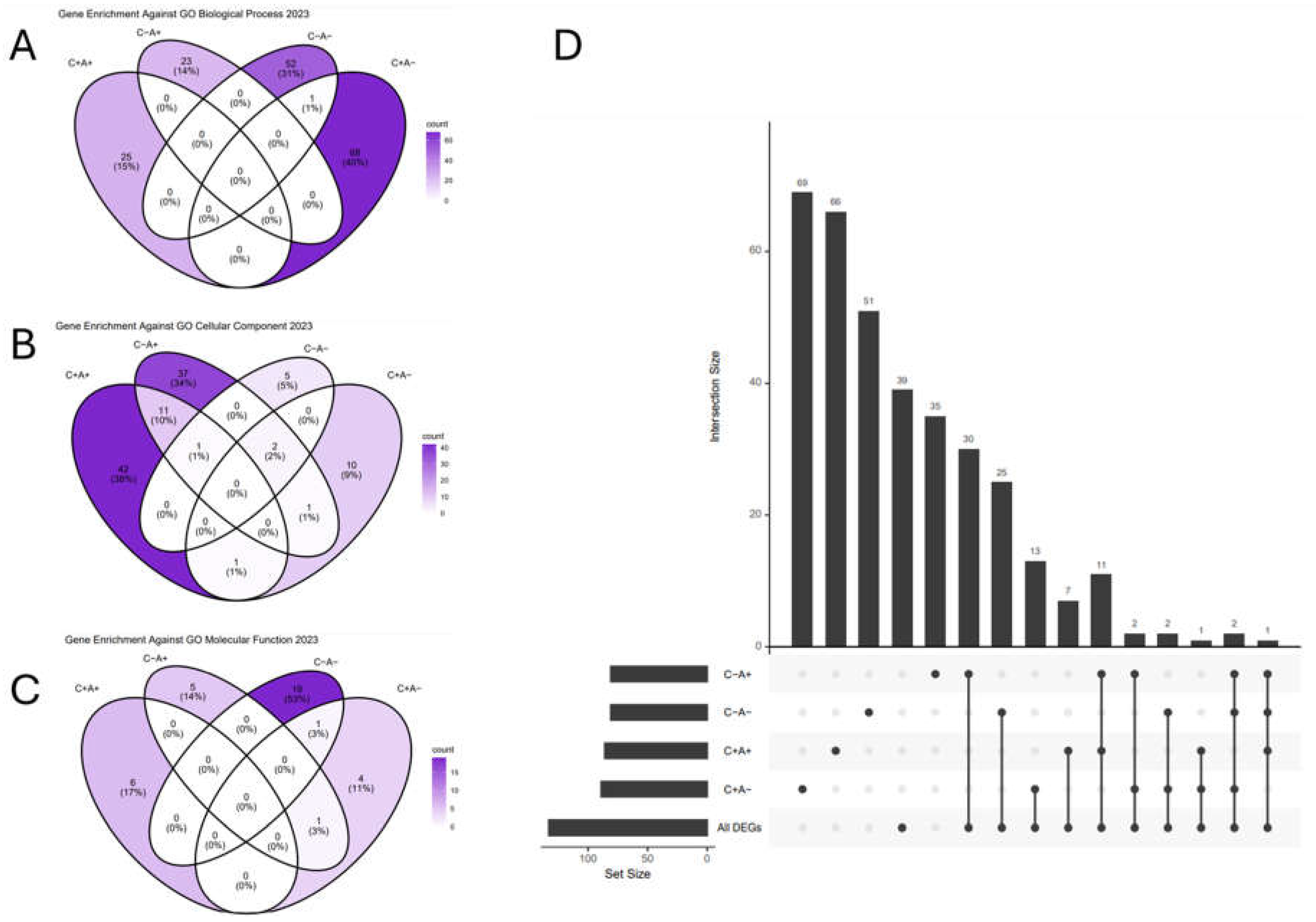
3. Results
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tian, J.; Zhang, D.; Yao, X.; Huang, Y.; Lu, Q. Global Epidemiology of Systemic Lupus Erythematosus: A Comprehensive Systematic Analysis and Modelling Study. Ann. Rheum. Dis. 2023, 82, 351–356. [Google Scholar] [CrossRef]
- Dörner, T.; Furie, R. Novel Paradigms in Systemic Lupus Erythematosus. Lancet 2019, 393, 2344–2358. [Google Scholar] [CrossRef]
- Lleo, A.; Selmi, C.; Invernizzi, P.; Podda, M.; Gershwin, M.E. The Consequences of Apoptosis in Autoimmunity. J. Autoimmun. 2008, 31, 257–262. [Google Scholar] [CrossRef]
- Take a Look at the Eyes in Systemic Lupus Erythematosus: A Novel Point of View. Autoimmun. Rev. 2019, 18, 247–254. [CrossRef]
- Zonana-Nacach, A.; Barr, S.G.; Magder, L.S.; Petri, M. Damage in Systemic Lupus Erythematosus and Its Association with Corticosteroids. Arthritis Rheum. 2000, 43, 1801–1808. [Google Scholar] [CrossRef]
- Gladman, D.D.; Urowitz, M.B.; Rahman, P.; Ibañez, D.; Tam, L.-S. Accrual of Organ Damage over Time in Patients with Systemic Lupus Erythematosus. J. Rheumatol. 2003, 30, 1955–1959. [Google Scholar]
- Basta, F.; Fasola, F.; Triantafyllias, K.; Schwarting, A. Systemic Lupus Erythematosus (SLE) Therapy: The Old and the New. Rheumatol Ther 2020, 7, 433–446. [Google Scholar] [CrossRef]
- Jamil, A.; Mukkamalla, S.K.R. Lymphoma. In StatPearls [Internet]; StatPearls Publishing, 2023. [Google Scholar]
- Global Burden of Disease Cancer Collaboration; Fitzmaurice, C. ; Akinyemiju, T.F.; Al Lami, F.H.; Alam, T.; Alizadeh-Navaei, R.; Allen, C.; Alsharif, U.; Alvis-Guzman, N.; Amini, E.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 2018, 4, 1553–1568. [Google Scholar]
- Shankland, K.R.; Armitage, J.O.; Hancock, B.W. Non-Hodgkin Lymphoma. Lancet 2012, 380, 848–857. [Google Scholar] [CrossRef]
- Ma, J.S.Y.; Kim, J.Y.; Kazane, S.A.; Choi, S.-H.; Yun, H.Y.; Kim, M.S.; Rodgers, D.T.; Pugh, H.M.; Singer, O.; Sun, S.B.; et al. Versatile Strategy for Controlling the Specificity and Activity of Engineered T Cells. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, E450–E458. [Google Scholar] [CrossRef]
- Zintzaras, E.; Voulgarelis, M.; Moutsopoulos, H.M. The Risk of Lymphoma Development in Autoimmune Diseases: A Meta-Analysis. Arch. Intern. Med. 2005, 165, 2337–2344. [Google Scholar] [CrossRef]
- The Resolution of Inflammation: Principles and Challenges. Semin. Immunol. 2015, 27, 149–160. [CrossRef]
- Rogovskii, V. Immune Tolerance as the Physiologic Counterpart of Chronic Inflammation. Front. Immunol. 2020, 11, 2061. [Google Scholar] [CrossRef]
- Abdulkhaleq, L.A.; Assi, M.A.; Abdullah, R.; Zamri-Saad, M.; Taufiq-Yap, Y.H.; Hezmee, M.N.M. The Crucial Roles of Inflammatory Mediators in Inflammation: A Review. Vet World 2018, 11, 627–635. [Google Scholar] [CrossRef]
- Buckley, C.D.; Gilroy, D.W.; Serhan, C.N.; Stockinger, B.; Tak, P.P. The Resolution of Inflammation. Nat. Rev. Immunol. 2012, 13, 59–66. [Google Scholar] [CrossRef]
- Delogu, L.G.; Deidda, S.; Delitala, G.; Manetti, R. Infectious Diseases and Autoimmunity. J. Infect. Dev. Ctries. 2011, 5, 679–687. [Google Scholar] [CrossRef]
- Kim-Hellmuth, S.; Bechheim, M.; Pütz, B.; Mohammadi, P.; Nédélec, Y.; Giangreco, N.; Becker, J.; Kaiser, V.; Fricker, N.; Beier, E.; et al. Genetic Regulatory Effects Modified by Immune Activation Contribute to Autoimmune Disease Associations. Nat. Commun. 2017, 8, 266. [Google Scholar] [CrossRef]
- Kim, R.; Emi, M.; Tanabe, K. Cancer Immunosuppression and Autoimmune Disease: Beyond Immunosuppressive Networks for Tumour Immunity. Immunology 2006, 119, 254–264. [Google Scholar] [CrossRef]
- Ghorani, E.; Swanton, C.; Quezada, S.A. Cancer Cell-Intrinsic Mechanisms Driving Acquired Immune Tolerance. Immunity 2023, 56, 2270–2295. [Google Scholar] [CrossRef]
- Makkouk, A.; Weiner, G.J. Cancer Immunotherapy and Breaking Immune Tolerance: New Approaches to an Old Challenge. Cancer Res. 2015, 75, 5–10. [Google Scholar] [CrossRef]
- Menter, T.; Tzankov, A. Mechanisms of Immune Evasion and Immune Modulation by Lymphoma Cells. Front. Oncol. 2018, 8, 344644. [Google Scholar] [CrossRef]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. Methods Mol. Biol. 2016, 1418, 93–110. [Google Scholar]
- Rapier-Sharman, N.; Clancy, J.; Pickett, B.E. Joint Secondary Transcriptomic Analysis of Non-Hodgkin’s B-Cell Lymphomas Predicts Reliance on Pathways Associated with the Extracellular Matrix and Robust Diagnostic Biomarkers. J Bioinform Syst Biol 2022, 5, 119–135. [Google Scholar] [CrossRef]
- Faramand, R.; Jain, M.; Staedtke, V.; Kotani, H.; Bai, R.; Reid, K.; Lee, S.B.; Spitler, K.; Wang, X.; Cao, B.; et al. Tumor Microenvironment Composition and Severe Cytokine Release Syndrome (CRS) Influence Toxicity in Patients with Large B-Cell Lymphoma Treated with Axicabtagene Ciloleucel. Clin. Cancer Res. 2020, 26, 4823–4831. [Google Scholar] [CrossRef]
- Koues, O.I.; Kowalewski, R.A.; Chang, L.-W.; Pyfrom, S.C.; Schmidt, J.A.; Luo, H.; Sandoval, L.E.; Hughes, T.B.; Bednarski, J.J.; Cashen, A.F.; et al. Enhancer Sequence Variants and Transcription-Factor Deregulation Synergize to Construct Pathogenic Regulatory Circuits in B-Cell Lymphoma. Immunity 2015, 42, 186–198. [Google Scholar] [CrossRef]
- Raju, S.; Kretzmer, L.Z.; Koues, O.I.; Payton, J.E.; Oltz, E.M.; Cashen, A.; Polic, B.; Schreiber, R.D.; Shaw, A.S.; Markiewicz, M.A. NKG2D-NKG2D Ligand Interaction Inhibits the Outgrowth of Naturally Arising Low-Grade B Cell Lymphoma In Vivo. J. Immunol. 2016, 196, 4805–4813. [Google Scholar] [CrossRef]
- Teater, M.; Dominguez, P.M.; Redmond, D.; Chen, Z.; Ennishi, D.; Scott, D.W.; Cimmino, L.; Ghione, P.; Chaudhuri, J.; Gascoyne, R.D.; et al. AICDA Drives Epigenetic Heterogeneity and Accelerates Germinal Center-Derived Lymphomagenesis. Nat. Commun. 2018, 9, 222. [Google Scholar] [CrossRef]
- Porpaczy, E.; Tripolt, S.; Hoelbl-Kovacic, A.; Gisslinger, B.; Bago-Horvath, Z.; Casanova-Hevia, E.; Clappier, E.; Decker, T.; Fajmann, S.; Fux, D.A.; et al. Aggressive B-Cell Lymphomas in Patients with Myelofibrosis Receiving JAK1/2 Inhibitor Therapy. Blood 2018, 132, 694–706. [Google Scholar] [CrossRef]
- Li, M.; Chiang, Y.-L.; Lyssiotis, C.A.; Teater, M.R.; Hong, J.Y.; Shen, H.; Wang, L.; Hu, J.; Jing, H.; Chen, Z.; et al. Non-Oncogene Addiction to SIRT3 Plays a Critical Role in Lymphomagenesis. Cancer Cell 2019, 35, 916–931.e9. [Google Scholar] [CrossRef] [PubMed]
- Rouhigharabaei, L.; Ferreiro, J.F.; Tousseyn, T.; van der Krogt, J.-A.; Put, N.; Haralambieva, E.; Graux, C.; Maes, B.; Vicente, C.; Vandenberghe, P.; et al. Non-IG Aberrations of FOXP1 in B-Cell Malignancies Lead to an Aberrant Expression of N-Truncated Isoforms of FOXP1; 2015. [Google Scholar]
- Panwar, B.; Schmiedel, B.J.; Liang, S.; White, B.; Rodriguez, E.; Kalunian, K.; McKnight, A.J.; Soloff, R.; Seumois, G.; Vijayanand, P.; et al. Multi-Cell Type Gene Coexpression Network Analysis Reveals Coordinated Interferon Response and Cross-Cell Type Correlations in Systemic Lupus Erythematosus. Genome Res. 2021, 31, 659–676. [Google Scholar] [CrossRef] [PubMed]
- Andreoletti, G.; Lanata, C.M.; Trupin, L.; Paranjpe, I.; Jain, T.S.; Nititham, J.; Taylor, K.E.; Combes, A.J.; Maliskova, L.; Ye, C.J.; et al. Transcriptomic Analysis of Immune Cells in a Multi-Ethnic Cohort of Systemic Lupus Erythematosus Patients Identifies Ethnicity- and Disease-Specific Expression Signatures. Commun Biol 2021, 4, 488. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, S.; Ehrenberg, P.K.; Geretz, A.; Yum, L.; Kundu, G.; May, K.; Fourati, S.; Nganou-Makamdop, K.; Williams, L.D.; Sawant, S.; et al. Monocyte-Derived Transcriptome Signature Indicates Antibody-Dependent Cellular Phagocytosis as a Potential Mechanism of Vaccine-Induced Protection against HIV-1. Elife 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Gao, C.; Zhao, K.; Yang, Y.; Rassadkina, Y.; Fajnzylber, J.; Regan, J.; Li, J.Z.; Lichterfeld, M.; Yu, X.G. Immune-Profiling of SARS-CoV-2 Viremic Patients Reveals Dysregulated Innate Immune Responses. Front. Immunol. 2022, 13, 984553. [Google Scholar] [CrossRef] [PubMed]
- ENCODE Project Consortium An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 2012, 489, 57–74. [CrossRef] [PubMed]
- Wang, X.; Campbell, M.R.; Cho, H.-Y.; Pittman, G.S.; Martos, S.N.; Bell, D.A. Epigenomic Profiling of Isolated Blood Cell Types Reveals Highly Specific B Cell Smoking Signatures and Links to Disease Risk. Clin. Epigenetics 2023, 15, 90. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J.M.; Pyfrom, S.C.; Schmidt, J.A.; Koues, O.I.; Kowalewski, R.A.; Grams, N.R.; Sun, J.J.; Berman, L.R.; Duncavage, E.J.; Lee, Y.-S.; et al. Loss of Synergistic Transcriptional Feedback Loops Drives Diverse B-Cell Cancers. EBioMedicine 2021, 71, 103559. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Ren, Z.; Zhang, B.; Mao, L.; Zhu, G.; Gao, L.; Su, J.; Ye, J.; Long, Z.; Zhu, Y.; et al. Insufficient Epitope-Specific T Cell Clones Are Responsible for Impaired Cellular Immunity to Inactivated SARS-CoV-2 Vaccine in Older Adults. Nat Aging 2023, 3, 418–435. [Google Scholar] [CrossRef] [PubMed]
- Rapier-Sharman, N.; Krapohl, J.; Beausoleil, E.J.; Gifford, K.T.L.; Hinatsu, B.R.; Hoffmann, C.S.; Komer, M.; Scott, T.M.; Pickett, B.E. Preprocessing of Public RNA-Sequencing Datasets to Facilitate Downstream Analyses of Human Diseases. Data 2021, 6, 75. [Google Scholar] [CrossRef]
- Transcriptomics Secondary Analysis of Severe Human Infection with SARS-CoV-2 Identifies Gene Expression Changes and Predicts Three Transcriptional Biomarkers in Leukocytes. Comput. Struct. Biotechnol. J. 2023, 21, 1403–1413. [CrossRef]
- Moreno, C.; Bybee, E.; Tellez Freitas, C.M.; Pickett, B.E.; Weber, K.S. Meta-Analysis of Two Human RNA-Seq Datasets to Determine Periodontitis Diagnostic Biomarkers and Drug Target Candidates. Int. J. Mol. Sci. 2022, 23, 5580. [Google Scholar] [CrossRef]
- GitHub - Ncbi/sra-Tools: SRA Tools. Available online: https://github.com/ncbi/sra-tools (accessed on 19 July 2024).
- Orjuela, S.; Huang, R.; Hembach, K.M.; Robinson, M.D.; Soneson, C. ARMOR: An Automated Reproducible MOdular Workflow for Preprocessing and Differential Analysis of RNA-Seq Data. G3 Genes|Genomes|Genetics 2019, 9, 2089–2096. [Google Scholar] [CrossRef] [PubMed]
- Köster, J.; Rahmann, S. Snakemake--a Scalable Bioinformatics Workflow Engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef] [PubMed]
- Babraham Bioinformatics - Trim Galore! Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 8 March 2023).
- Babraham Bioinformatics - FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 8 March 2023).
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular Signatures Database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Smyth, G.K. Camera: A Competitive Gene Set Test Accounting for Inter-Gene Correlation. Nucleic Acids Research 2012, 40, e133–e133. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer Science & Business Media, 2009; ISBN 9780387981413. [Google Scholar]
- Shaffer, J.P. Multiple Hypothesis Testing. Annu. Rev. Psychol. 1995, 46, 561–584. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinformatics 2013, 14, 128. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr Protoc 2021, 1, e90. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-Scale Gene Function Analysis with the PANTHER Classification System. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Website Available online. [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The Harmonizome: A Collection of Processed Datasets Gathered to Serve and Mine Knowledge about Genes and Proteins. Database 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.-H.; Chen, C.; Akyol, T.; Dusa, A.; Yu, G.; Cao, B.; Cai, P. ggVennDiagram: Intuitive Venn Diagram Software Extended. Imeta 2024, 3, e177. [Google Scholar] [CrossRef]
- Gao, C.-H.; Yu, G.; Cai, P. ggVennDiagram: An Intuitive, Easy-to-Use, and Highly Customizable R Package to Generate Venn Diagram. Front. Genet. 2021, 12, 706907. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R Package for the Visualization of Intersecting Sets and Their Properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping Identifiers for the Integration of Genomic Datasets with the R/Bioconductor Package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A Powerful Link between Biological Databases and Microarray Data Analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef]
- Ochoa, D.; Hercules, A.; Carmona, M.; Suveges, D.; Baker, J.; Malangone, C.; Lopez, I.; Miranda, A.; Cruz-Castillo, C.; Fumis, L.; et al. The next-Generation Open Targets Platform: Reimagined, Redesigned, Rebuilt. Nucleic Acids Res. 2023, 51, D1353–D1359. [Google Scholar] [CrossRef]
- Davis, J.; Handunnetti, S.M.; Sharpe, C.; Turner, G.; Anderson, M.A.; Roberts, A.W.; Seymour, J.F.; Tam, C.S.; Ritchie, D.; Koldej, R. Long Term Responses to Venetoclax and Ibrutinib in Mantle Cell Lymphoma Are Associated with Immunological Recovery and Prognostic Changes in Inflammatory Biomarkers. Blood 2019, 134, 2791–2791. [Google Scholar] [CrossRef]
- Guo, J.; Gao, Y.; Wang, Y.; Zou, Y.; Du, Y.; Luo, C.; Shi, Y.; Yang, Y.; Wu, X.; Su, Y.; et al. Investigation of C1-Complex Regions Reveals New C1Q Variants Associated with Protection from Systemic Lupus Erythematosus, and Affect Its Transcript Abundance. Sci. Rep. 2018, 8, 8048. [Google Scholar] [CrossRef] [PubMed]
- Sontheimer, R.D.; Racila, E.; Racila, D.M. C1q: Its Functions within the Innate and Adaptive Immune Responses and Its Role in Lupus Autoimmunity. J. Invest. Dermatol. 2005, 125, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Suomela, S.; Cao, L.; Bowcock, A.; Saarialho-Kere, U. Interferon Alpha-Inducible Protein 27 (IFI27) Is Upregulated in Psoriatic Skin and Certain Epithelial Cancers. J. Invest. Dermatol. 2004, 122, 717–721. [Google Scholar] [CrossRef]
- Sagou, K.; Sato, Y.; Okuno, Y.; Watanabe, T.; Inagaki, T.; Motooka, Y.; Toyokuni, S.; Murata, T.; Kiyoi, H.; Kimura, H. Epstein-Barr Virus Lytic Gene BNRF1 Promotes B-Cell Lymphomagenesis via IFI27 Upregulation. PLoS Pathog. 2024, 20, e1011954. [Google Scholar] [CrossRef] [PubMed]
- Skov, V.; Larsen, T.S.; Thomassen, M.; Riley, C.H.; Jensen, M.K.; Bjerrum, O.W.; Kruse, T.A.; Hasselbalch, H.C. Whole-Blood Transcriptional Profiling of Interferon-Inducible Genes Identifies Highly Upregulated IFI27 in Primary Myelofibrosis. Eur. J. Haematol. 2011, 87, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, L.; Wang, J.; Zhang, M.; Song, Z.; Ni, B.; You, Y. Identification of Key Biomarkers and Immune Infiltration in Systemic Lupus Erythematosus by Integrated Bioinformatics Analysis. J. Transl. Med. 2021, 19, 1–17. [Google Scholar]
- Gallagher, K.M.; Roderick, J.E.; Tan, S.H.; Tan, T.K.; Murphy, L.; Yu, J.; Li, R.; O’Connor, K.W.; Zhu, J.; Green, M.R.; et al. ESRRB Regulates Glucocorticoid Gene Expression in Mice and Patients with Acute Lymphoblastic Leukemia. Blood Adv 2020, 4, 3154–3168. [Google Scholar]
- Dobbs Spendlove, M.; M Gibson, T.; McCain, S.; Stone, B.C.; Gill, T.; Pickett, B.E. Pathway2Targets: An Open-Source Pathway-Based Approach to Repurpose Therapeutic Drugs and Prioritize Human Targets. PeerJ 2023, 11, e16088. [Google Scholar] [CrossRef]
- Adil, S.; Paracha, R.Z.; Tariq, S.; Nisar, M.; Ijaz, S.; Siddiqa, A.; Hussain, Z.; Amir, A. A Computational Systems Analyses to Identify Biomarkers and Mechanistic Link in Psoriasis and Cutaneous Squamous Cell Carcinoma. Front. Immunol. 2021, 12, 662528. [Google Scholar] [CrossRef]
- Liu, X.; Ji, J.; Forsti, A.; Sundquist, K.; Sundquist, J.; Hemminki, K. Autoimmune Disease and Subsequent Urological Cancer. J. Urol. 2013, 189, 2262–2268. [Google Scholar] [CrossRef] [PubMed]
- Hemminki, K.; Liu, X.; Ji, J.; Sundquist, K.; Sundquist, J. Subsequent COPD and Lung Cancer in Patients with Autoimmune Disease. Eur. Respir. J. 2011, 37, 463–465. [Google Scholar] [CrossRef]
- Hemminki, K.; Liu, X.; Ji, J.; Sundquist, J.; Sundquist, K. Autoimmune Disease and Subsequent Digestive Tract Cancer by Histology. Ann. Oncol. 2012, 23, 927–933. [Google Scholar] [CrossRef] [PubMed]
- Joseph, C.G.; Darrah, E.; Shah, A.A.; Skora, A.D.; Casciola-Rosen, L.A.; Wigley, F.M.; Boin, F.; Fava, A.; Thoburn, C.; Kinde, I.; et al. Association of the Autoimmune Disease Scleroderma with an Immunologic Response to Cancer. Science 2014, 343, 152–157. [Google Scholar] [CrossRef]
- Shiokawa, M.; Kodama, Y.; Yoshimura, K.; Kawanami, C.; Mimura, J.; Yamashita, Y.; Asada, M.; Kikuyama, M.; Okabe, Y.; Inokuma, T.; et al. Risk of Cancer in Patients with Autoimmune Pancreatitis. Am. J. Gastroenterol. 2013, 108, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Rahat, M.A.; Shakya, J. Parallel Aspects of the Microenvironment in Cancer and Autoimmune Disease. Mediators Inflamm. 2016, 2016, 4375120. [Google Scholar] [CrossRef] [PubMed]
- Rahat, M.A.; Coffelt, S.B.; Granot, Z.; Muthana, M.; Amedei, A. Macrophages and Neutrophils: Regulation of the Inflammatory Microenvironment in Autoimmunity and Cancer. Mediators Inflamm. 2016, 2016, 5894347. [Google Scholar] [CrossRef]
- Jiménez-Morales, S.; Ren, X.; Dean, M. Editorial: The Genetic Causes Underlying Immune Mediated Disease: A Focus on Autoimmunity and Cancer. Front. Genet. 2022, 13, 889160. [Google Scholar] [CrossRef]
- Medzhitov, R. Origin and Physiological Roles of Inflammation. Nature 2008, 454, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Demers-Mathieu, V. Optimal Selection of IFN-α-Inducible Genes to Determine Type I Interferon Signature Improves the Diagnosis of Systemic Lupus Erythematosus. Biomedicines 2023, 11. [Google Scholar] [CrossRef]
- Li, H.; Liang, J.; Gao, Y.; Liu, M.; Xia, N.; Kong, W.; Zheng, L.; Zhang, Y.; Li, Z.; Chen, H.; et al. IGFBP2 Function as a Novel Biomarker for Active Lupus Nephritis. J. Mol. Med. 2022, 100, 1479–1491. [Google Scholar] [CrossRef]
- Mok, C.C.; Ding, H.H.; Kharboutli, M.; Mohan, C. Axl, Ferritin, Insulin-Like Growth Factor Binding Protein 2, and Tumor Necrosis Factor Receptor Type II as Biomarkers in Systemic Lupus Erythematosus. Arthritis Care Res. 2016, 68, 1303–1309. [Google Scholar] [CrossRef]
- Iorgulescu, J.B.; Medeiros, L.J.; Patel, K.P. Predictive and Prognostic Molecular Biomarkers in Lymphomas. Pathology 2024, 56, 239–258. [Google Scholar] [CrossRef]
- Beck, D.B.; Werner, A.; Kastner, D.L.; Aksentijevich, I. Disorders of Ubiquitylation: Unchained Inflammation. Nat. Rev. Rheumatol. 2022, 18, 435–447. [Google Scholar] [CrossRef]
- Lu, D.; Song, J.; Sun, Y.; Qi, F.; Liu, L.; Jin, Y.; McNutt, M.A.; Yin, Y. Mutations of Deubiquitinase OTUD1 Are Associated with Autoimmune Disorders. J. Autoimmun. 2018, 94, 156–165. [Google Scholar] [CrossRef]
- Pardoll, D.M. Inducing Autoimmune Disease to Treat Cancer. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 5340–5342. [Google Scholar] [CrossRef]
- Pennell, C.A.; Barnum, J.L.; McDonald-Hyman, C.S.; Panoskaltsis-Mortari, A.; Riddle, M.J.; Xiong, Z.; Loschi, M.; Thangavelu, G.; Campbell, H.M.; Storlie, M.D.; et al. Human CD19-Targeted Mouse T Cells Induce B Cell Aplasia and Toxicity in Human CD19 Transgenic Mice. Mol. Ther. 2018, 26, 1423–1434. [Google Scholar] [CrossRef]
- Seligman, C.; Chang, Y.-M.; Luo, J.; Garden, O.A. Exploring the Role of Immune Checkpoint Inhibitors in the Etiology of Myasthenia Gravis and Lambert-Eaton Myasthenic Syndrome: A Systematic Review. Front. Neurol. 2022, 13, 1004810. [Google Scholar] [CrossRef]
- Young, A.; Quandt, Z.; Bluestone, J.A. The Balancing Act between Cancer Immunity and Autoimmunity in Response to Immunotherapy. Cancer Immunol Res 2018, 6, 1445–1452. [Google Scholar] [CrossRef]
- Hoos, A.; Eggermont, A.M.M.; Janetzki, S.; Hodi, F.S.; Ibrahim, R.; Anderson, A.; Humphrey, R.; Blumenstein, B.; Old, L.; Wolchok, J. Improved Endpoints for Cancer Immunotherapy Trials. J. Natl. Cancer Inst. 2010, 102, 1388–1397. [Google Scholar] [CrossRef] [PubMed]
- DePeaux, K.; Rivadeneira, D.B.; Lontos, K.; Dean, V.G.; Gunn, W.G.; Watson, M.J.; Yao, T.; Wilfahrt, D.; Hinck, C.; Wieteska, L.; et al. An Oncolytic Virus-Delivered TGFβ Inhibitor Overcomes the Immunosuppressive Tumor Microenvironment. J. Exp. Med. 2023, 220. [Google Scholar] [CrossRef]
- Katti, A.; Diaz, B.J.; Caragine, C.M.; Sanjana, N.E.; Dow, L.E. CRISPR in Cancer Biology and Therapy. Nat. Rev. Cancer 2022, 22, 259–279. [Google Scholar] [CrossRef]
- Wang, J.; Yang, X.; Zhang, Y.; Jiang, X.; Li, Y.; Cui, J.; Liao, Y. Single-Cell Analysis with Childhood and Adult Systemic Lupus Erythematosus. Autoimmunity 2024, 57, 2281228. [Google Scholar] [CrossRef]
- Kim, G.-D.; Das, R.; Goduni, L.; McClellan, S.; Hazlett, L.D.; Mahabeleshwar, G.H. Kruppel-like Factor 6 Promotes Macrophage-Mediated Inflammation by Suppressing B Cell Leukemia/Lymphoma 6 Expression. J. Biol. Chem. 2016, 291, 21271–21282. [Google Scholar] [CrossRef]
- Waku, T.; Katayama, H.; Hiraoka, M.; Hatanaka, A.; Nakamura, N.; Tanaka, Y.; Tamura, N.; Watanabe, A.; Kobayashi, A. NFE2L1 and NFE2L3 Complementarily Maintain Basal Proteasome Activity in Cancer Cells through CPEB3-Mediated Translational Repression. Mol. Cell. Biol. 2020, 40. [Google Scholar] [CrossRef]
- Dankers, W.; Northcott, M.; Bennett, T.; D’Cruz, A.; Sherlock, R.; Gearing, L.J.; Hertzog, P.; Russ, B.; Miceli, I.; Scheer, S.; et al. Type 1 Interferon Suppresses Expression and Glucocorticoid Induction of Glucocorticoid-Induced Leucine Zipper (GILZ). Front. Immunol. 2022, 13, 1034880. [Google Scholar] [CrossRef] [PubMed]
- Letter to the Editor: Protein Phosphatase 1 Subunit Ppp1r15a/GADD34 Is Overexpressed in Systemic Lupus Erythematosus and Related to the Expression of Type I Interferon Response Genes. Autoimmun. Rev. 2019, 18, 211–213. [CrossRef]
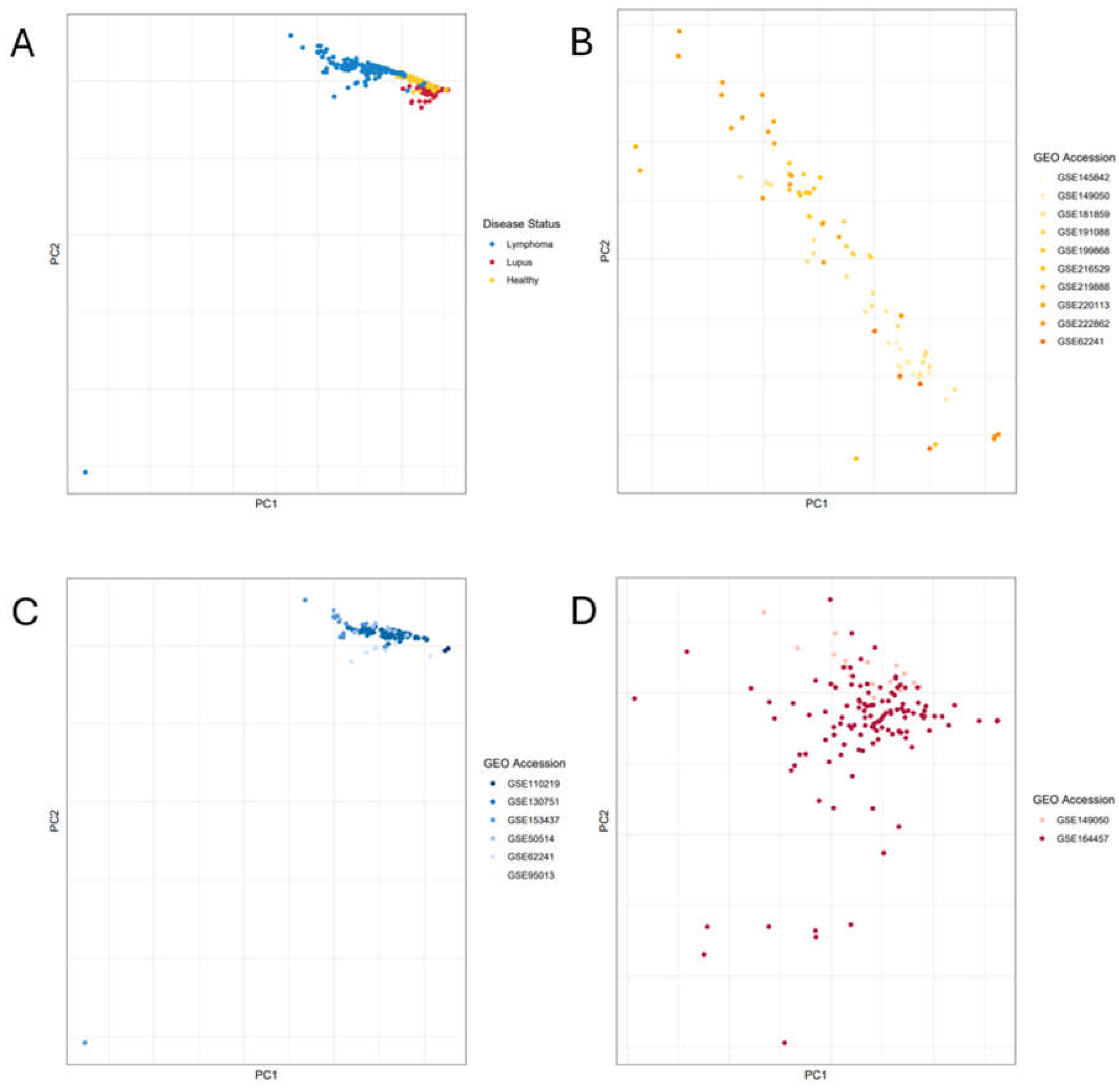
| Sample Phenotype | Type of Sequencing Reads | GEO Identifier | # Relevant Samples Included in Current Study |
|---|---|---|---|
| large B-cell lymphoma | paired end | GSE153437 [25] | 25 |
| follicular lymphoma | paired end | * GSE62241 [26,27] | 10 |
| diffuse large B-cell lymphoma | paired end | GSE95013 [28] | 29 |
| B-cell lymphoma | single end | GSE110219 [29] | 2 |
| diffuse large B-cell lymphoma | paired end | GSE130751 [30] | 63 |
| diffuse large B-cell lymphoma | paired end | GSE50514 [31] | 7 |
| lupus B-cells | single end | * GSE149050 [32] | 18 |
| lupus B-cells | paired end | GSE164457 [33] | 120 |
| healthy B-cells | paired end | GSE145842 [38] | 6 |
| healthy B-cells | single end | * GSE149050 [32] | 14 |
| healthy B-cells | paired end | GSE181859 [34] | 20 |
| healthy B-cells | paired end | * GSE62241 [26,27] | 4 |
| healthy B-cells | paired end | GSE191088 [39] | 6 |
| healthy B-cells | paired end | GSE199868 (currently unpublished) | 13 |
| healthy B-cells | paired end | GSE216529 [35] | 2 |
| healthy B-cells | single end | GSE219888 [36] | 2 |
| healthy B-cells | paired end | GSE220113 [37] | 17 |
| healthy B-cells | single end | GSE222862 (currently unpublished) | 3 |
| Gene Symbol | lupus* log2FC | lupus* FDR | lymphoma** log2FC | lymphoma** FDR | IIT score | IIT corrected p-value*** | |
|---|---|---|---|---|---|---|---|
|
C+A+ Quad I |
C1QB | 4.85 | 1.98E-31 | 9.69 | 7.78E-47 | 81.5 | 0 |
| IFI27 | 6.74 | 4.35E-34 | 7.31 | 2.88E-44 | 78.96 | 0 | |
| IGFBP2 | 1.99 | 3.13E-33 | 5.42 | 2.67E-31 | 38.67 | 0 | |
| TCN2 | 3.27 | 1.21E-22 | 4.29 | 1.07E-33 | 37.22 | 0 | |
|
C+A- Quad II |
PTMS | -2.59 | 1.83E-17 | 3.72 | 8.08E-27 | 25.21 | 0 |
| TPM2 | -2.48 | 2.23E-15 | 3.88 | 1.65E-25 | 21.69 | 0 | |
| NFE2L3 | -1.42 | 4.61E-21 | 1.99 | 3.27E-37 | 18.03 | 0 | |
| PLXNA1 | -1.38 | 5.76E-21 | 2.75 | 1.04E-34 | 14.91 | 0 | |
|
C-A- Quad III |
TAPT1 | -1.57 | 3.70E-30 | -2.21 | 6.12E-37 | 28.45 | 0 |
| PNRC1 | -1.72 | 4.13E-43 | -2.11 | 1.16E-43 | 27.03 | 0 | |
| OTUD1 | -1.82 | 5.75E-34 | -2.24 | 5.46E-37 | 25.54 | 0 | |
| MAP3K1 | -1.64 | 7.47E-35 | -2.15 | 1.69E-36 | 24.68 | 0 | |
|
C-A+ Quad IV |
TSC22D3 | 2.36 | 5.27E-34 | -3.22 | 2.83E-45 | 47.78 | 0 |
| KLF6 | 2.7 | 1.48E-38 | -3.13 | 2.23E-46 | 42.09 | 0 | |
| PPP1R15A | 2.93 | 1.23E-29 | -3.17 | 1.38E-40 | 38.84 | 0 | |
| ARL4A | 1.9 | 9.03E-34 | -2.69 | 1.67E-40 | 35.81 | 0 |
| Term | Overlap | Bonferroni p-value | Odds Ratio | Combined Score | GO DAG* |
|---|---|---|---|---|---|
| RNA Binding (GO:0003723) | 489/1411 | 7.58E-10 | 1.53 | 42.71 | Molecular Function |
| Protein Serine/Threonine Kinase Activity (GO:0004674) | 141/342 | 7.27E-07 | 1.98 | 40.43 | Molecular Function |
| Cytoplasmic Translation (GO:0002181) | 50/93 | 9.63E-05 | 3.26 | 58.06 | Biological Process |
| Macromolecule Biosynthetic Process (GO:0009059) | 81/183 | 3.23E-04 | 2.23 | 35.48 | Biological Process |
| Ubiquitin-Like Protein Transferase Activity (GO:0019787) | 97/240 | 4.95E-04 | 1.91 | 25.7 | Molecular Function |
| Pathway | Overlap | FDR P-Value | Odds Ratio | Combined Score | Database |
|---|---|---|---|---|---|
| Neutrophil Degranulation | 190/468 | 1.66E-08 | 1.94 | 49.11 | Reactome |
| Immune System | 638/1943 | 1.66E-08 | 1.41 | 34.88 | Reactome |
| Adaptive Immune System | 271/733 | 5.95E-08 | 1.67 | 38.41 | Reactome |
| Signaling By Rho GTPases | 242/644 | 6.34E-08 | 1.71 | 38.42 | Reactome |
| Signaling By Rho GTPases, Miro GTPases And RHOBTB3 | 247/660 | 6.34E-08 | 1.7 | 38.14 | Reactome |
| Eukaryotic Translation Elongation | 52/90 | 9.11E-08 | 3.84 | 84.12 | Reactome |
| Innate Immune System | 356/1035 | 7.98E-07 | 1.5 | 29.25 | Reactome |
| Formation Of A Pool Of Free 40S Subunits | 53/98 | 1.18E-06 | 3.31 | 62.96 | Reactome |
| Peptide Chain Elongation | 48/86 | 1.23E-06 | 3.54 | 66.49 | Reactome |
| Cellular Responses To Stress | 259/722 | 1.23E-06 | 1.59 | 29.75 | Reactome |
| Gene Symbol | Quadrant | Immune Imbalance Score | IIT corrected p-value* | Number Unique Drugs | Number Approved Drugs | Weighted Target Score |
| C3 | Quad I | 33.57 | 0 | 3 | 1 | 1357 |
| CXCR4 | Quad IV | 28.73 | 0 | 7 | 1 | 1313 |
| MAP3K1 | Quad III | 24.68 | 0 | 1 | 0 | 698 |
| IDO1 | Quad I | 22.02 | 0 | 2 | 0 | 913.5 |
| CLU | Quad I | 21.67 | 0 | 2 | 0 | 725.5 |
| CD276 | Quad I | 20.17 | 0 | 2 | 0 | 436 |
| COL1A1 | Quad I | 16.39 | 0 | 2 | 1 | 1299 |
| EPHB2 | Quad I | 16.07 | 0 | 1 | 1 | 316 |
| NR1H2 | Quad IV | 15.45 | 0 | 5 | 0 | 186 |
| TUBA1A | Quad IV | 15.17 | 0 | 24 | 1 | 295 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





