Introduction
Systemic lupus erythematosus (SLE), commonly known as lupus, is a chronic autoimmune disease that can affect various parts of the body, including the skin, joints, kidneys, heart, lungs, blood vessels, and brain [
1]. Characterized by periods of illness (flares) and remission, SLE is caused by a dysfunction in the immune system, where it mistakenly attacks healthy tissues [
2]. This leads to widespread inflammation and tissue damage [
1]. Symptoms of SLE can range widely, from mild to severe, and can include fatigue, joint pain, skin rashes, fever, and organ damage [
3]. Treatment often involves medications to suppress the immune system, such as corticosteroids, antimalarial drugs, immunosuppressive agents, and biologics, and it requires careful management to control symptoms and reduce the risk of flares [
4].
B cells play a crucial role in the pathogenesis of SLE, contributing to the disease through various mechanisms [
5,
6]. In SLE, B cells are involved in the production of autoantibodies, antigen presentation, and immune regulation [
7,
8]. Upon activation, B cells secret high affinity antibodies such as IgG to boost systemic immune response [
9]. Therapy targeting B cells in SLE has shown promise, with belimumab, a monoclonal antibody that targets BAFF, being the first FDA-approved treatment for SLE [
10,
11]. However, the overall clinical efficacy of B cell depletion has been modest, indicating the complexity of B cell roles in SLE [
12,
13,
14]. Therefore, exploring the underlying mechanisms that involved in B cell activation and SLE progression is an urgent issue.
Long non-coding RNAs (lncRNAs) play a significant role in SLE, contributing to the disease's pathogenesis through various mechanisms [
15,
16]. LncRNAs are involved in immune response regulation and immune cell development, and their aberrant expression is associated with the dysregulation of these processes in SLE [
17,
18]. In SLE, several lncRNAs such as GAS5, NEAT1, TUG1, SNHG16, and linc0597 have been found to be dysregulated and may serve as potential biomarkers and therapeutic targets [
19,
20]. These lncRNAs may interact with proteins, DNA, and even other RNAs, influencing the immune response and the development of autoimmune diseases [
21,
22]. The mechanisms by which lncRNAs contribute to SLE involve their ability to modulate gene expression at the epigenetic level, affecting DNA methylation and histone modifications, regulating the expression of genes involved in immune function, thereby influencing the disease's progression [
23].
TCONS_00483150 is reported as a novel diagnostic biomarker that recently reported as a potential for SLE, distinguishing patients with SLE from healthy controls and those with rheumatoid arthritis and patients with active/stable SLE from healthy controls. [
24]. In this study, we aimed to investigate the effects of a lncRNA TCONS_00483150 on the function of self-reactive B cells in systemic lupus erythematosus and explored the potential regulatory mechanisms.
Materials and Methods
B Cell Isolation
B cells were purified from spleens of C57B6/L mice by using a B-cell isolation kit (Miltenyi Biotec GmbH) according to manufacturer’s protocol.
Cell Transfection
The lncRNA overexpression vectors were synthesized by GenePharma (Shanghai, China). B cells were transfected with the overexpression vectors and negative conrtol using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. After transfection for 24 h, cells were collected for subsequent experiments.
B Cell Activation Analysis
After lncRNA transfection and stimulation with STAT activator (Colivelin, MCE, USA) for 24 h. B cells were collected and centrifuged for 5 min at 1,000 × g at 4°C; the cell pellet was resuspended in PBS at a cell density of 1×106 cells/ml, labeled with PE-conjugated anti-CD19 and FITC-conjugated anti-CD138 antibody (Biolegend, CHina) for analysis of CD19+CD138+ B cells using a Flow Cytometer (BD Biosciences, USA).
Western Blot
Cells were collected and lysed with RIPA lysis buffer containing protease and phosphorylase (Thermo, USA). The total protein was extracted after centrifuge. The concentration of cell lysates was identified using the BCA protein Quantification kit (Beyotime, China). Then an equal amount of proteins was separated by electrophoresis, transferred to PVDF membrane (Millipore, MA, USA), and blocked with 5% skim milk. The protein bands were incubated with primary antibodies against Blipm-1, JAK, STAT3, Bcl-2, BAX, BAFF, APRIL, and β-actin overnight at 4°C. Next, the protein bands were incubated with the corresponding anti-mouse or anti-rabbit secondary antibody. The images of indicated protein bands were visualized using ECL reagent (Millipore, Germany).
qPCR
Total RNA was extracted from peripheral blood mononuclear cells (PBMCs) using Trizol Reagent (TaKaRa, Japan). Following the addition of chloroform and centrifugation, isopropyl alcohol (400 μL) was introduced into a new enzyme-free EP tube. The upper layer of RNA was carefully transferred post-centrifugation. Reverse transcription of RNA to complementary DNA (cDNA) was conducted using the Thermo Scientific RevertAid RT kit (Thermo Scientific, USA). Quantitative real-time polymerase chain reaction (qRT-PCR) was carried out with TB Green Premix Ex Taq II (Tli RNaseH Plus, TaKaRa, Japan) and analyzed on a LightCycler 96 PCR instrument (Roche, Tusem, China). U6 or GAPDH served as endogenous controls, and the relative expression levels of miRNAs or mRNAs were determined using the 2−ΔΔCt method.
Enzyme-Linked Immunosorbent Assay (ELISA)
After indicated treatment, B cells were collected by centrifugation at 12,000 × g for 10 minutes at 4°C. B cell supernatants were then assessed for IgG and IgM levels using respective ELISA kits from Abcam (USA), following the provided instructions. Each experiment was conducted three times to ensure reliability. Optical density measurements at 450 nm were taken using an automated ELISA reader.
CCK-8
Cell viability was measured by cell counting kit 8 (CCK-8) assay kit (Beyotime, China). In brief, cells were seeded into 96-well plates and incubated for 24 h, then CCK-8 reagent was added into each well and incubated for another 2 h. The optical density value at 450 nm was then measured using the microplate reader (Thermo, USA).
Cell Apoptosis and Cell Cycle
Cell apoptosis was detected by Annexin V-FITC/PI apoptosis kit (Beyotime, China) and flow cytometry. After indicated treatment, B cells were collected after centrifuge and incubated with staining buffer that added with Annexin V-FITC and PI in dark for 30 min. B cells were analyzed using Flow Cytometer (BD Biosciences, USA). For cell cycle analysis, the cells were stained with PI regent that in dark for 30 min and analyzed using the Flow Cytometer (BD Biosciences, USA).
Statistics
Analysis was conducted with GraphPad Prism Version 8.0, presenting results as mean ± standard deviation. The Student's t-test was applied for pairwise comparisons, and one-way ANOVA was used for multiple group comparisons. Significance was determined at a P value <0.05.
Results
TCONS_00483150 Regulates B Cell Phenotype
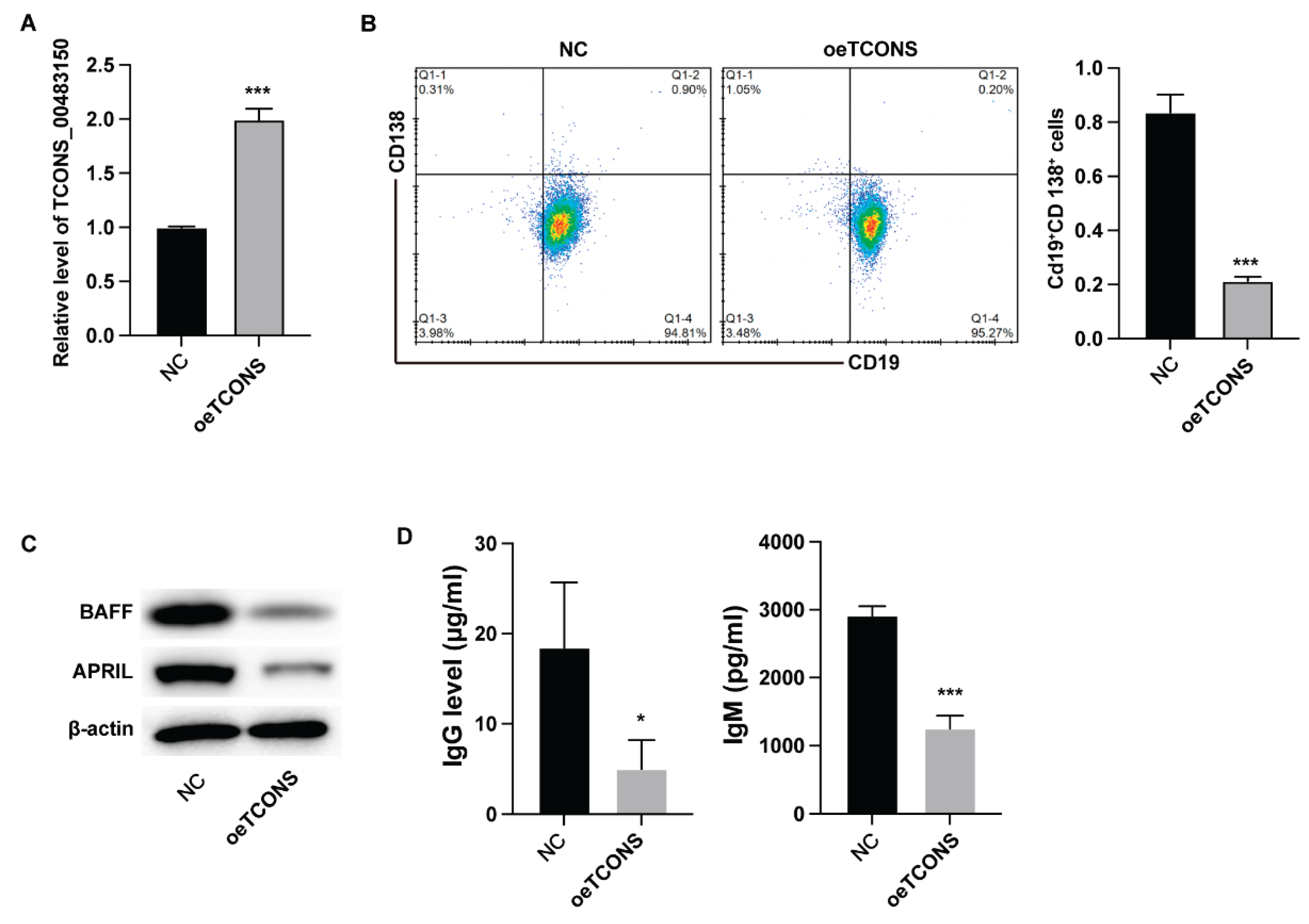
We first analyzed the impact of TCONS_00483150 on B cell functions via ectopic expression of TCONS_00483150. The results from qPCR verified the successful expression of TCONS_00483150 in B cells (
Figure 1A). Overexpression of TCONS_00483150 led to deceased portion of CD19+CD138+ B cells (
Figure 1B) and suppressed the expression of BAFF and APRIL (
Figure 1C). Moreover, the production of IgG and IgM was significantly suppressed by TCONS_00483150 overexpression (
Figure 1D). These data indicated that TCONS_00483150 repressed the function of B cells.
TCONS_00483150 Regulates B Cell Functions
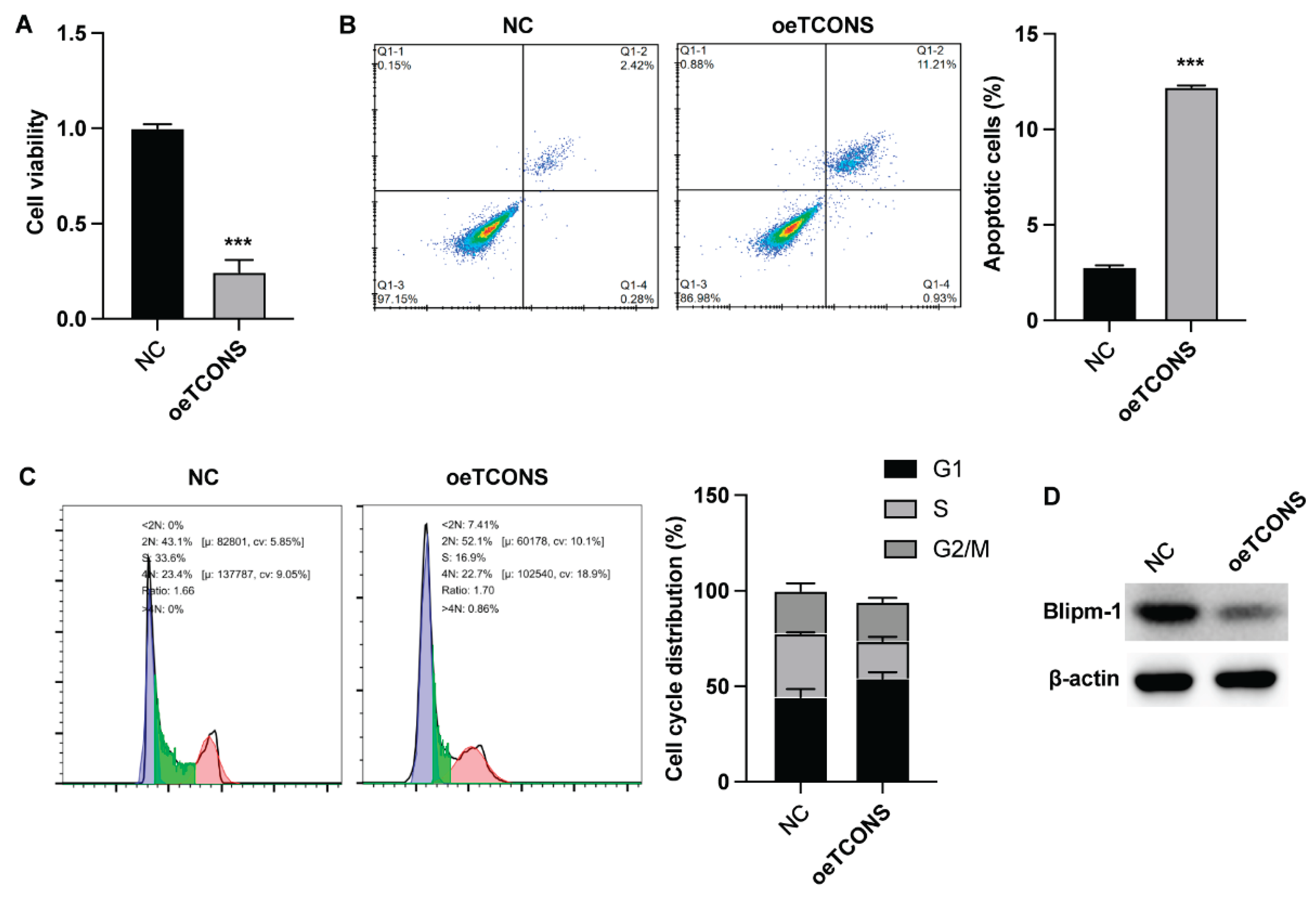
The results from CCK-8 assay indicated that TCONS_00483150 suppressed the viability of B cells (
Figure 2A). TCONS_00483150 overexpression also caused a notable elevation of B cell apoptosis (
Figure 2B). Moreover, the assessment of cell cycle indicated that TCONS_00483150 led to decreased cell portion in S phase, suggesting the repressed cell cycle progression (
Figure 2C). The protein level of Blipm-1 was also downregulated by TCONS_00483150 (
Figure 2D).
TCONS_00483150 Regulates JSK/STAT3 Signaling Pathway in B Cell
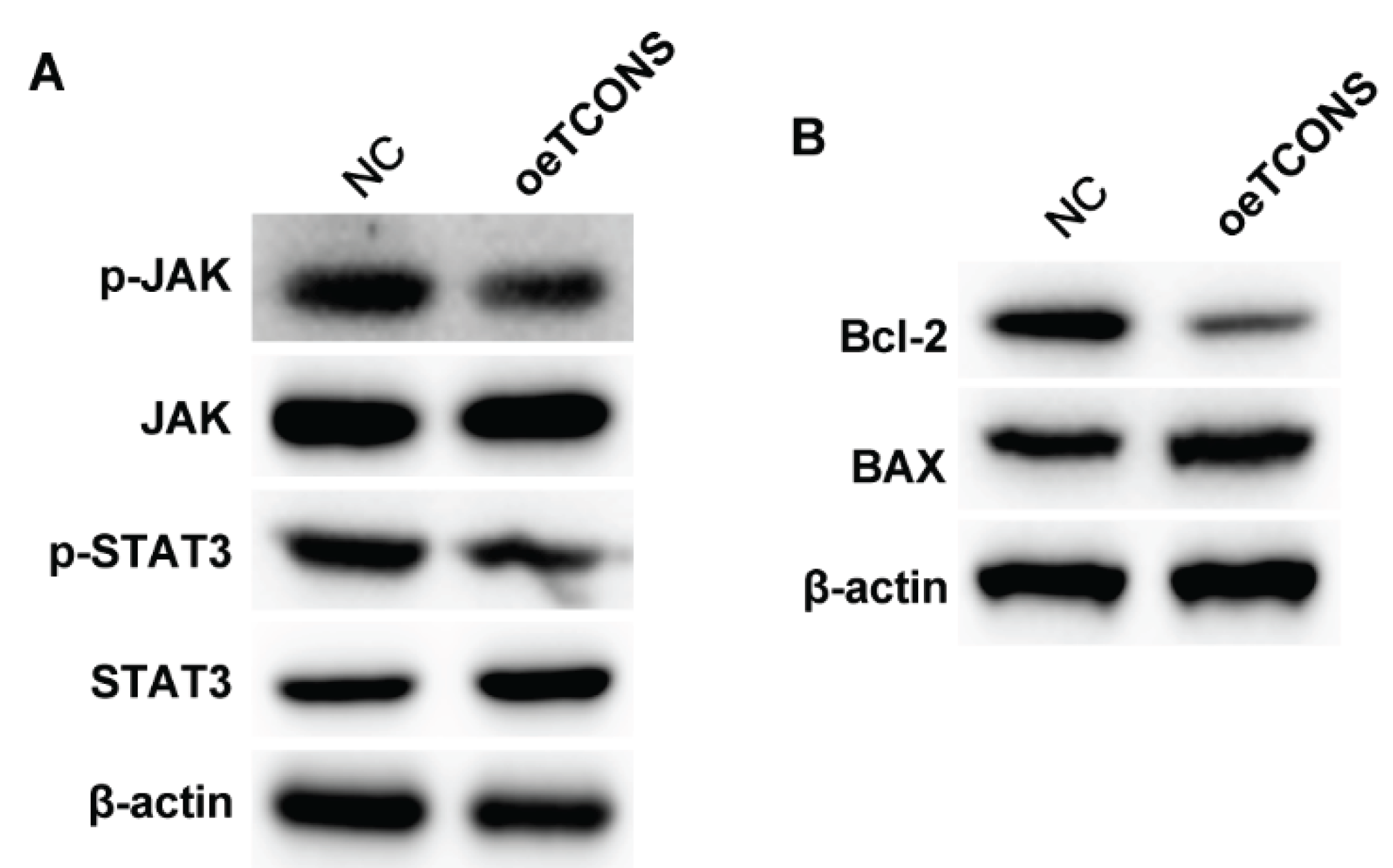
Next, we analyzed the potential downstream regulatory mechanisms underlying the TCONS_00483150-regulated B cell behaviors. Overexpression of TCONS_00483150 suppressed the phosphorylation of the JAK and STAT3 protein, but not altered the level of total protein (
Figure 3A). Besides, TCONS_00483150 downregulated the level of anti-apoptotic factor Bcl-2 and upregulated the expression of pro-apoptotic BAX (
Figure 3B).
TCONS_00483150 Regulates B Cell Function Through the JAK/STAT3 Signaling Pathway
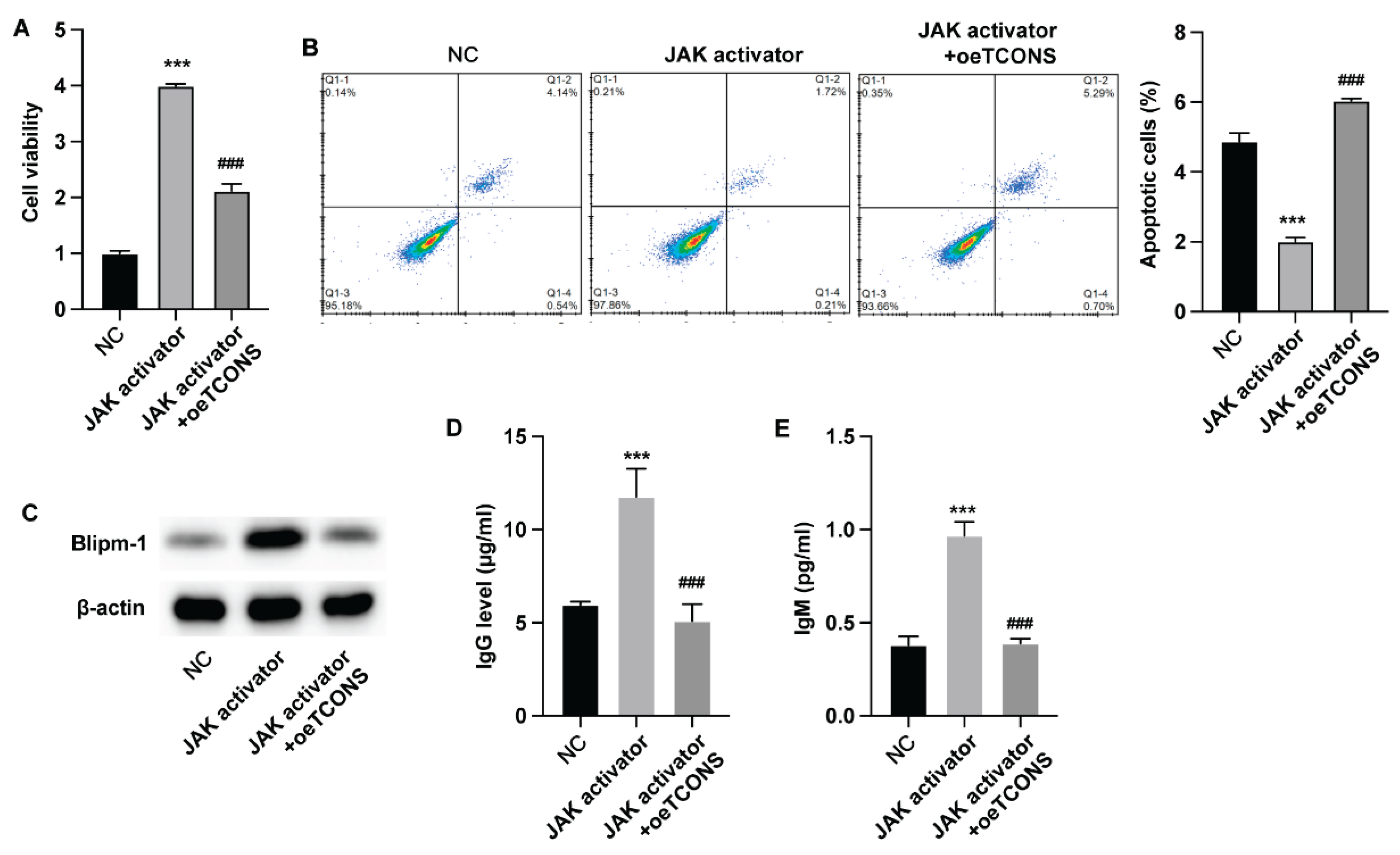
To investigate whether TCONS_00483150 modulated the functions of B cells via regulating JAK/STAT3 signaling pathway, we adopted the STST3 activator for cell treatment. Treatment with STAT3 activator notably increased the viability of B cells and suppressed cell apoptosis, whereas overexpression of TCONS_00483150 repressed these effects (
Figure 4A,B). Moreover, overexpression of TCONS_00483150 abolished the elevated Blipm-1 (
Figure 4C) and production of IgM and IgG (
Figure 4D,F) that activated by STAT3 activator.
Discussion
Systemic lupus erythematosus (SLE) is a severe autoimmune disease characterized by dysregulated B cell functions, which contribute to the production of numerous autoantibodies [
25]. The aberrant expression of non-coding RNAs, particularly long non-coding RNAs (lncRNAs), has been implicated in SLE pathogenesis [
26]. Recent studies have revealed substantial differences in the expression of autoimmune-associated lncRNAs between SLE patients and healthy individuals, offering valuable insights into disease mechanisms and potential biomarkers for diagnostic and therapeutic advancements [
15,
27]. Investigations into the mechanisms by which specific lncRNAs regulate B lymphocyte development in SLE are ongoing, shedding light on the complex interplay of genetic and molecular factors in this disease process. for example, lncRNA MALAT-1 is reported to be associated with the regulation of IL-21 levels and the SIRT1 signaling pathway, which are involved in the inflammatory response and oxidative stress in SLE [
28]. The expression of lncRNA GAS5 is linked to the modulation of inflammatory cytokines, and it may act as a protective factor by targeting miR-21 and PTEN, affecting the development of SLE [
19]. Besides, the lncRNA NEAT1 expression was abnormally increased in SLE patients and predominantly expressed in human monocytes. NEAT1 participates in TLR4-mediated inflammatory process through affecting the activation of the late MAPK signaling pathway [
29].
LncRNA TCONS_00483150 is a long non-coding RNA that has been identified in the context of SLE. The influence of TCONS_00483150 on the function of these self-reactive B cells and the underlying mechanisms is a topic of ongoing research. It is reported that TCONS_00483150 is a potential diagnostic biomarker of SLE [
24]. However, the specific mechanisms by which TCONS_00483150 affects self-reactive B cells in SLE are not yet fully understood and likely involve complex interactions with other genetic and epigenetic factors. In current study, we performed overexpression of TCONS_00483150 in B cells and analyzed the B cell activation and viability. The results indicated that TCONS_00483150 overexpression reduced the portion of CD19+/CD138+ B cells, along with decreased production of IgG and IgM and expression of BAFF. Meanwhile, TCONS_00483150 overexpression also repressed B cell proliferation and promoted cell apoptosis. The analysis of signaling pathways revealed repressed activation of JAK/STAT3 signaling upon TCONS_00483150 overexpression. Activation of the JAK/STAT signaling pathway, particularly leading to the phosphorylation of STAT3, is a key event in the immune response, with STAT3 being an essential downstream transcription factor [
30]. In B cells, STAT3 phosphorylation is crucial for the formation of germinal centers (GCs) and the accumulation of plasma cells, as observed in MRLlpr/lpr lupus mice [
31,
32]. Notably, MRLlpr/lpr mice with a targeted knockout of STAT3 in B cells show a significant reduction in spontaneous GC formation and plasma cell generation [
33]. Our findings suggested that activation of JAK using specific activator could stimulate the viability and activation of B cells, which was repressed by TCONS_00483150.
Conclusion
In current study, we demonstrated that TCONS_00483150 overexpression could suppress the proliferation and activation of B cells, potentially via suppressing the activation of JAK/STAT3. Our study uncovered a novel mechanism underlying the dysfunction of B cells in SLE, offering a potential therapeutic target for the treatment of this disease.
Funding
This study was supported by the Ningbo Natural Science Foundation, No. 2022J257.
References
- Zucchi, D.; Silvagni, E.; Elefante, E.; Signorini, V.; Cardelli, C.; Trentin, F.; Schilirò, D.; Cascarano, G.; Valevich, A.; Bortoluzzi, A.; Tani, C. Systemic lupus erythematosus: one year in review 2023. Clin Exp Rheumatol 2023, 41, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, K.V.; Nocton, J.J. Unusual Presentations of Systemic Lupus Erythematosus. Med Clin North Am 2024, 108, 43–57. [Google Scholar] [CrossRef]
- Aringer, M.; Toro-Domínguez, D.; Alarcón-Riquelme, M.E. Classification of systemic lupus erythematosus: From the development of classification criteria to a new taxonomy? Best Pract Res Clin Rheumatol 2023, 37, 101949. [Google Scholar] [CrossRef] [PubMed]
- Durcan, L.; O'Dwyer, T.; Petri, M. Management strategies and future directions for systemic lupus erythematosus in adults. Lancet 2019, 393, 2332–2343. [Google Scholar] [CrossRef] [PubMed]
- Domeier, P.P.; Rahman, Z.S.M. Regulation of B Cell Responses in SLE by Three Classes of Interferons. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef]
- Raza, I.G.A.; Clarke, A.J. B Cell Metabolism and Autophagy in Autoimmunity. Frontiers in immunology 2021, 12, 681105. [Google Scholar] [CrossRef]
- Yap, D.Y.H.; Chan, T.M. B Cell Abnormalities in Systemic Lupus Erythematosus and Lupus Nephritis-Role in Pathogenesis and Effect of Immunosuppressive Treatments. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J.; Kumar, V.; Karnell, J.L.; Naiman, B.; Gross, P.S.; Rahman, S.; Zerrouki, K.; Hanna, R.; Morehouse, C.; Holoweckyj, N.; Liu, H.; Manna, Z.; Goldbach-Mansky, R.; Hasni, S.; Siegel, R.; Sanjuan, M.; Streicher, K.; Cancro, M.P.; Kolbeck, R.; Ettinger, R. IL-21 drives expansion and plasma cell differentiation of autoreactive CD11c(hi)T-bet(+) B cells in SLE. Nat Commun 2018, 9, 1758. [Google Scholar] [CrossRef]
- Krustev, E.; Clarke, A.E.; Barber, M.R.W. B cell depletion and inhibition in systemic lupus erythematosus. Expert Rev Clin Immunol 2023, 19, 55–70. [Google Scholar] [CrossRef]
- Möckel, T.; Basta, F.; Weinmann-Menke, J. A. Schwarting, B cell activating factor (BAFF): Structure, functions, autoimmunity and clinical implications in Systemic Lupus Erythematosus (SLE). Autoimmun Rev 2021, 20, 102736. [Google Scholar] [CrossRef]
- Taubmann, J.; Müller, F.; Mutlu, M.Y.; Völkl, S.; Aigner, M.; Bozec, A.; Mackensen, A.; Grieshaber-Bouyer, R.; Schett, G. CD19 Chimeric Antigen Receptor T Cell Treatment: Unraveling the Role of B Cells in Systemic Lupus Erythematosus. Arthritis Rheumatol 2024, 76, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Favas, C.; Isenberg, D.A. B-cell-depletion therapy in SLE--what are the current prospects for its acceptance? , Nat Rev Rheumatol 2009, 5, 711–716. [Google Scholar] [CrossRef]
- Barnas, J.L.; Looney, R.J.; Anolik, J.H. B cell targeted therapies in autoimmune disease. Curr Opin Immunol 2019, 61, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Liossis, S.C.; Staveri, C. B Cell-Based Treatments in SLE: Past Experience and Current Directions. Curr Rheumatol Rep 2017, 19, 78. [Google Scholar] [CrossRef]
- Wu, H.; Chen, S.; Li, A.; Shen, K.; Wang, S.; Wang, S.; Wu, P.; Luo, W.; Pan, Q. LncRNA Expression Profiles in Systemic Lupus Erythematosus and Rheumatoid Arthritis: Emerging Biomarkers and Therapeutic Targets. Frontiers in immunology 2021, 12, 792884. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; You, J.P.; Liu, X.R.; Zhao, Y.F.; Cui, Y.; Zhao, Z.Z.; Qi, Y.Y. PRDX6 AS1 gene polymorphisms and SLE susceptibility in Chinese populations. Frontiers in immunology 2022, 13, 987385. [Google Scholar] [CrossRef]
- Chen, X.; Luo, X.; Wei, Y.; Sun, H.; Dai, L.; Tangzhou, Y.; Jin, H.; Yin, Z. LncRNA H19 induces immune dysregulation of BMMSCs, at least partly, by inhibiting IL-2 production. Mol Med 2021, 27, 61. [Google Scholar] [CrossRef]
- Xiang, M.; Wang, Y.; Chen, Q.; Wang, J.; Gao, Z.; Liang, J.; Xu, J. LncRNA NEAT1 promotes IL-6 secretion in monocyte-derived dendritic cells via sponging miR-365a-3p in systemic lupus erythematosus. Epigenetics 2023, 18, 2226492. [Google Scholar] [CrossRef]
- Liu, C.H.; Lu, Y.L.; Huang, H.T.; Wang, C.F.; Luo, H.C.; Wei, G.J.; Lei, M.; Tan, T.; Wang, Y.; Huang, Y.Y.; Wei, Y.S.; Lan, Y. Association of LncRNA-GAS5 gene polymorphisms and PBMC LncRNA-GAS5 level with risk of systemic lupus erythematosus in Chinese population. J Cell Mol Med 2021, 25, 3548–3559. [Google Scholar] [CrossRef]
- Hsieh, Y.T.; Chen, Y.C.; Chou, Y.C.; Kuo, P.Y.; Yen, Y.T.; Tsai, H.W.; Wang, C.R. Long noncoding RNA SNHG16 regulates TLR4-mediated autophagy and NETosis formation in alveolar hemorrhage associated with systemic lupus erythematosus. J Biomed Sci 2023, 30, 78. [Google Scholar] [CrossRef]
- Lu, J.; Ma, H.; Wang, Q.; Song, Z.; Wang, J. Chemotherapy-mediated lncRNA-induced immune cell plasticity in cancer immunopathogenesis. International immunopharmacology 2024, 141, 112967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, H.; Wu, W.; Zhou, R.; Li, S.; Wang, Z.; Dai, Z.; Zhang, L.; Liu, F.; Liu, Z.; Zhang, J.; Luo, P.; Liu, Z.; Cheng, Q. Machine learning-based identification of tumor-infiltrating immune cell-associated lncRNAs for improving outcomes and immunotherapy responses in patients with low-grade glioma. Theranostics 2022, 12, 5931–5948. [Google Scholar] [CrossRef]
- Crawford, J.D.; Wang, H.; Trejo-Zambrano, D.; Cimbro, R.; Talbot, C.C., Jr.; Thomas, M.A.; Curran, A.M.; Girgis, A.A.; Schroeder, J.T.; Fava, A.; Goldman, D.W.; Petri, M.; Rosen, A.; Antiochos, B.; Darrah, E. The XIST lncRNA is a sex-specific reservoir of TLR7 ligands in SLE. JCI Insight 2023, 8. [Google Scholar] [CrossRef]
- Guo, G.; Chen, A.; Ye, L.; Wang, H.; Chen, Z.; Yan, K.; Shi, X.; Li, B.; Lin, Q.; You, X.; Jiang, C.; Zhang, Q.; Ding, X.; Xue, X.; Zhang, H. TCONS_00483150 as a novel diagnostic biomarker of systemic lupus erythematosus. Epigenomics 2020, 12, 973–988. [Google Scholar] [CrossRef]
- Kiriakidou, M.; Ching, C.L. Systemic Lupus Erythematosus. Ann Intern Med 2020, 172, Itc81–itc96. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Chen, W.; Zheng, F.; Tang, D.; Liu, D.; Wang, G.; Xu, Y.; Yin, L.; Zhang, X.; Dai, Y. Reconstruction and analysis of the aberrant lncRNA-miRNA-mRNA network in systemic lupus erythematosus. Lupus 2020, 29, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Nezhad, M.T.N.; Rajabi, M.; Nekooeizadeh, P.; Sanjari, S.; Pourvirdi, B.; Heidari, M.M.; Esfahani, P.V.; Abdoli, A.; Bagheri, S.; Tobeiha, M. Systemic lupus erythematosus: From non-coding RNAs to exosomal non-coding RNAs. Pathol Res Pract 2023, 247, 154508. [Google Scholar] [CrossRef]
- Mao, Y.M.; He, Y.S.; Wu, G.C.; Hu, Y.Q.; Xiang, K.; Liao, T.; Yan, Y.L.; Yang, X.K.; Shuai, Z.W.; Wang, G.H.; Pan, H.F.; Ye, D.Q. Association of MALAT-1 gene single nucleotide polymorphisms with genetic susceptibility to systemic lupus erythematosus. Lupus 2021, 30, 1923–1930. [Google Scholar] [CrossRef]
- Zhang, F.; Wu, L.; Qian, J.; Qu, B.; Xia, S.; La, T.; Wu, Y.; Ma, J.; Zeng, J.; Guo, Q.; Cui, Y.; Yang, W.; Huang, J.; Zhu, W.; Yao, Y.; Shen, N.; Tang, Y. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun 2016, 75, 96–104. [Google Scholar] [CrossRef]
- Avery, D.T.; Deenick, E.K.; Ma, C.S.; Suryani, S.; Simpson, N.; Chew, G.Y.; Chan, T.D.; Palendira, U.; Bustamante, J.; Boisson-Dupuis, S.; Choo, S.; Bleasel, K.E.; Peake, J.; King, C.; French, M.A.; Engelhard, D.; Al-Hajjar, S.; Al-Muhsen, S.; Magdorf, K.; Roesler, J.; Arkwright, P.D.; Hissaria, P.; Riminton, D.S.; Wong, M.; Brink, R.; Fulcher, D.A.; Casanova, J.L.; Cook, M.C.; Tangye, S.G. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. Journal of Experimental Medicine 2010, 207, 155–171. [Google Scholar] [CrossRef]
- Ettinger, R.; Sims, G.P.; Fairhurst, A.M.; Robbins, R.; Da Silva, Y.S.; Spolski, R.; Leonard, W.J.; Lipsky, P.E. IL-21 induces differentiation of human naive and memory B cells into antibody-secreting plasma cells. Journal of Immunology 2005, 175, 7867–7879. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Chen, X.; Dascani, P.; Hu, X.; Bolli, R.; Zhang, H.G.; McLeish, K.R.; Yan, J. STAT3 signaling in b cells is critical for germinal center maintenance and contributes to the pathogenesis of murine models of lupus. Journal of Immunology 2016, 196, 4477–4486. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.M.; Chen, W.; Nyuydzefe, M.S.; Trzeciak, A.; Flynn, R.; Tonra, J.R.; Marusic, S.; Blazar, B.R.; Waksal, S.D.; Zanin-Zhorov, A. ROCK2 signaling is required to induce a subset of T follicular helper cells through opposing effects on STATs in autoimmune settings. Science Signaling 2016, 9, ra73–ra73. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).