1. Introduction
Cancer is the second cause of death in the United States and the leading cause among people younger than 85 years [
1]. Due to the high mortality and high possibility of recurrence, cancer has been a major health issue in recent century [
2]. Lymphoma is a highly heterogeneous hematological tumor comprising Hodgkin lymphoma and non-Hodgkin lymphoma (NHL). In US, non-Hodgkin lymphoma collectively account for 38.7 per 100,000 in incidence and 10.4 per 100,000 in mortality [
1]. Even though chemotherapy, radiotherapy, targeted therapy and cell therapy have dramatically improved the outcomes of lymphoma, the prognosis of some lymphoma patients was much worse than it had been predicted [
3,
4]. Therefore, to explore an accurate prediction of a cancer patient’s survival and response to individualized treatment therapy, a reliable biomarker for lymphoma is required.
Protein arginine methyltransferase 5 (PRMT5) is the main type-II arginine methyltransferase and forms a tight hetero-octameric complex with its substrate-binding partner MEP50 [
5]. PRMT5 symmetrically demethylate histone [
6] and chromatin remodeling factors in nuclei [
7], and thus directly or indirectly modulate oncogenic genes [
8,
9,
10]. At pan-cancer level, PRMT5 up-regulates cancer cell survival, proliferation and migration via promoting DNA repairing, destabilizing p53, and boosting E2F1 activity [
11,
12,
13]. In lymphoma, preclinical studies strongly indicated that overexpression of PRMT5 is closely related to lymphomagenesis [
14], resistance to Bruton tyrosine kinase (BTK) inhibitors [
11] and Richter’s transformation [
15]. These data indicated that PRMT5 might be a biomarker to predict the prognosis of lymphoma patients. However, the relationship between PRMT5 and cancer remains to be solidified, and the gap between preclinical and clinical data of PRMT5 remains to be bridged.
In this study, we collected clinical and genetic information from The Cancer Genome Atlas (TCGA), and identified the overall clinical correlation of PRMT5 with cancer, including malignant lymphoma. Additionally, we evaluated the association of PRMT5 with tumor-infiltrating immune cells in lymphoma using CIBERSORT. Coincident with previous preclinical findings, our retrospective analysis data demonstrated a possible clinical correlation and mechanism among PRMT5, tumor immune microenvironment, and tumor progression signaling. Based on the accordant pre-clinical and clinical data showing tumor-supporting effect of PRMT5, LASSO-COX regression was used to evaluate if PRMT5 is a credible survival prediction index for lymphoma.
2. Materials and Methods
Data Acquisition
Cancer patient datasets with gene expression profiles and clinical information were downloaded from the publicly available TCGA [
16], in which included 4 normal and 180 tumor tissues. Subsequent processing excluded cases with insufficient or missing data on overall survival time. Then, RNA sequencing data were utilized to explore the influence of PRMT5 expression on tumorigenesis and immune microenvironment.
Survival and expression analysis by GEPIA
Gene Expression Profiling Interactive Analysis (GEPIA) (
http://gepia.cancer-pku.cn/index.html), an online database was utilized to investigate the correlation between PRMT5 expression and clinicopathologic information in lymphoma. GEPIA [
17] was a webserver with a standard pipeline that analyzes 9,736 tumors from the RNA sequencing expression data and 8,587 normal samples from projects known as the GTEx and TCGA. Survival curve of differential PRMT5 expression were analyzed by GEPIA to explore the correlation of the gene expression with prognosis of lymphoma patients. Additionally, boxplots using disease state (Tumor or Normal) as variable was graphed to calculate differential expression of PRMT5.
Gene set enrichment analysis (GSEA)
GSEA generated an initial list on the classification of the genes according to their correlation with PRMT5 expression. Then this computational method was used to detect the pathways in which genes are primarily enriched. Nominal p < 0.05 and the FDR < 0.25 were considered statistically significant [
18].
Functional enrichment analysis of DEGs
Significant differentially expressed genes (DEGs) in cancer were analyzed by the clusterProfiler of R package (version 4.0) [
19], including Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis [
20,
21]. The biological processes, cell components and molecular functions were evaluated. Significant pathways were statistically identified by KEGG analysis, in which
p < 0.05 was applied.
Immune response of TIICs in lymphoma via CIBERSORT
CIBERSORT [
22] (
http://cibersort.stanford.edu/), a deconvolution algorithm based on gene expression can evaluate the changes in the expression of one set of genes relative to all other genes in the sample. TIIC concentration can therefore be precisely estimated via this process. CIBERSORT’s consistent performance encourages a growing focus on cell heterogeneity studies [
23,
24]. Our current analysis gauged the immune response of 22 TIICs in lymphoma via CIBERSORT, to assess its correlation with survival and molecular subpopulation. Shortly, gene expression datasets were set out using standard annotation files and uploaded to CIBERSORT web portal, with the algorithm running with its default signature matrix at one thousand permutations. CIBERSORT estimated a P-value for deconvolution via Monte Carlo sampling, establishing a measure of confidence in the results. In order to assess the influence of PRMT5 expression, we used 183 samples from the TCGA, where the full range of genes are represented. We divided 183 samples into one half with low expression and another half with high expression, and then used the data to make vioplot.
LASSO-COX regression
LASSO-COX regression with multicollinearity control was utilized to identify the most probable prognostic factors on survival of lymphoma [
25]. We established LASSO-COX regression based on training dataset to select prognostic variables for overall survival evaluation. LASSO-COX method coefficients can be calculated equivalently from the following equation:
where Coefi represented the risk coefficient and Expi represented the expression of each gene. According to the median risk score, patients were divided into two groups for Kaplan-Meier survival analysis and receiver operating characteristic (ROC) curve. The test set and all sets were equally divided into high-risk and low-risk groups, and Kaplan-Meier survival analysis and ROC curve were conducted for each group to verify the accuracy of the model.
Statistical analysis
Statistics from TCGA were conducted by R software (version 4.2.1). Overall survivals were calculated with the Kaplan-Meier method and compared with the log-rank test. Statistical significance of differences was determined by Student’s t test with p < 0.05. Correlation between various immune cells and PRMT5 expression was evaluated by Spearman’s correlation coefficients, and correlation between each target gene and PRMT5 expression was evaluated by Pearson’s correlation coefficients.
3. Results
3.1. Patient Characteristics and Multivariate Analysis
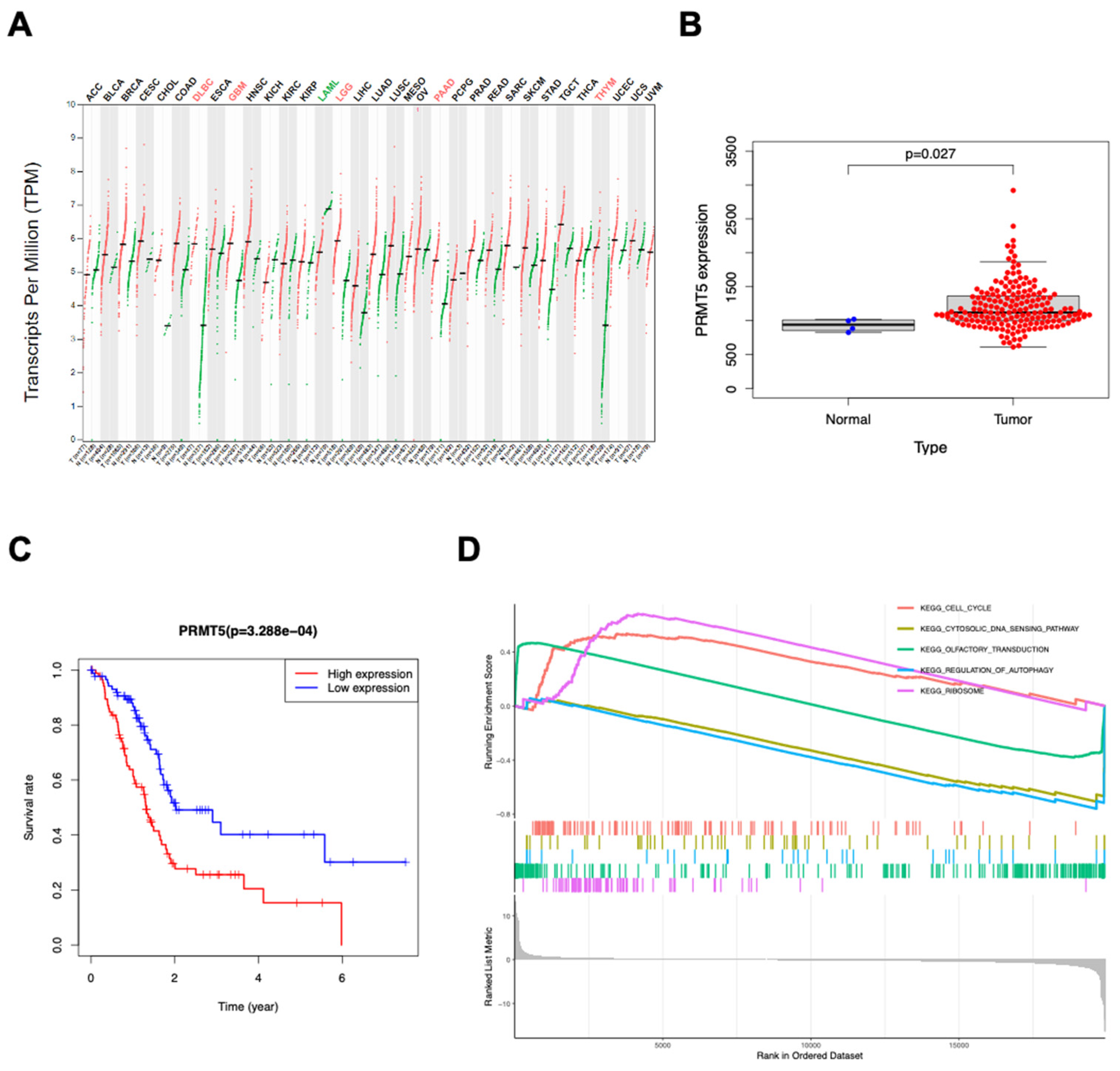
Clinical information and gene expression data on 180 primary tumors were acquired from TCGA database through available downloads. Pan-cancer analysis demonstrated that PRMT5 was upregulated in multiple cancers including diffuse large B cell lymphoma (DLBCL) compared with normal tissues (
Figure 1A). Additionally, PRMT5 expression in tumor samples was obviously higher than in normal samples (
Figure 1B). Furthermore, increased expression of PRMT5 in lymphomas was also significantly associated with poor overall survival (
Figure 1C). To investigate the biological consequence of differential PRMT5 expression, we performed GSEA algorithm which inferred that high PRMT5 was correlated with cell cycle, ribosome, cytosolic DNA sensing pathway, autophagy regulation and olfactory transduction (
Figure 1D). Except for olfactory transduction, all the other pathways above are closely related to cancer, suggesting that PRMT5 probably reshapes human tumors via multiple aspects.
3.2. PRMT5 Likely Regulates Tumor Immune Microenvironment and Tumor Per SE
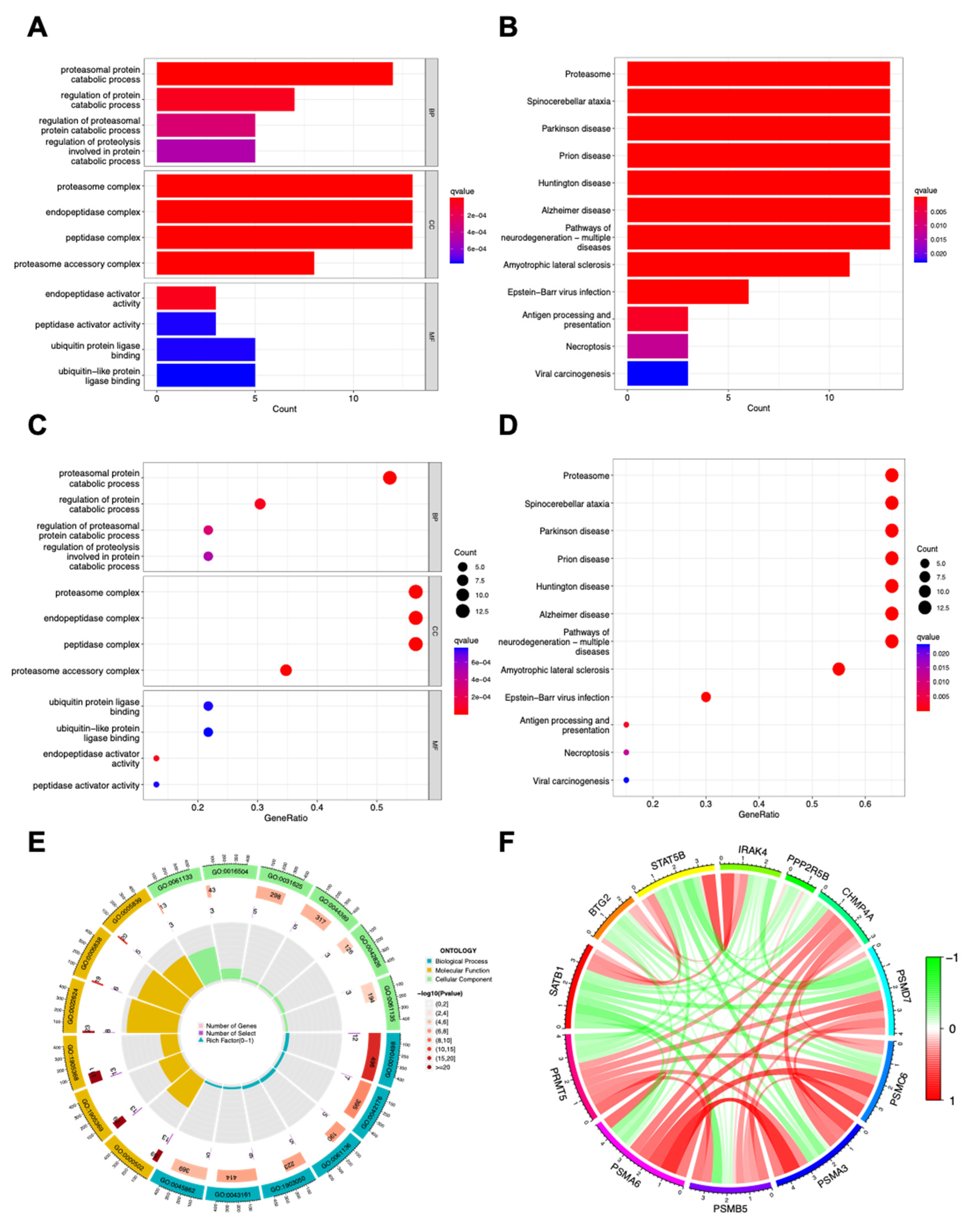
In pan-cancer DEGs analysis, a broad spectrum of proteasomal proteins were positively correlated with PRMT5 expression. Therefore, protein degradation related pathways and diseases (GO pathways: proteasomal and regulation of protein catabolic progress, proteasome complex and ubiquitin protein ligase binding (
Figure 2A,C,E); KEGG pathways: proteasome, spinocerebellar ataxia, Parkinson disease, Epstein-Barr virus infection and viral carcinogenesis (
Figure 2B,D)) were found in the top lists. Proteasome activity was fine adjusted to maintain cell cycle and survival-apoptosis balance, and, for this reason, proteasome inhibitors were developed for cancer treatment [
26,
27]. Resent findings also supported that proteasome activity is crucial for immune cell and anti-tumor immunity [
28,
29], indicating that PRMT5 likely regulate cancer immunity via proteasome activity. Besides proteasomal proteins, other cancer (SATB1, STAT5B and PPP2R5B) or immune (BTG2, IRAK4 and CHMP4A) related genes were also found related to PRMT5 abundance. Interactions among PRMT5 and its related genes were shown in
Figure 2F, demonstrating the cooperation of PRMT5 related genes reinforce each other to amplify the overall effect of PRMT5 overexpression on cancer. Those GO and KEGG data implied that PRMT5 likely regulates both tumor immune microenvironment and tumor per se.
3.3. PRMT5 Correlates with Immune Infiltration in Lymphoma
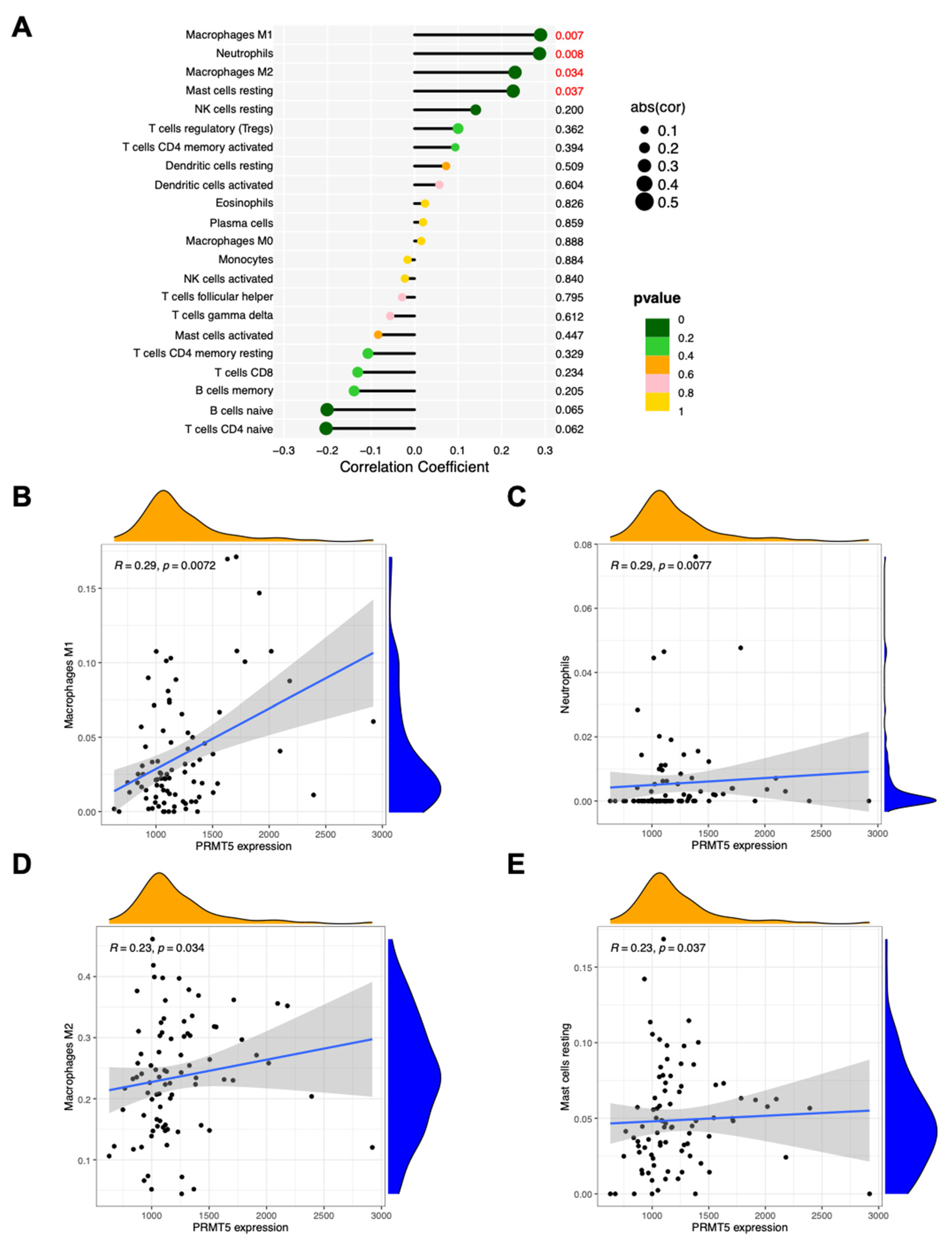
Tumor-infiltrating lymphocytes independently predict the overall survival and sentinel lymph node status among cancer patients. Since GO and KEGG data showed possible relationship between PRMT5 and immunity, the proportions of immune cells in lymphoma with different PRMT5 abundance were analyzed (
Figure 3A). Four types of immune cells, namely, macrophages M1 (R = 0.29, p = 0.0072), neutrophils (R = 0.29, p = 0.0077), macrophages M2 (R = 0.23, p = 0.034), and resting mast cells (R = 0.23, p = 0.037) in lymphoma showed correlation with PRMT5 expression (
Figure 3B–E). In general, macrophages M2, neutrophils, and mast cells infiltrated in tumors are tumor-supporting whereas macrophages M1 in tumors are tumor inhibiting [
30,
31,
32]. The overall effect of PRMT5 overexpression on tumor immune microenvironment is likely beneficial to tumor growth.
3.4. PRMT5 Related Genes in Lymphoma Overlapped with Those Found in Pan-Cancer
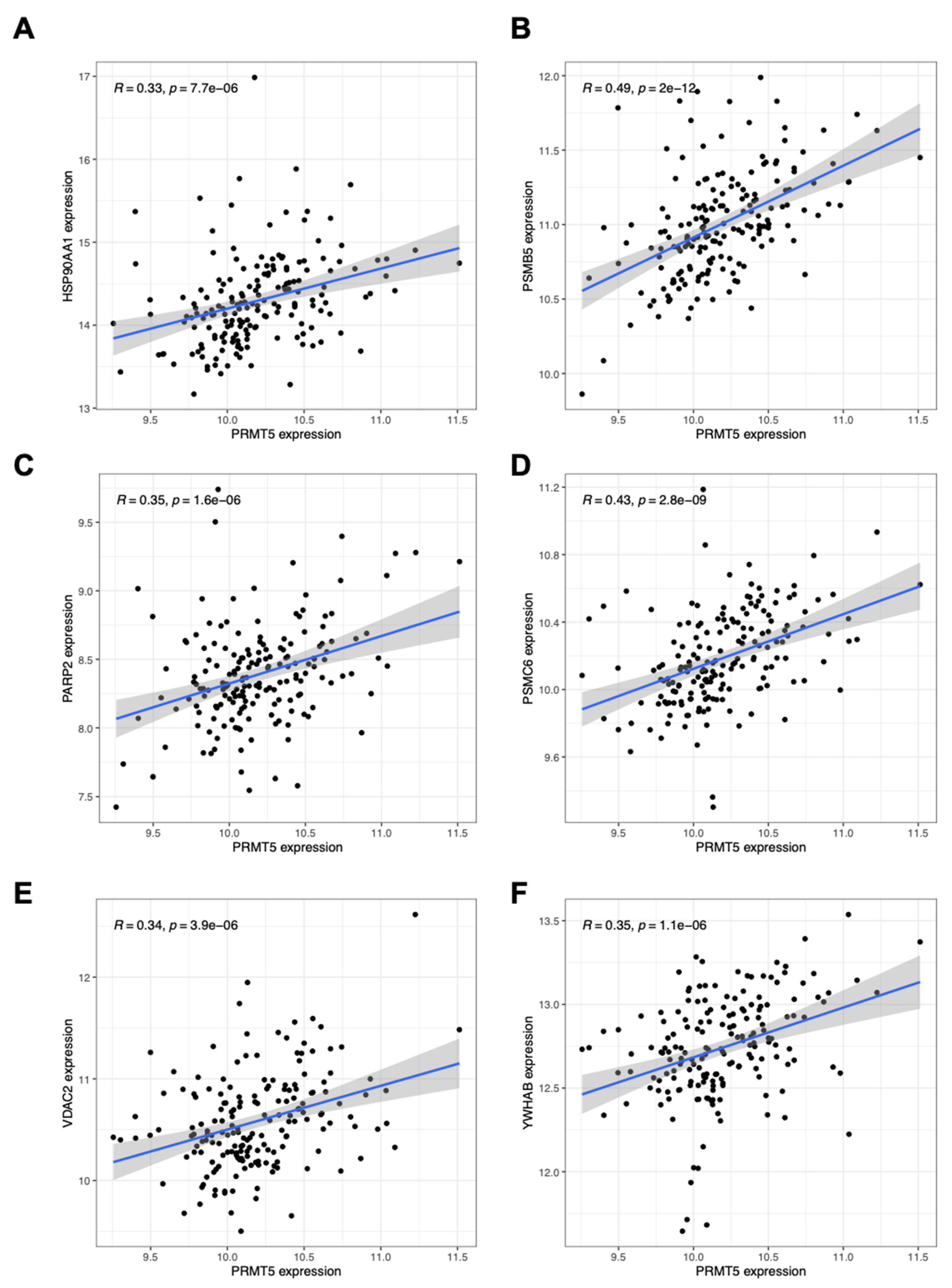
After analyzing pan-cancer PRMT5 related genes with GO and KEGG related pathways, we further investigated the correlation between PRMT5 expression and key genes expression in lymphoma. Almost all PRMT5 related DEGs found in lymphoma are overlapped with those found in pan-cancer (
Figure 4A–F and
Figure S1). This result indicated that PRMT5 function was likely conserved in lymphoma and other cancer types. Since PRMT5 is evolutionally conserved and critical for tumor progression in many aspects, we considered if PRMT5 is a reliable survival indicator for at least lymphoma.
3.5. PRMT5 is a Reliable Survival Prediction Index for Lymphoma Patients
The 10-fold cross-validation method was applied to the iterative analysis, and a model with optimal performance with minimum number of variables was obtained (
Figure 5A). Lasso regression was used to screen parameters, and the variation characteristics of the coefficient of these variables were shown in
Figure 5B. Additionally, ROC curve analysis was utilized to predict the overall survival at 1, 3, and 5 years in lymphoma patients. Notably, overall survival at 1, 3 and 5 years showed adequate predictive ability based on the model (AUC 0.741, 0.767 and 0.794 respectively) (
Figure 5C). Then Kaplan-Meier curves were plotted based on the risk classes which showed that high PRMT5 expression could predict poor survival (
Figure 5D).
4. Discussion
PRMT5 changes stability and/or interaction preference of its substrates by methylation of their arginine residuals, and these substrates amplify the effect of PRMT5 by regulating their downstream pathways. Accumulated preclinical studies showed tumor-promoting effects of PRMT5 via multiple pathways [
33,
34,
35]. Indeed, clinical data proved that PRMT5 is overexpressed in a variety of cancer tissues, including breast cancer [
36], cervical cancer [
37], prostate cancer [
38] and mantle cell lymphoma [
11]. By deep analyzing clinical data at pan-cancer level and in lymphoma, we found many connections between preclinical and clinical data about PRMT5, and it gave us many interesting inspirations.
PRMT5 methylates mRNA splicing proteins and alters splicing pattern of a subset of genes including MDM4, a p53 inactivator. Alternative spliced MDM4 during PRMT5 deficiency showed decreased p53 binding capability and thus activate p53 [
13]. p53 stops tumor progression in many aspects, such as arresting cell cycle, inhibiting ribosome biogenesis, facilitating cytosolic DNA sensing and inducing autophagy [
39]. All these aspects were found in PRMT5 related pathways concluded from GSEA analysis. In addition, PRMT5 interacts with a majority proportion of ribosome proteins and directly methylate RPS10, a protein member of ribosome small subunit. Correct arginine methylation of RPS10 is required for ribosome biogenesis [
40], coincident with our GSEA analysis results using clinical data. Furthermore, arginine methylation of E2F1 by PRMT5 promotes cell cycle progression, migration, and invasion by decreasing the affinity of E2F1 for retinoblastoma protein [
13], allowing for enhanced E2F1 target gene transcription. PRMT5 also methylates and thus enable activation of AKT1, which stimulates cell cycle and suppresses autophagy [
33]. All these pre-clinical findings and the bioinformatic summary of clinical data in this study solidified the conclusion that PRMT5 promotes cancer progression by regulating cell cycle, ribosome biogenesis, and autophagy.
PRMT5 regulates transcription of a subset of genes via histone arginine methylation (H3R2, H3R8, H4R3, and H2AR3). PRMT5 enhanced the transcription of STAT1 through symmetric dimethylation of histone H3R2 and promoted PD-L1 expression in cervical cancer cells. In addition, knockdown of PRMT5 can increase the expression of IFN-γ, TNF-α and granzyme B in T cells [
37]. Similarly, we found several immune cells regulating genes corelated with PRMT5 expression and overexpression of PRMT5 showed an impact on infiltrated immune cells in tumor. Previous pre-clinical findings [
41] and our results from clinical data all strongly supports the hypothesis that PRMT5 regulating tumor immune microenvironment. Interestingly, by comparing CHIP-Sequencing data of PRMT5 with 1 kb distance from transcription starting site (TSS) on CHIP-atlas database and PRMT5 related genes (DEGs between high and low PRMT5 samples) in this study, only PSMD14 was present in both data lists. How PRMT5 regulates transcription of proteasome proteins as well as other related genes is still to be answered. Since PRMT5 CHIP-atlas data were based on certain cell lines, transcription of those PRMT5 related genes is likely either only directly regulated under certain signaling background, or indirectly regulated by PRMT5 substrates.
In summary, bioinformatic data in this study and previous pre-clinical studies coincidently showed that PRMT5 regulates both tumor immune microenvironment and tumor per se. Based on this conclusion and the result of survival prediction, PRMT5 is qualified for a reliable survival prediction index of lymphoma patients.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org, Figure S1: PRMT5 correlates with multiple gene expression in lymphoma.
Author Contributions
Conceptualization, M.L., X.S. and Y.C.; methodology, Y.L. and Y.C.; software, Y.C.; formal analysis, Y.C., Y.L. and M.Z.; investigation, Y.C. and M.Z.; original draft preparation, Y.C.; review and editing, M.L. and X.S.; supervision, M.L.; project administration, X.S.; funding acquisition, M.L. and X.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Dalian Medical University (Grant No. JCHZ2023009) and Wu Jieping Funding (No.2022LCYJZD01).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| TCGA |
The Cancer Genome Atlas |
| GEPIA |
Gene Expression Profiling Interactive Analysis |
| DLBCL |
Diffuse Large B Cell Lymphoma |
| NHL |
Non-Hodgkin Lymphoma |
| PRMT5 |
Protein Arginine Methyltransferase 5 |
| BTK |
Bruton Tyrosine Kinase |
References
- Siegel, R.L., A.N. Giaquinto, and A. Jemal, Cancer statistics, 2024. CA Cancer J Clin, 2024. 74(1): p. 12-49.
- Holohan, C., et al., Cancer drug resistance: an evolving paradigm. Nat Rev Cancer, 2013. 13(10): p. 714-26. [CrossRef]
- Elstrom, R.L., et al., Response to second-line therapy defines the potential for cure in patients with recurrent diffuse large B-cell lymphoma: implications for the development of novel therapeutic strategies. Clin Lymphoma Myeloma Leuk, 2010. 10(3): p. 192-6. [CrossRef]
- Van Den Neste, E., et al., Outcomes of diffuse large B-cell lymphoma patients relapsing after autologous stem cell transplantation: an analysis of patients included in the CORAL study. Bone Marrow Transplant, 2017. 52(2): p. 216-221.
- Antonysamy, S., et al., Crystal structure of the human PRMT5:MEP50 complex. Proc Natl Acad Sci U S A, 2012. 109(44): p. 17960-5.
- Migliori, V., et al., Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol, 2012. 19(2): p. 136-44. [CrossRef]
- Girardot, M., et al., PRMT5-mediated histone H4 arginine-3 symmetrical dimethylation marks chromatin at G + C-rich regions of the mouse genome. Nucleic Acids Res, 2014. 42(1): p. 235-48. [CrossRef]
- Liu, M., et al., PRMT5-dependent transcriptional repression of c-Myc target genes promotes gastric cancer progression. Theranostics, 2020. 10(10): p. 4437-4452. [CrossRef]
- Wei, T.Y., et al., Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade. Cancer Sci, 2012. 103(9): p. 1640-50.
- Karkhanis, V., et al., Protein arginine methyltransferase 5 represses tumor suppressor miRNAs that down-regulate CYCLIN D1 and c-MYC expression in aggressive B-cell lymphoma. J Biol Chem, 2020. 295(5): p. 1165-1180.
- Che, Y., et al., Exploiting PRMT5 as a target for combination therapy in mantle cell lymphoma characterized by frequent ATM and TP53 mutations. Blood Cancer J, 2023. 13(1): p. 27. [CrossRef]
- Pastore, F., et al., PRMT5 Inhibition Modulates E2F1 Methylation and Gene-Regulatory Networks Leading to Therapeutic Efficacy in JAK2(V617F)-Mutant MPN. Cancer Discov, 2020. 10(11): p. 1742-1757. [CrossRef]
- Gerhart, S.V., et al., Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing. Sci Rep, 2018. 8(1): p. 9711. [CrossRef]
- Zhu, F., et al., PRMT5 is upregulated by B-cell receptor signaling and forms a positive-feedback loop with PI3K/AKT in lymphoma cells. Leukemia, 2019. 33(12): p. 2898-2911.
- Hing, Z.A., et al., Dysregulation of PRMT5 in chronic lymphocytic leukemia promotes progression with high risk of Richter's transformation. Nat Commun, 2023. 14(1): p. 97.
- Blum, A., P. Wang, and J.C. Zenklusen, SnapShot: TCGA-Analyzed Tumors. Cell, 2018. 173(2): p. 530.
- Tang, Z., et al., GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res, 2017. 45(W1): p. W98-w102.
- Subramanian, A., et al., Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A, 2005. 102(43): p. 15545-50.
- Yu, G., et al., clusterProfiler: an R package for comparing biological themes among gene clusters. Omics, 2012. 16(5): p. 284-7. [CrossRef]
- Lu, Y., et al., A probabilistic generative model for GO enrichment analysis. Nucleic Acids Res, 2008. 36(17): p. e109. [CrossRef]
- Kanehisa, M., et al., KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res, 2017. 45(D1): p. D353-d361. [CrossRef]
- Gentles, A.J., et al., The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med, 2015. 21(8): p. 938-945.
- Ali, H.R., et al., Patterns of Immune Infiltration in Breast Cancer and Their Clinical Implications: A Gene-Expression-Based Retrospective Study. PLoS Med, 2016. 13(12): p. e1002194.
- Bense, R.D., et al., Relevance of Tumor-Infiltrating Immune Cell Composition and Functionality for Disease Outcome in Breast Cancer. J Natl Cancer Inst, 2017. 109(1).
- Tibshirani, R., The lasso method for variable selection in the Cox model. Stat Med, 1997. 16(4): p. 385-95.
- Manasanch, E.E. and R.Z. Orlowski, Proteasome inhibitors in cancer therapy. Nat Rev Clin Oncol, 2017. 14(7): p. 417-433. [CrossRef]
- Zhou, C., et al., Anti-tumor efficacy of HRS-4642 and its potential combination with proteasome inhibition in KRAS G12D-mutant cancer. Cancer Cell, 2024. 42(7): p. 1286-1300.e8. [CrossRef]
- Javitt, A., et al., The proteasome regulator PSME4 modulates proteasome activity and antigen diversity to abrogate antitumor immunity in NSCLC. Nat Cancer, 2023. 4(5): p. 629-647. [CrossRef]
- Çetin, G., et al., The Ubiquitin-Proteasome System in Immune Cells. Biomolecules, 2021. 11(1).
- Xiong, S., L. Dong, and L. Cheng, Neutrophils in cancer carcinogenesis and metastasis. J Hematol Oncol, 2021. 14(1): p. 173.
- Li, M., et al., Metabolism, metabolites, and macrophages in cancer. J Hematol Oncol, 2023. 16(1): p. 80.
- Liu, X., et al., Mast cells in colorectal cancer tumour progression, angiogenesis, and lymphangiogenesis. Front Immunol, 2023. 14: p. 1209056.
- Huang, L., et al., PRMT5 activates AKT via methylation to promote tumor metastasis. Nat Commun, 2022. 13(1): p. 3955.
- Sloan, S.L., et al., PRMT5 supports multiple oncogenic pathways in mantle cell lymphoma. Blood, 2023. 142(10): p. 887-902.
- Sun, Y., et al., MST2 methylation by PRMT5 inhibits Hippo signaling and promotes pancreatic cancer progression. Embo j, 2023. 42(23): p. e114558.
- Poulard, C., et al., Nuclear PRMT5 is a biomarker of sensitivity to tamoxifen in ERα(+) breast cancer. EMBO Mol Med, 2023. 15(8): p. e17248.
- Jiang, Y., et al., PRMT5 disruption drives antitumor immunity in cervical cancer by reprogramming T cell-mediated response and regulating PD-L1 expression. Theranostics, 2021. 11(18): p. 9162-9176. [CrossRef]
- Yao, B., et al., The circSPON2/miR-331-3p axis regulates PRMT5, an epigenetic regulator of CAMK2N1 transcription and prostate cancer progression. Mol Cancer, 2022. 21(1): p. 119. [CrossRef]
- Vousden, K.H. and D.P. Lane, p53 in health and disease. Nat Rev Mol Cell Biol, 2007. 8(4): p. 275-83. [CrossRef]
- Ren, J., et al., Methylation of ribosomal protein S10 by protein-arginine methyltransferase 5 regulates ribosome biogenesis. J Biol Chem, 2010. 285(17): p. 12695-705. [CrossRef]
- Hu, R., et al., PRMT5 Inhibition Promotes PD-L1 Expression and Immuno-Resistance in Lung Cancer. Front Immunol, 2021. 12: p. 722188. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).