Submitted:
02 July 2024
Posted:
03 July 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Methods
Cell Culture
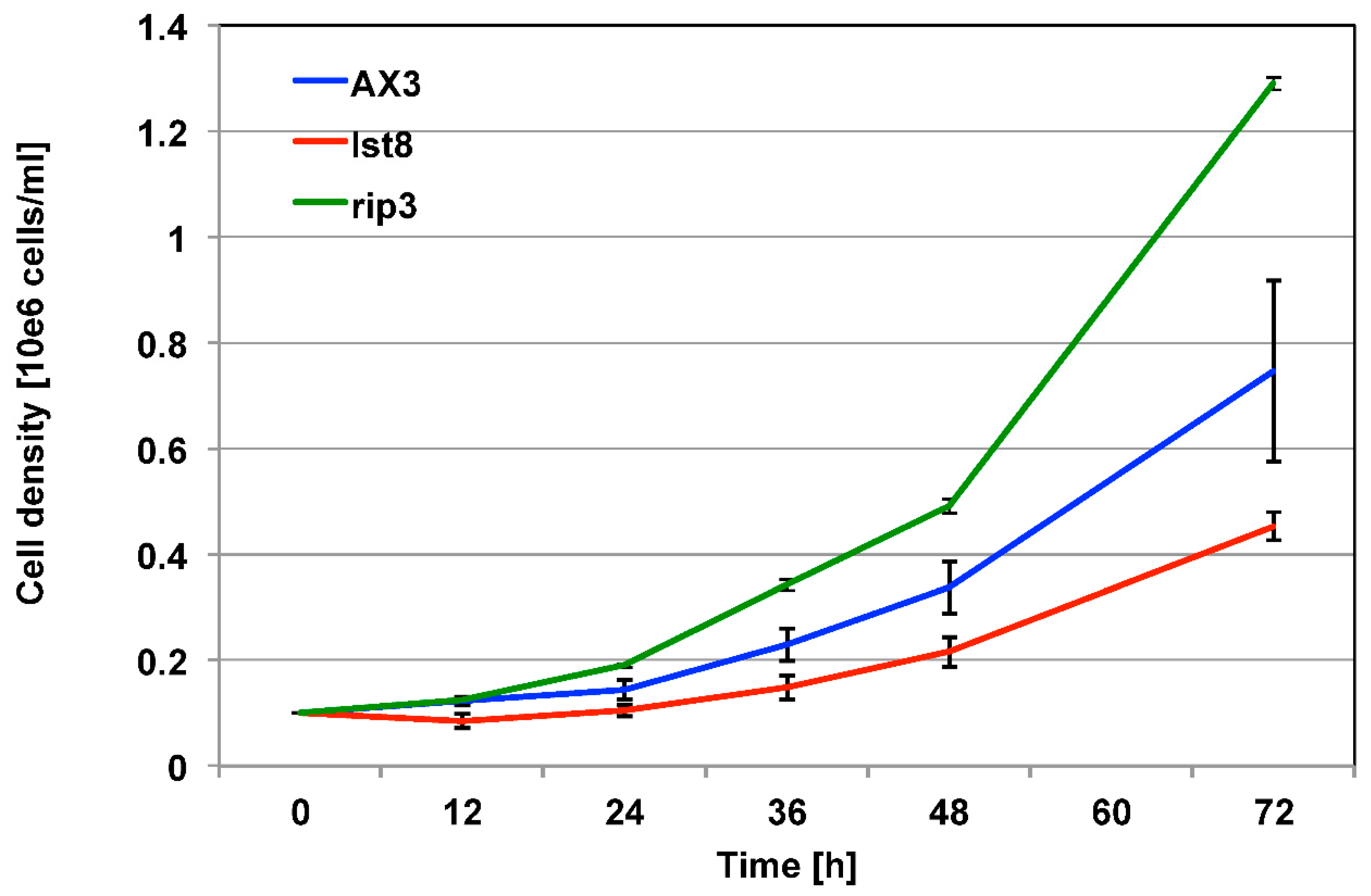
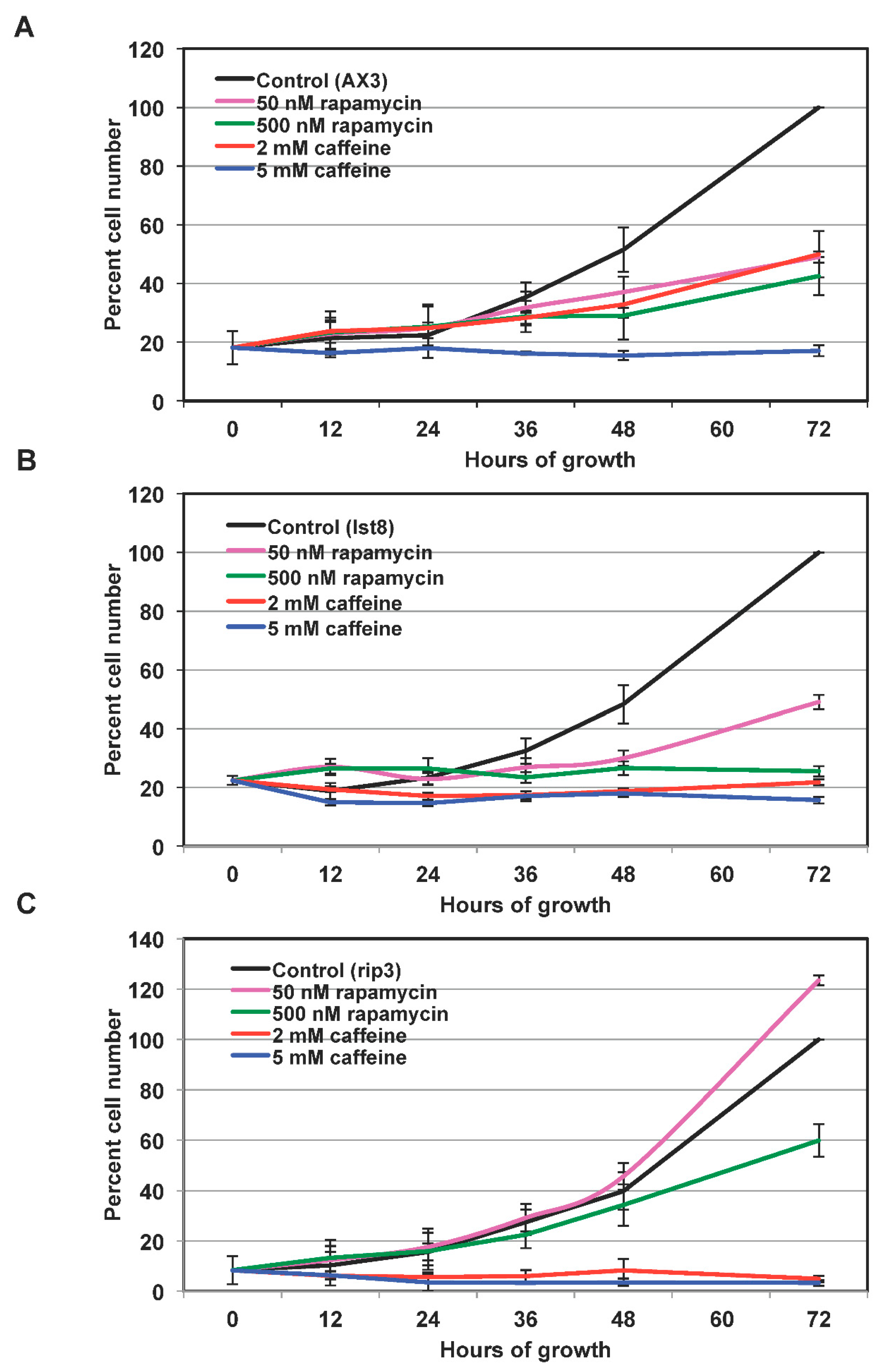
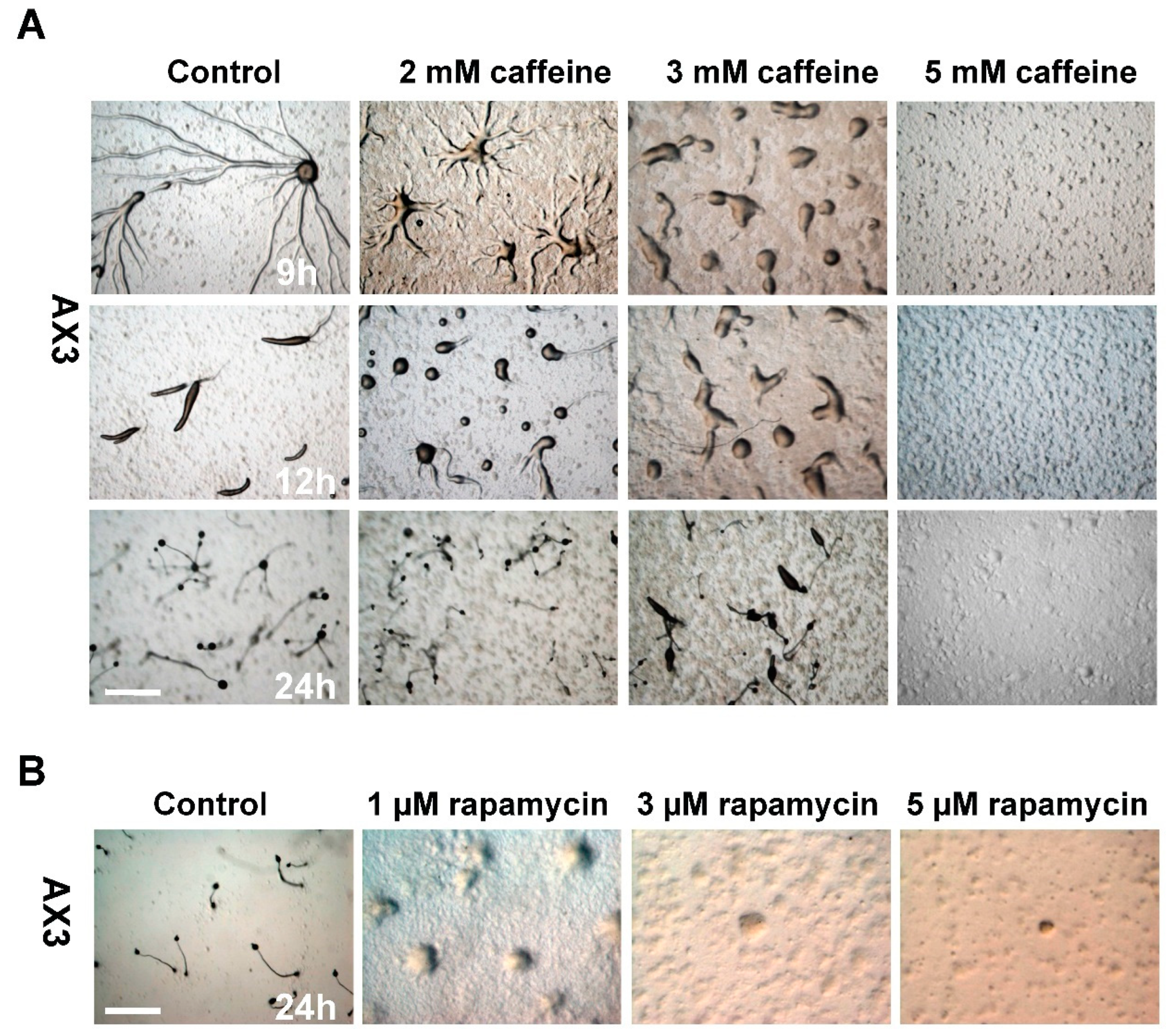
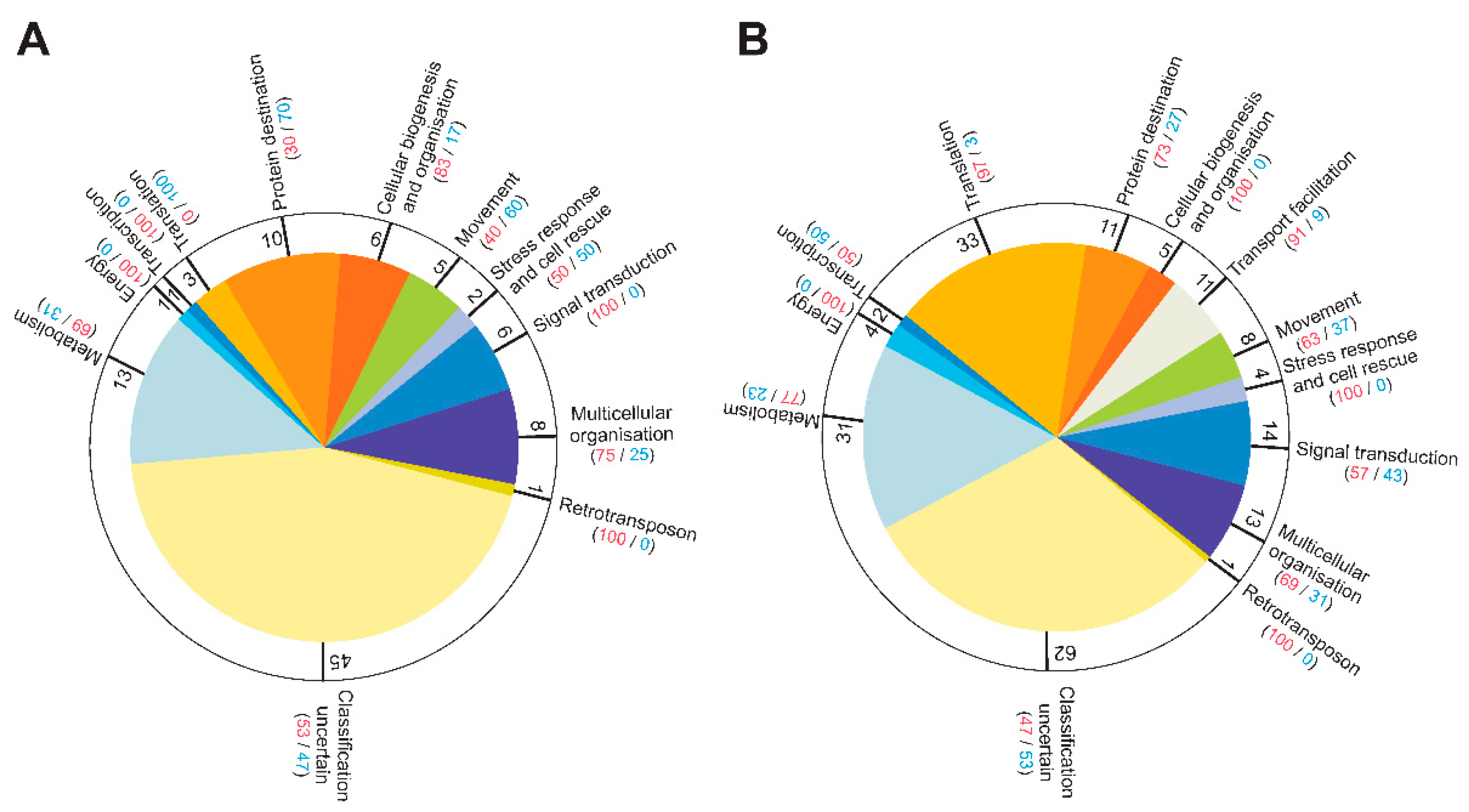
Results
Discussion
Conclusions
Supplementary Materials
References
- Bonner, J.T.; Princeton University Press: Princeton, 1967.
- Jaiswal, P.; Kimmel, A.R. mTORC1/AMPK responses define a core gene set for developmental cell fate switching. BMC Biology 2019, 17, 58. [CrossRef]
- Jaiswal, P.; Majithia, A.; Rosel, D.; Liao, X.; Khurana, T.; Kimmel, A. Integrated actions of mTOR complexes 1 and 2 for growth and development of Dictyostelium. The International Journal of Developmental Biology 2019, 63, 521-527.
- Soulard, A.; Cohen, A.; Hall, M.N. TOR signaling in invertebrates. Curr Opin Cell Biol 2009, 21, 825-836. [CrossRef]
- De Virgilio, C.; Loewith, R. The TOR signalling network from yeast to man. Int J Biochem Cell Biol 2006, 38, 1476-1481. [CrossRef]
- Jaiswal, P.K.; Kimmel, A.R. Nutrient/Starvation sensing for Reciprocal mTORC1/AMPK response in Dictyostelium, at the junction between Growth and Development. The FASEB Journal 2018, 32, lb141-lb141. [CrossRef]
- Powers, T.; Dilova, I.; Chen, C.Y.; Wedaman, K. Yeast TOR signaling: a mechanism for metabolic regulation. Curr Top Microbiol Immunol 2004, 279, 39-51. [CrossRef]
- Mayer, C.; Grummt, I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 2006, 25, 6384-6391. [CrossRef]
- Kuranda, K.; Leberre, V.; Sokol, S.; Palamarczyk, G.; François, J. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Molecular Microbiology 2006, 61, 1147-1166. [CrossRef]
- Saiki, S.; Sasazawa, Y.; Imamichi, Y.; Kawajiri, S.; Fujimaki, T.; Tanida, I.; Kobayashi, H.; Sato, F.; Sato, S.; Ishikawa, K.; et al. Caffeine induces apoptosis by enhancement of autophagy via PI3K/Akt/mTOR/p70S6K inhibition. Autophagy 2011, 7, 176-187. [CrossRef]
- Rosel, D.; Khurana, T.; Majithia, A.; Huang, X.; Bhandari, R.; Kimmel, A.R. TOR complex 2 (TORC2) in Dictyostelium suppresses phagocytic nutrient capture independently of TORC1-mediated nutrient sensing. J Cell Sci 2012, 125, 37-48. [CrossRef]
- Jaiswal, P.; Meena, N.P.; Chang, F.-S.; Liao, X.-H.; Kim, L.; Kimmel, A.R. An integrated, cross-regulation pathway model involving activating/adaptive and feed-forward/feed-back loops for directed oscillatory cAMP signal-relay/response during the development of Dictyostelium</i>. Frontiers in cell and developmental biology 2023, 11, 1263316. [CrossRef]
- Lee, S.; Comer, F.I.; Sasaki, A.; McLeod, I.X.; Duong, Y.; Okumura, K.; Yates, J.R., 3rd; Parent, C.A.; Firtel, R.A. TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol Biol Cell 2005, 16, 4572-4583. [CrossRef]
- Liu, L.; Parent, C.A. Review series: TOR kinase complexes and cell migration. J Cell Biol 2011, 194, 815-824. [CrossRef]
- Yu, L.; Coelho, J.E.; Zhang, X.; Fu, Y.; Tillman, A.; Karaoz, U.; Fredholm, B.B.; Weng, Z.; Chen, J.F. Uncovering multiple molecular targets for caffeine using a drug target validation strategy combining A 2A receptor knockout mice with microarray profiling. Physiol Genomics 2009, 37, 199-210. [CrossRef]
- Jaiswal, P.; Soldati, T.; Thewes, S.; Baskar, R. Regulation of aggregate size and pattern by adenosine and caffeine in cellular slime molds. BMC Developmental Biology 2012, 12, 5. [CrossRef]
- Jaiswal, P.; Singh, S.P.; Aiyar, P.; Akkali, R.; Baskar, R. Regulation of multiple tip formation by caffeine in cellular slime molds. BMC Developmental Biology 2012, 12, 26. [CrossRef]
- van Haastert, P.J.; De Wit, R.J.; Konijn, T.M. Antagonists of chemoattractants reveal separate receptors for cAMP, folic acid and pterin in Dictyostelium. Exp Cell Res 1982, 140, 453-456. [CrossRef]
- Gonzalez, C.; Klein, G.; Satre, M. Caffeine, an inhibitor of endocytosis in Dictyostelium discoideum amoebae. J Cell Physiol 1990, 144, 408-415. [CrossRef]
- Brenner, M.; Thoms, S.D. Caffeine blocks activation of cyclic AMP synthesis in Dictyostelium discoideum. Dev Biol 1984, 101, 136-146. [CrossRef]
- Reinke, A.; Chen, J.C.Y.; Aronova, S.; Powers, T. Caffeine Targets TOR Complex I and Provides Evidence for a Regulatory Link between the FRB and Kinase Domains of Tor1p*. Journal of Biological Chemistry 2006, 281, 31616-31626. [CrossRef]
- Foukas, L.C.; Daniele, N.; Ktori, C.; Anderson, K.E.; Jensen, J.; Shepherd, P.R. Direct Effects of Caffeine and Theophylline on p110δ and Other Phosphoinositide 3-Kinases: DIFFERENTIAL EFFECTS ON LIPID KINASE AND PROTEIN KINASE ACTIVITIES*. Journal of Biological Chemistry 2002, 277, 37124-37130. [CrossRef]
- Singh, S.P.; Dhakshinamoorthy, R.; Jaiswal, P.; Schmidt, S.; Thewes, S.; Baskar, R. The thyroxine inactivating gene, type III deiodinase, suppresses multiple signaling centers in Dictyostelium discoideum. Developmental Biology 2014, 396, 256-268. [CrossRef]
- Meena, N.P.; Jaiswal, P.; Chang, F.-S.; Brzostowski, J.; Kimmel, A.R. DPF is a cell-density sensing factor, with cell-autonomous and non-autonomous functions during Dictyostelium growth and development. BMC Biology 2019, 17, 97. [CrossRef]
- Mehto, S.; Jena, K.K.; Yadav, R.; Priyadarsini, S.; Samal, P.; Krishna, S.; Dhar, K.; Jain, A.; Chauhan, N.R.; Murmu, K.C.; et al. Selective autophagy of RIPosomes maintains innate immune homeostasis during bacterial infection. The EMBO Journal 2022, 41, e111289. [CrossRef]
- Farbrother, P.; Wagner, C.; Na, J.; Tunggal, B.; Morio, T.; Urushihara, H.; Tanaka, Y.; Schleicher, M.; Steinert, M.; Eichinger, L. Dictyostelium transcriptional host cell response upon infection with Legionella. Cell Microbiol 2006, 8, 438-456. [CrossRef]
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 2001, 98, 5116-5121. [CrossRef]
- Lee, S.; Parent, C.A.; Insall, R.; Firtel, R.A. A novel Ras-interacting protein required for chemotaxis and cyclic adenosine monophosphate signal relay in Dictyostelium. Mol Biol Cell 1999, 10, 2829-2845. [CrossRef]
- Wu, L.; Hansen, D.; Franke, J.; Kessin, R.H.; Podgorski, G.J. Regulation of Dictyostelium early development genes in signal transduction mutants. Dev Biol 1995, 171, 149-158. [CrossRef]
- Ekim, B.; Magnuson, B.; Acosta-Jaquez, H.A.; Keller, J.A.; Feener, E.P.; Fingar, D.C. mTOR kinase domain phosphorylation promotes mTORC1 signaling, cell growth, and cell cycle progression. Mol Cell Biol 2011, 31, 2787-2801. [CrossRef]
- Vilella-Bach, M.; Nuzzi, P.; Fang, Y.; Chen, J. The FKBP12-rapamycin-binding domain is required for FKBP12-rapamycin-associated protein kinase activity and G1 progression. J Biol Chem 1999, 274, 4266-4272. [CrossRef]
- Chauhan, S.; Jena, K.K.; Mehto, S.; Chauhan, N.R.; Sahu, R.; Dhar, K.; Yadav, R.; Krishna, S.; Jaiswal, P.; Chauhan, S. Innate immunity and inflammophagy: balancing the defence and immune homeostasis. The FEBS Journal 2022, 289, 4112-4131. [CrossRef]
- Urushihara, H.; Morio, T.; Saito, T.; Kohara, Y.; Koriki, E.; Ochiai, H.; Maeda, M.; Williams, J.G.; Takeuchi, I.; Tanaka, Y. Analyses of cDNAs from growth and slug stages of Dictyostelium discoideum. Nucleic Acids Res 2004, 32, 1647-1653. [CrossRef]

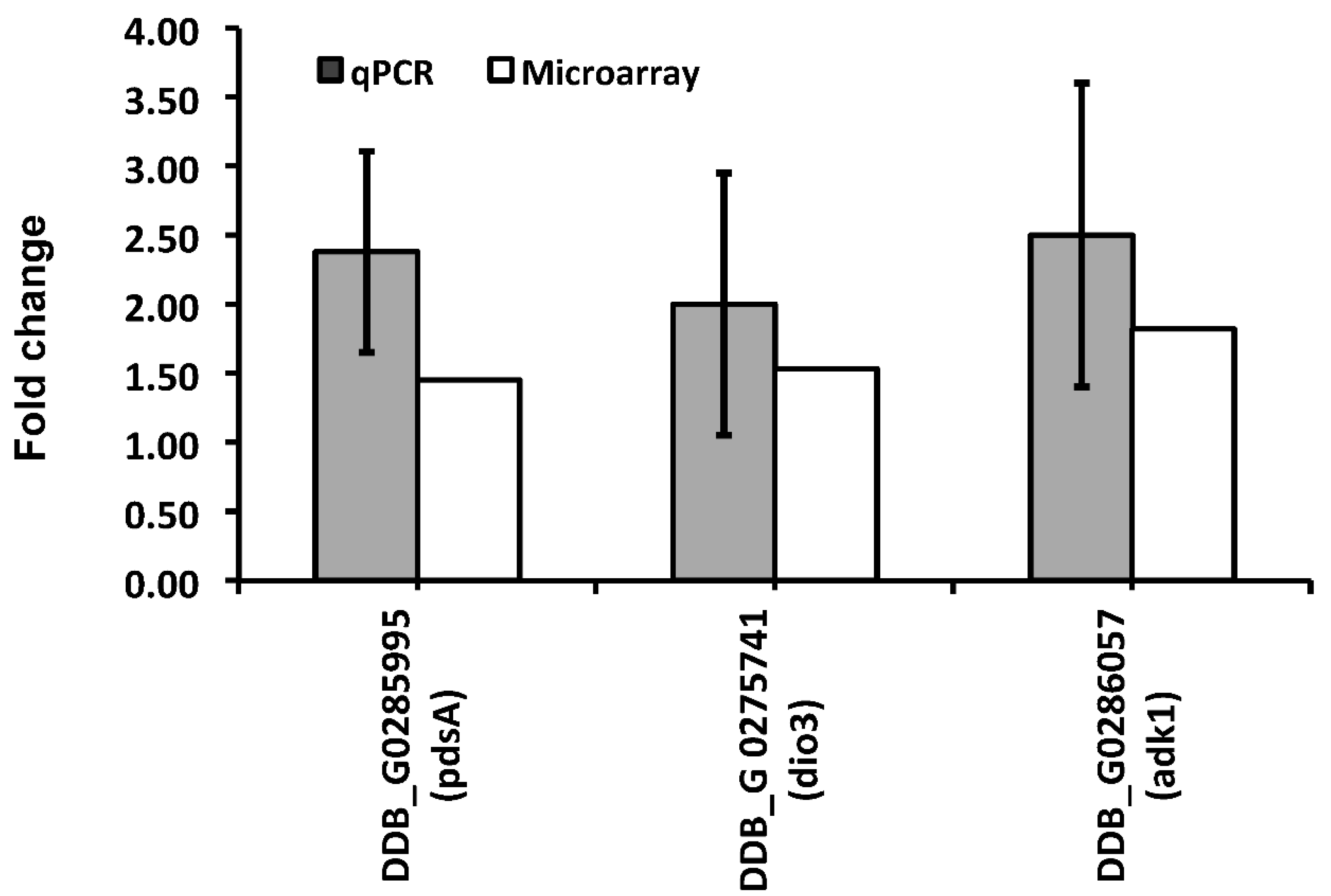

| DDB_G ID | FC 3h | FC 6h | Gene Name | Gene Product |
|---|---|---|---|---|
| DDB_G0281843 | 2,36 | 2,69 | DDB_G0281843 | PAN-1 domain-containing protein |
| DDB_G0273397 | 1,72 | 1,61 | carA-2 | cAMP receptor 1 |
| DDB_G0289393 | 1,69 | 1,95 | psiH | PA14 domain-containing protein |
| DDB_G0287035 | 1,66 | 2,22 | lmpB | lysosomal integral membrane glycoprotein (LmpB) |
| DDB_G0273919 | 1,59 | 1,40 | dscA-2 | discoidin I, alpha chain |
| DDB_G0277863 | 1,57 | 1,91 | pdiA | cAMP phosphodiesterase inhibitor |
| DDB_G0285617 | 1,57 | 1,58 | DDB_G0285617 | agglutinin domain-containing protein |
| DDB_G0285505 | 1,56 | 2,01 | ino1 | inositol-3-phosphate synthase, myo-inositol-1-phosphate synthase |
| DDB_G0292566 | 1,56 | 1,64 | hibA | 3-hydroxyisobutyrate dehydrogenase |
| DDB_G0287281 | 1,55 | 1,67 | DDB_G0287281 | citrate synthase |
| DDB_G0277501 | 1,53 | 1,71 | capB | cAMP binding protein B |
| DDB_G0274093 | 1,53 | 1,40 | adrm1-2 | adhesion regulating molecule family protein |
| DDB_G0272668 | 1,49 | 1,64 | ppr2 | protein phosphatase 4 regulatory subunit 2 |
| DDB_G0282817 | 1,47 | 2,42 | patB | P-type ATPase |
| DDB_G0273805 | 1,46 | 2,10 | ndkC-2 | nucleosidediphosphate kinase, NDP kinase |
| DDB_G0282051 | 1,45 | 1,66 | DDB_G0282051_RTE | TRE5-A ORF1 |
| DDB_G0285995 | 1,45 | 1,47 | pdsA | cAMP/cGMP phosphodiesterase |
| DDB_G0272819 | 1,42 | 1,76 | hspC | heat shock protein |
| DDB_G0272969 | 1,42 | 1,42 | psmB1 | proteasome subunit beta type 1, 20S proteasome subunit beta-1 |
| DDB_G0272875 | 1,42 | 1,58 | cpnB-2 | copine B |
| DDB_G0269122 | 1,41 | 1,42 | cysA | cystathionine gamma-lyase |
| DDB_G ID | FC 3h | FC 6h | Gene Name | Gene Product |
|---|---|---|---|---|
| DDB_G0269136 | 0,97 | 1,59 | efaAII | elongation factor 1 alpha |
| DDB_G0284035 | 0,97 | 1,45 | efa1B | elongation factor 1 beta |
| DDB_G0282979 | 0,86 | 1,53 | efa1G | elongation factor 1 gamma |
| DDB_G0288373 | 1,05 | 2,13 | efbA | elongation factor 2 |
| DDB_G0281403 | 0,99 | 1,72 | nacA | putative nascent polypeptide-associated complex alpha subunit |
| DDB_G0270316 | 1,16 | 1,41 | rpsA | 40S ribosomal protein SA |
| DDB_G0293742 | 1,23 | 1,80 | rps2 | 40S ribosomal protein S2 |
| DDB_G0293000 | 0,92 | 1,57 | rps3 | 40S ribosomal protein S3 |
| DDB_G0277345 | 1,19 | 1,58 | rps3a | 40S ribosomal protein S3a |
| DDB_G0272825 | 1,27 | 1,49 | rps4 | 40S ribosomal protein S4 |
| DDB_G0286075 | 1,19 | 1,46 | rps5 | 40S ribosomal protein S5 |
| DDB_G0280823 | 1,04 | 1,54 | rps6 | 40S ribosomal protein S6 |
| DDB_G0291864 | 1,02 | 1,45 | rps8 | 40S ribosomal protein S8 |
| DDB_G0284533 | 1,14 | 1,60 | rps13 | 40S ribosomal protein S13 |
| DDB_G0273743 | 1,16 | 1,49 | rps30-2 | 40S ribosomal protein S30 |
| DDB_G0291862 | 1,11 | 1,94 | rpl3 | 60S ribosomal protein L3 |
| DDB_G0277803 | 1,29 | 1,66 | rpl4 | 60S ribosomal protein L4 |
| DDB_G0278539 | 1,38 | 2,27 | rpl5 | 60S ribosomal protein L5 |
| DDB_G0276441 | 1,24 | 1,50 | rpl7 | S60 ribosomal protein L7 |
| DDB_G0274113 | 1,15 | 1,75 | rpl8 | 60S ribosomal protein L8 |
| DDB_G0278959 | 1,15 | 1,41 | rpl9 | 60S ribosomal protein L9 |
| DDB_G0291870 | 1,21 | 1,62 | rpl13 | S60 ribosomal protein L13 |
| DDB_G0275881 | 1,08 | 1,78 | rpl13a | S60 ribosomal protein L13a |
| DDB_G0277975 | 0,99 | 1,46 | rpl14 | S60 ribosomal protein L14 |
| DDB_G0273983 | 1,31 | 2,14 | rpl15-2 | S60 ribosomal protein L15 |
| DDB_G0279997 | 1,01 | 1,45 | rpl18 | S60 ribosomal protein L18 |
| DDB_G0281565 | 1,17 | 1,68 | rpl19 | S60 ribosomal protein L19 |
| DDB_G0290315 | 0,91 | 1,62 | rpl23 | S60 ribosomal protein L23 |
| DDB_G0293502 | 1,08 | 1,55 | rpl23a | S60 ribosomal protein L23a |
| DDB_G0292388 | 1,21 | 1,53 | rpl27a | S60 ribosomal protein L27a |
| DDB_G0282853 | 1,17 | 1,61 | DDB_G0282853 | TCTP family protein 1 |
| DDB_G0276871 | 0,66 | 1,28 | rpl10a | S60 ribosomal protein L10a |
| DDB_G0277823 | 0,65 | 1,35 | alas | alanyl-tRNAsynthetase |
| DDB_G0285451 | 0,71 | 0,90 | leuS | leucyl-tRNAsynthetase |
| DDB_G0279113 | 0,88 | 0,68 | metS | methionyl-tRNAsynthetase |
| DDB_G ID | FC 3h | FC 6h | Gene Name | Gene Product |
|---|---|---|---|---|
| DDB_G0273397 | 1,72 | 1,61 | carA-2 | cAMP receptor 1 |
| DDB_G0277829 | 1,36 | 1,47 | carC | cAMP receptor 3 |
| DDB_G0274023 | 1,35 | 1,77 | capA-2 | cAMP-binding protein |
| DDB_G0277501 | 1,53 | 1,71 | capB | cAMP-binding protein |
| DDB_G0285995 | 1,45 | 1,47 | pdsA | cAMP/cGMP phosphodiesterase |
| DDB_G0277863 | 1,57 | 1,91 | pdiA | cAMP phosphodiesterase inhibitor |
| DDB_G0268620 | 1,30 | 1,67 | pkbA | AKT/PKB protein kinase PkbA |
| DDB_G0272875 | 1,42 | 1,58 | cpnB-2 | copine B |
| DDB_G0287587 | 1,69 | 1,37 | smlA | unknown |
| DDB_G0284363 | 1,30 | 1,65 | cf60 | component of the counting factor (CF) complex |
| DDB_G0274597 | 0,87 | 2,95 | ctnA | component of the counting factor (CF) complex, countin |
| DDB_G0285425 | 1,14 | 1,65 | gpaD | G-protein subunit alpha 4 |
| DDB_G0283419 | 1,43 | 0,93 | gpaI | G-protein subunit alpha 9 |
| DDB_G0275045 | 1,14 | 1,70 | gpbB | GTP-binding protein subunit beta-like protein |
| DDB_G0273817 | 1,40 | 0,81 | ptpA1-2 | protein-tyrosine phosphatase 1 |
| DDB_G0272833 | 2,62 | 0,72 | csbA | contact site B protein |
| DDB_G0276883 | 1,09 | 0,68 | canA | calcineurin A |
| DDB_G0285845 | 0,95 | 0,62 | phdA | PH domain-containing protein; Developmental Gene |
| DDB_G0291253 | 1,06 | 0,50 | dia2 | unknown |
| DDB_G0275439 | 1,19 | 0,50 | cad2 | putative adhesion molecule |
| DDB_G0285793 | 1,18 | 0,36 | cadA | calcium-dependent cell adhesion molecule-1 |
| DDB_G0281605 | 0,67 | 1,22 | cfaD | counting factor associated protein |
| DDB_G0290261 | 0,77 | 1,66 | cfrC | cell surface glycoprotein GP138 C |
| DDB_G0269202 | 0,43 | 2,07 | gdcA | gp64 and disintegrin-like, cysteine-rich protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




