1. Introduction
Rice blast is one of the most dangerous diseases [
1,
2], and the causative agent of this disease is the parasitic fungus
Magnaporthe oryzae. Defeat of rice by pathogen in the early stages of development leads to a decrease in seed germination, sparseness of crops and death of seedlings. The greatest harm from infection occurs during the heading and flowering, which is associated with the formation of underdeveloped or feeble seeds, which significantly reduces the quality of the grain [
3]. Blast reduces rice production in the world, causing crop losses from 15-40% and reaching up to 80-100% in epiphytotic years [
4].
Global climate change affects the evolution of pathogen biotypes, which challenges breeders and forces them to increase rice productivity by creating and introducing new cultivars resistant to changing races or isolates of
Magnaporthe oryzae [
5]. Currently, more than 100 blast resistance genes have been discovered [
5,
6,
7,
8,
9,
10,
11,
12,
13,
14,
15]. Most of them with broad spectrum resistance to pathogen [
5,
6,
16,
17,
18,
19], there are some major genes with a wide spectrum of resistance, such as:
Pi-z5 (
Pi-2) [
20],
Pi-9 (
t) [
21],
Pigm [
22,
23,
24],
Pi-ta [
25],
Pi-1 and
Pi-33 [
7,
8,
9,
10,
11,
12,
13,
14,
26], which serve as indicators of blast resistance gene clusters, which is important when choosing parental pairs for hybridization. Most blast resistance genes are dominant, some of them are quantitative [
27], and most of them, such as
Pita,
Pi-1,
Pi25,
Pigm and
Pia, encode proteins that recognize the signal and inhibit the growth of the phytopathogen, maintaining plant immunity [
23,
28,
29,
30]. For example, the
Pita gene encodes a cytoplasmic membrane receptor protein that leads to resistance. [
31]. And the
Pigm gene encodes two proteins, PigmR and PigmS, which regulate resistance and yield of rice plants [
23]. Recently, the
Pigm-1 gene has been found, encoding the Pigm-1 protein, which is an allelic variant of PigmR [
32]. Moreover, many resistance genes are clustered on chromosomes 6, 9, 11, and 12 [
33,
34]. Disease resistance is controlled by one or two [
35,
36], three [
37] or more pairs of genes [
38]. Typically, rice has one or two dominant resistance genes that are effective against one race of the fungus [
39].
Determination of resistance genes in genetically diverse rice material is important to identify new sources of blast resistance. DNA markers are used to identify resistance genes [
26]. The most effective way to combat blast is to grow resistant rice cultivars. However, after a few years, the cultivar loses resistance due to the high variability of rice pathogens [
40,
41].
The selection of disease-resistant cultivars based on conventional breeding methods is complicated due to the difficulty of determining the presence of the desired allele of a particular gene. The use of molecular markers closely linked to genes that provide resistance to the pathogen greatly facilitates breeding work [
42]. Therefore, the main factor in rice breeding for blast resistance is the selection of resistance donors based on the identification of genes that control this trait in rice and the creation of new cultivars with a high level of resistance [
43]. Success in combat with rice blast has been achieved by introducing and pyramiding major blast resistance genes using MAS [
6,
7,
8,
9,
10,
11,
12,
13,
14]. Most resistance genes provide rice immunity only to certain races of the pathogen. It is known that such genes, when pyramided in one genotype, can provide stable resistance to the disease.
Pyramiding of multiple resistance genes in a single genotype is the most efficient strategy in breeding for resistance to a variable population of
Magnaporthe oryzae. Cultivars that combine combinations of 3-5 resistance genes are more resistant, as they show an increase and expansion of the spectrum of resistance to blast. Thus, the introduction of genes by combining (pyramiding) several blast resistance genes in one genotype is the most promising direction of rice selection [
7,
8,
9,
10,
11,
12,
13,
14,
43]. Screening cultivars for the presence of genes for resistance to
Magnaporthe oryzae, as well as the introduction and pyramid of resistance genes to the pathogen, contribute to a significant reduction in the time for rice improvement. Introduction of resistance genes into local cultivars is a preferred strategy in a rice breeding program for disease prevention. The aim of the work is to improve the resistance of rice to
Magnaporthe oryzae based on the introduction and pyramid of the major blast resistance genes using MAS.
3. Discussion
There are most of the research works focused on the creation of pyramidal rice lines carrying two or more blast resistance genes using MAS and hybridization methods. MAS analysis facilitates the identification of genes, their introduction and pyramiding into new lines [
44,
45,
46], that significantly speeds up rice breeding. The main goal of rice breeders in combating blast is to develop new cultivars and lines with broad-spectrum and durable resistance [
7,
8,
9,
10,
11,
12,
13,
14,
43]. Rice lines carrying only one major resistance gene cannot withstand a changing population of the fungus [
8]. In this case, the development of cultivars carrying not only one gene, but the pyramid of several genes of resistance to blast will help overcome this disease [
47,
48,
49].
However, there is some research on assessing the resistance of created lines to phytopathogens in a natural or artificial infectious background which identify the actual degree of resistance of newly created lines. Jiang, H. et al. [
8] developed several lines containing two blast resistance genes (
Pi 37/Pid 3), (
Pi 5/Pi54), (
Pi 54/Pid3) and (
Pigm/Pid 37), which showed high resistance to leaf and neck blast under natural infection conditions. Dubina et al. created pyramidal rice lines carrying five blast resistance genes (
Pi-1,
Pi-2,
Pi-33,
Pi-ta and
Pi-b) in a homozygous state, and obtained cultivars (Pentagen, Magnate, Pirouette, Argamac, Kapitan, Lenaris) [
48]. Kostylev, P.I. et al. [
49] obtained pyramided rice lines with 2–6 genes of resistance to blast (
Pi-1,
Pi-2,
Pi-33,
Pi-ta,
Pi-b and
Pi-40). Evaluation of the resistance of pyramided lines to fungal infection showed that lines containing 2 or more genes are more resistant compared to monogenic lines. Ji, Z.J. et al. [
50] created a pyramidal line with the
Pi1 and
Pi2 genes, which demonstrated resistance to blast. Positive results obtained by different researchers confirm the effectiveness of using these gene combinations (
Pi-1,
Pi-2,
Pi-33 and
Pi-ta) in improving rice protection against blast. The combination of several genes in one genotype has shown the greatest efficiency in the strategy of durable resistance of rice to blast [
7,
8,
9,
10,
11,
12,
13,
14,
43]. However, there is evidence that rice lines carrying both individual resistance genes [
44,
51] and lines with pyramidal genes can have a broad spectrum of resistance to
Magnaporthe oryzae [
45,
52]. This mainly depends on weather conditions and the degree of pathogenicity of the fungal strains.
It is well known that resistance of rice cultivars is enhanced by pyramiding of several resistance genes [
51]. Wang, X. et al. [
52] reported the creation of 74 lines by introgression and pyramiding of genes for resistance to biotic factors such as leafhopper (BPH), blast, fire blight (BLB) and aroma gene. These improved rice lines (ILs), containing combinations of 1 to 4 resistance genes to BPH, blast, BLB and aroma gene, were characterized by moderate or high resistance to multiple biotic stresses [
52].
It should be noted that the introduction and pyramiding of several genes in lines sometimes does not lead to an increase in the resistance of the lines and can even sometimes have a negative effect. The introduction of several genes into one line, they may be incompatible, suppressing each other's action and leading to sensitivity to the phytopathogen or to a decrease in the yield of the lines [
53].
The results of molecular screening of parental samples for the presence of genes showed that locally zoned varieties contain one of the four resistance genes studied – the Pi-2 gene, which is present in only three varieties (Bakanasski, Aisaule and Aru). Introgression of blast resistance genes into local zoned cultivars from differentiation varieties was successful. Seven pyramidal lines of rice are created. Three of them were found to show improved resistance to blast according to the results of the phytopathological test: 2 lines - F2 Bakanasski /7667 var. vulgaris and F2 Aisaule /7664 var. italica, containing 2 resistance genes (Pi-1 and Pi-ta) and 1-line F2 Aisaule /7689 var. zeravschanica, carrying 3 genes (Pi-1, Pi-33 and Pi-ta).
The resistance of pyramidal lines was assessed under infectious nursery conditions, which made it possible to identify the level of resistance of the created lines. The results of the phytopathological testing demonstrated that these three pyramidal lines with pyramidal 2 and 3 resistance genes showed moderately resistance to a mixture of fungal conidia of different isolates of M. oryzae, while 3 local varieties (Bakanasski, Aisaule and Aru) containing 1 resistance gene (Pi-2) – sensitive to phytopathogen. Thus, disease resistance of rice was enhanced by creating pyramidal lines. The test results showed that half of the pyramidal lines created were moderately resistant, and the other half were sensitive. This indicates that the level of resistance of the lines is revealed when encountering a phytopathogen in the conditions of an infectious nursery.
4. Materials and Methods
4.1. Plant Materials
The 35 genotypes of local and foreign selection from the rice collection of the Institute of Plant Biology and Biotechnology (IPBB, Almaty, Kazakhstan): 04636, 19-14, 57-14, 03-27, 04468, 04470, 04469, 95-06, 25-14, 04888, 212-05, 7653, 7662, 7663, 7664, 7666, 7667, 7668, 7679, 7683, 7684, 7686, 7689, 7690, 7695, 7698, 7701, 7702, 7712, 7824, Don 7712, Bakanasski, Fatima, Aisaule, Aru were used in the crossing as parents. 54 hybrid lines of the F2 generation (Bakanasski/7668 var. subuzbekistanica Kond., Bakanasski / 04470 var. vulgaris Koern., Bakanasski/7653 var. subuzbekistanica Kond., Bakanasski/7653 var. subjanthoseros Brsch., Bakanasski/7684 var. subvulgaris Brsch., Bakanasski/7684 var. vulgaris Koern., 7824/Aisaule var. italica Alef., 7824/Aisaule var. subuzbekistanica Kond., Aru/04468 var. subvulgaris Brsch., 7698/Bakanasski var. subvulgaris Brsch., 7698/Bakanasski var. italica Alef., Aru/7701 var. vulgaris Koern., Bakanasski/0327 var. vulgaris Koern., Aisaule/7679 var. subvulgaris Brsch., 7698/Aisaule var. italica Alef., 7698/Aisaule var. suberythroseros Kan., Aru/7702 var. erythroceros Koern., Aisaule/7689 var. zeravschanica Brsch., Aru/0327 var. vulgaris Koern., Aru/0327 var. subpyrocarpa Alef., Bakanasski/7667 var. subjanthoseros Brsch., Bakanasski/7667 var. vulgaris Koern., Bakanasski/7701 var. vulgaris Koern., Aisaule/7664 var. italica Alef., Aisaule/7664 var. subvulgaris Brsch., Aisaule/04470 var. subvulgaris Brsch., Aisaule/04470 var. italica Alef., Aisaule/Don 7712 var. sundensis Koern., Aisaule/Don 7712 var. italica Alef., Fatima/7695 var. italica Alef., Fatima/7695 var. breviaristata Vav., Bakanasski/7683 var. subjanthoseros Brsch., 7698/Aru var. erythroceros Koern., 7698/Aru var. suberythroseros Kan., Bakanasski/Don 7712, Bakanasski/Don 7712 var. Desvauxii Koern., Aru/212-05 var. vulgaris Koern., Aru/7668 var. vulgaris Koern., Aisaule/04468 var. italica Alef., Aru/04636 var. vulgaris Koern., Bakanasski/7690 var. subjanthoseros Brsch., Bakanasski/7690 var. vulgaris Koern., Bakanasski/04468 var. subvulgaris Brsch., Bakanasski/04468 var. vulgaris Koern., Aru/7666 var. suberythroseros Kan., Fatima/7653 var. persica Kan., Fatima/7689 var. italica Alef., Aru/Don 7712 var. erythroceros Koern., Aru/Don 7712 var. Desvauxii Koern., Aru/Don 7712 var. vulgaris Koern., Aru/Don 7712 var. Desvauxii Koern. semi-awned, ♀ Aru/7683 var. janthoceros Koern., Aru/7683 var. vulgaris Koern., Aisaule/7663 var. italica Alef.) obtained by crossing were used for analysis.
4.2. Hybridization Method
Hybridization was carried out using pneumocastration (three-channel pneumocastration) and the "TWEL" plant pollination method [
54]. 31 genotypes were used as donors, the genome of which contains genes of resistance to rice blast (
Pi-1,
Pi-2,
Pi-33 and
Pi-ta), and recipients were highly productive zoned local cultivars, such as Bakanasski, Fatima, Aisaule and Aru.
4.3. DNA Extraction and Multiplex PCR Analysis
Genomic DNA was extracted from two-week-old seedlings using the cetyltrimethylammonium bromide (CTAB) method [
55]. CTAB reagent was obtained from Pan-Reac AppliChem GmbH (Darmstadt, Germany). All PCR analysis were performed using a T100 thermal cycler (BioRad, Germany). Multiplex PCR analysis was performed by adding several primers in PCR mix to determine the presence of two or more genes in a genotype. Each microtube contained 15 µl of PCR reaction mixture consisting of 40–50 ng of template DNA, 1.2 µl of each forward and reverse primer, 1.2 µl of
Taq buffer, 0.05 mmol/L dNTPs and 1.0 U of
Taq polymerase. The following PCR conditions were used: 94°C for 5 min; 35 cycles of 35 sec at 94°C, 45 sec at 60°C, and 30 sec at 72°C; and an extension of 5 min at 72°C. SSR (simple sequence repeat) markers were used to identify genes. The list of SSR markers (Syntol, Russia) for
Pi complex genes is given in
Table 3. The annealing temperature is the same for all primers, 60º C.
The following genotypes were used as differentiator cultivars that are carriers of resistance genes: C101-Lac (+) for Pi-1, Pi-33 genes, C101-A51 (+) for Pi-2 gene and IR 36 (+) for Pi-ta gene. The cultivar Flagman (-) was used as a negative control for all studied genes.
4.5. Gel Electrophoresis
The amplification products were separated in 8% polyacrylamide gel (PAGE) in 1.0× TBE buffer by vertical electrophoresis chamber (Helicon, Russia). Electrophoretic gels were examined using the GelDoc XR (Bio-Rad, USA). The GelAnalyzer program was used to analyze gels.
4.6. Phytopathological Screening
Testing of 54 hybrid rice lines for resistance to Magnaporthe oryzae populations was carried out in 2023 in the artificial infectious nursery at the FSBSI “Federal Scientific Rice Centre” (Russia, Krasnodar). The most resistant cv. Avangard served as a positive control, and the highly susceptible cv. Pobeda 65 was used as a negative control.
Rice plants were infected with the fungal culture at the booting stage by spraying with a suspension of a mixture of conidia of 15 isolates of M. oryzae.
The degree of plant damage (in percentage) was considered on the 14th day after inoculation, according to the express method for assessing rice cultivars resistance to blast. The assessment was carried out considering the type of reaction (in points) on a ten-point scale (0-9) of the Standard Evaluation System for Rice [
56]. The degree of resistance was indicated using the following designations: resistant lines - R, moderately resistant - MR and susceptible - S.
The intensity of disease development (IDD, %) was calculated using the formula:
where - ∑ (a*b) – sum products of the number of infected plants multiplied by the corresponding damage score, n - the number of recorded plants, pieces.
4.7. Analysis of Structural Elements of Yield of Hybrid Rice Lines
Evaluation of the productivity of hybrid lines in comparison with the standard cv. Bakanasski was carried out according to the biometric parameters of the elements of yield structure (“bushiness, pc”, “plant height, cm”, “panicle length, cm”, “number of grains from the main panicle, pc”, “weight of seeds from the main panicle, g”, “weight of 1000 seeds, g” and etc.) [
57]
4.8. Statistical Analyses
To characterize the productivity of hybrid lines in comparison with the cv. Bakanasski, which is the standard for the Almaty region, statistical indicators such as the mean value, standard deviation for the main traits characterizing rice productivity, “weight of 1000 seeds, g” were used. Analysis of variance (one-way ANOVA) was performed to compare the mean values of the data groups. Data processing was carried out in Microsoft Excel 2021 using a data analysis package.
Figure 1.
Electropherogram of PCR products of samples for the presence of the Pi-1 and Pi-2 genes. (a): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-1 gene at the Rm144 and RM224 loci), 2 – Flagman (control "-" for the Pi-1 gene at the Rm144 and RM224 loci), 3 – C101-A51 (control "+" for the Pi-2 gene at loci RM-527 and SSR-140), 4 – Flagman (control "-" for the Pi-2 gene at loci RM-527 and SSR-140), 5 – 04636, 6 – 03-27, 7 – 04468, 8 – 04470, 9 – 04469, 10 – 25-14, 11 – 04888, 12 – 212-05, 13 – 7653, 14 – 7662, 15 – 7663, 16 – 7664, 17 – 7666, 18 – 7667, 19 – 7668; (b): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-1 gene at the Rm144 and RM224 loci), 2 – Flagman (control "-" for the Pi-1 gene at the Rm144 and RM224 loci), 3 – C101-A51 (control "+" for the Pi-2 gene at loci RM-527 and SSR-140), 4 – Flagman (control "-" for the Pi-2 gene at loci RM-527 and SSR-140), 5 – 7679, 6 – 7683, 7 – 7684, 8 – 7689, 9 – 7690, 10 – 7695, 11 – 7701, 12 – 7702, 13 – 7712, 14 – 7824, 15 – Don 7712, 16 – Bakanasski, 17 – Fatima, 18 – Aisaule, 19 – Aru.
Figure 1.
Electropherogram of PCR products of samples for the presence of the Pi-1 and Pi-2 genes. (a): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-1 gene at the Rm144 and RM224 loci), 2 – Flagman (control "-" for the Pi-1 gene at the Rm144 and RM224 loci), 3 – C101-A51 (control "+" for the Pi-2 gene at loci RM-527 and SSR-140), 4 – Flagman (control "-" for the Pi-2 gene at loci RM-527 and SSR-140), 5 – 04636, 6 – 03-27, 7 – 04468, 8 – 04470, 9 – 04469, 10 – 25-14, 11 – 04888, 12 – 212-05, 13 – 7653, 14 – 7662, 15 – 7663, 16 – 7664, 17 – 7666, 18 – 7667, 19 – 7668; (b): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-1 gene at the Rm144 and RM224 loci), 2 – Flagman (control "-" for the Pi-1 gene at the Rm144 and RM224 loci), 3 – C101-A51 (control "+" for the Pi-2 gene at loci RM-527 and SSR-140), 4 – Flagman (control "-" for the Pi-2 gene at loci RM-527 and SSR-140), 5 – 7679, 6 – 7683, 7 – 7684, 8 – 7689, 9 – 7690, 10 – 7695, 11 – 7701, 12 – 7702, 13 – 7712, 14 – 7824, 15 – Don 7712, 16 – Bakanasski, 17 – Fatima, 18 – Aisaule, 19 – Aru.
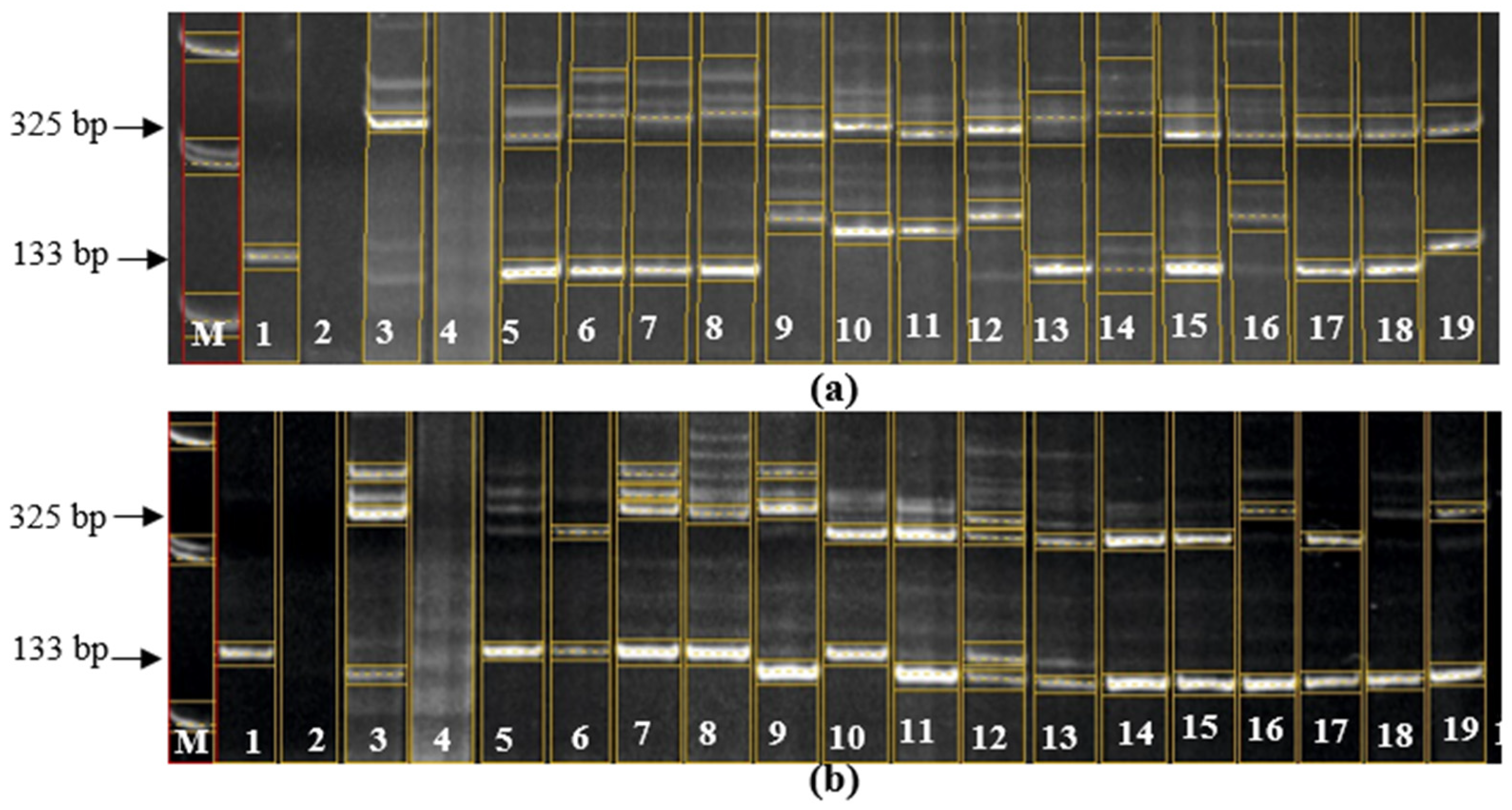
Figure 2.
Electropherogram of PCR products of samples for the presence of Pi-33 and Pi-ta genes. (a): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-33 gene at the Rm310 and RM72 loci); 2 – Flagman (control "-" for the Pi-33 gene at the Rm310 and RM72 loci); 3 – IR36 (control "+" for the Pi-ta gene at the Pita loci); 4 – Flagman (control "-" for the Pi-ta gene at the Pita loci); 5 – 04636; 6 – 19-14; 7 – 57-14; 8 – 03-27; 9 – 04468; 10 – 04470; 11 – 04469; 12 – 25-14; 13 – 04888; 14 – 212-05; 15 – 7653; 16 – 7662; 17 – 7663; 18 – 7664; 19 – 7666; (b): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-33 gene at the Rm310 and RM72 loci), 2 – Flagman (control "-" for the Pi-33 gene at the Rm310 and RM72 loci), 3 – IR 36 (control "+" for the Pi-ta gene at the Pita loci), 4 – Flagman (control "-" for the Pi-ta gene at the Pita loci), 5 – 7667, 6 – 7668, 7 – 7679, 8 – 7683, 9 – 7684, 10 – 7686, 11 – 7689, 12 – 7690, 13 – 7695, 14 – 7698, 15 – 7701, 16 – 7702, 17 – 7712, 18 – 7824; (c): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-33 gene at the Rm310 and RM72 loci), 2 – Flagman (control "-" for the Pi-33 gene at the Rm310 and RM72 loci), 3 – IR36 (control "+" for the Pi-ta gene at the Pita loci), 4 – Flagman (control "-" for the Pi-ta gene at the Pita loci), 5 – Don 7712, 6 – Bakanasski, 7 – Fatima, 8 - Aisaule, 9 – Aru.
Figure 2.
Electropherogram of PCR products of samples for the presence of Pi-33 and Pi-ta genes. (a): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-33 gene at the Rm310 and RM72 loci); 2 – Flagman (control "-" for the Pi-33 gene at the Rm310 and RM72 loci); 3 – IR36 (control "+" for the Pi-ta gene at the Pita loci); 4 – Flagman (control "-" for the Pi-ta gene at the Pita loci); 5 – 04636; 6 – 19-14; 7 – 57-14; 8 – 03-27; 9 – 04468; 10 – 04470; 11 – 04469; 12 – 25-14; 13 – 04888; 14 – 212-05; 15 – 7653; 16 – 7662; 17 – 7663; 18 – 7664; 19 – 7666; (b): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-33 gene at the Rm310 and RM72 loci), 2 – Flagman (control "-" for the Pi-33 gene at the Rm310 and RM72 loci), 3 – IR 36 (control "+" for the Pi-ta gene at the Pita loci), 4 – Flagman (control "-" for the Pi-ta gene at the Pita loci), 5 – 7667, 6 – 7668, 7 – 7679, 8 – 7683, 9 – 7684, 10 – 7686, 11 – 7689, 12 – 7690, 13 – 7695, 14 – 7698, 15 – 7701, 16 – 7702, 17 – 7712, 18 – 7824; (c): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-33 gene at the Rm310 and RM72 loci), 2 – Flagman (control "-" for the Pi-33 gene at the Rm310 and RM72 loci), 3 – IR36 (control "+" for the Pi-ta gene at the Pita loci), 4 – Flagman (control "-" for the Pi-ta gene at the Pita loci), 5 – Don 7712, 6 – Bakanasski, 7 – Fatima, 8 - Aisaule, 9 – Aru.
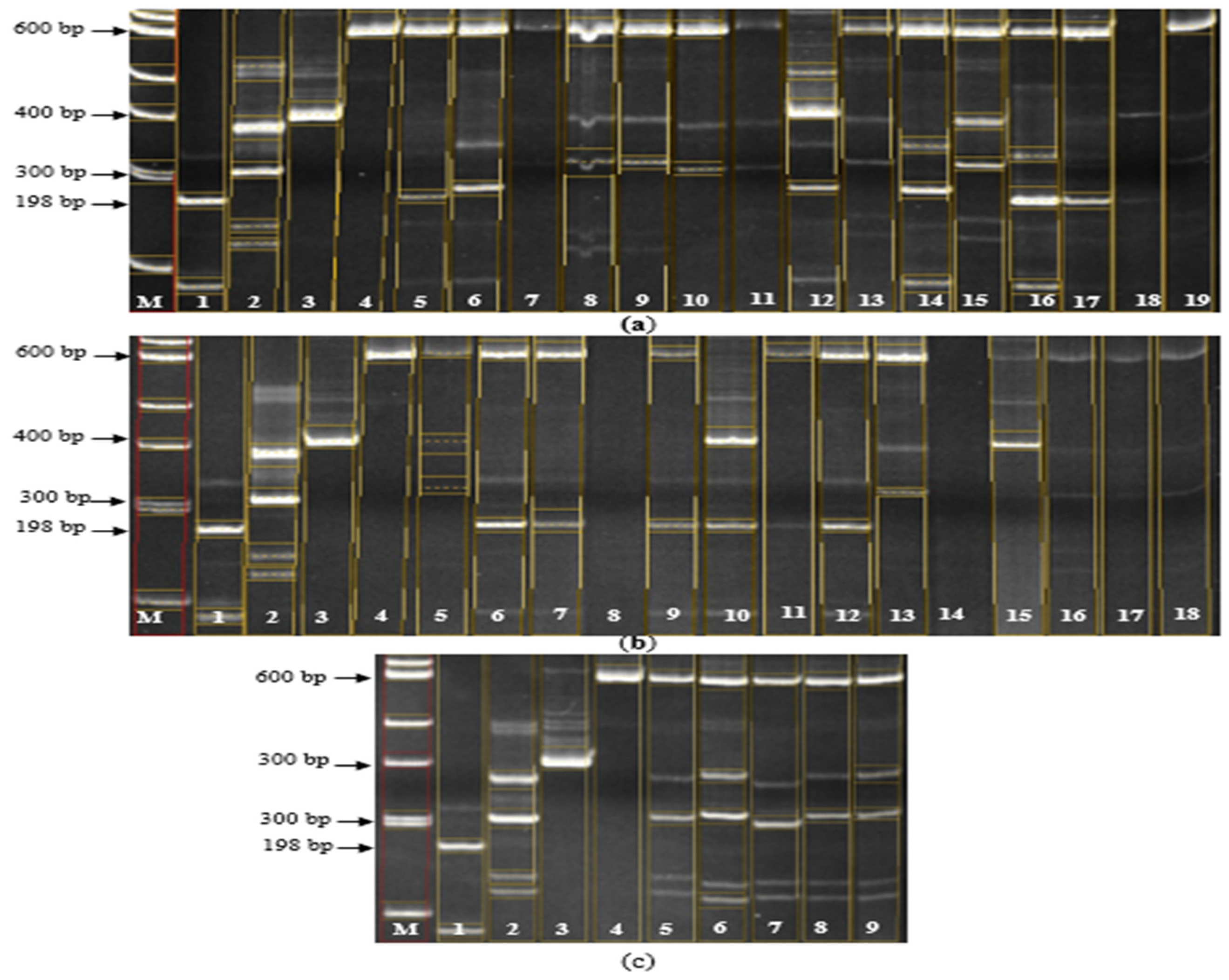
Figure 3.
Electropherogram of PCR products of hybrids for the presence of Pi-1 and Pi-2 genes. (a): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-1 gene at the Rm144 and RM224 loci), 2 – Flagman (control "-" for the Pi-1 gene at the Rm144 and RM224 loci), 3 – C101-A51(control "+" for the Pi-2 gene at loci RM-527 and SSR-140), 4 – Flagman (control "-" for the Pi-2 gene at loci RM-527 and SSR-140), 5 – Bakanasski/7668 var. subuzbekistanica, 6 – Bakanasski/04470 var. vulgaris, 7 – Bakanasski/7653 var. subuzbekistanica, 8 – Bakanasski/7653 var. subjanthoseros, 9 – Bakanasski/7684 var. subvulgaris, 10 – Bakanasski/7684 var. vulgaris, 11 – 7824/Aisaule var. italica, 12 – 7824/Aisaule var. subuzbekistanica, 13 – Aru/04468 var. subvulgaris, 14 – 7698/Bakanasski var. subvulgaris, 15 – 7698/Bakanasski var. italica, 16 – Aru/7701 var. vulgaris, 17 – Bakanasski/0327 var. vulgaris, 18 – Aisaule/7679 var. subvulgaris, 19 –7698/Aisaule var. italica (b): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-1 gene at the Rm144 and RM224 loci), 2 – Flagman (control "-" for the Pi-1 gene at the Rm144 and RM224 loci), 3 – C101-A51(control "+" for the Pi-2 gene at loci RM-527 and SSR-140), 4 – Flagman (control "-" for the Pi-2 gene at loci RM-527 and SSR-140), 5 – 7698/Aisaule var. suberythroseros, 6 – Aru/7702 var. erythroceros, 7 – Aisaule/7689 var. zeravschanica, 8 – Aru/0327 var. vulgaris, 9 – Aru/0327 var. subpyrocarpa, 10 – Bakanasski/7667 var. subjanthoseros, 11 – Bakanasski/7667 var. vulgaris, 12 – Bakanasski/7701 var. vulgaris, 13 – Aisaule/7664 var. italica, 14 – Aisaule/7664 var. subvulgaris, 15 – Aisaule/04470 var. subvulgaris, 16 – Aisaule/04470 var. italica, 17 – Aisaule/Don 7712 var. sundensis, 18 – Aisaule/Don 7712 var. italica, 19 – Fatima/7695 var. italica (c): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-1 gene at the Rm144 and RM224 loci), 2 – Flagman (control "-" for the Pi-1 gene at the Rm144 and RM224 loci), 3 – C101-A51 (control "+" for the Pi-2 gene at loci RM-527 and SSR-140), 4 – Flagman (control "-" for the Pi-2 gene at loci RM-527 and SSR-140), 5 – Fatima/7695 var. breviaristata, 6 – Bakanasski/7683 var. subjanthoseros, 7 – 7698/Aru var. erythroceros, 8 – 7698/Aru var. suberythroseros, 9 – Bakanasski/Don 7712, 10 – Bakanasski/Don 7712 var. Desvauxii, 11 – Aru/212-05 var. vulgaris, 12 – Aru/7668 var. vulgaris, 13 – Aisaule/04468 var. italica, 14 – Aru/04636 var. vulgaris, 15 – Bakanasski/7690 var. subjanthoseros, 16 – Bakanasski/7690 var. vulgaris, 17 – Bakanasski/04468 var. subvulgaris, 18 – Bakanasski/04468 var. vulgaris.
Figure 3.
Electropherogram of PCR products of hybrids for the presence of Pi-1 and Pi-2 genes. (a): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-1 gene at the Rm144 and RM224 loci), 2 – Flagman (control "-" for the Pi-1 gene at the Rm144 and RM224 loci), 3 – C101-A51(control "+" for the Pi-2 gene at loci RM-527 and SSR-140), 4 – Flagman (control "-" for the Pi-2 gene at loci RM-527 and SSR-140), 5 – Bakanasski/7668 var. subuzbekistanica, 6 – Bakanasski/04470 var. vulgaris, 7 – Bakanasski/7653 var. subuzbekistanica, 8 – Bakanasski/7653 var. subjanthoseros, 9 – Bakanasski/7684 var. subvulgaris, 10 – Bakanasski/7684 var. vulgaris, 11 – 7824/Aisaule var. italica, 12 – 7824/Aisaule var. subuzbekistanica, 13 – Aru/04468 var. subvulgaris, 14 – 7698/Bakanasski var. subvulgaris, 15 – 7698/Bakanasski var. italica, 16 – Aru/7701 var. vulgaris, 17 – Bakanasski/0327 var. vulgaris, 18 – Aisaule/7679 var. subvulgaris, 19 –7698/Aisaule var. italica (b): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-1 gene at the Rm144 and RM224 loci), 2 – Flagman (control "-" for the Pi-1 gene at the Rm144 and RM224 loci), 3 – C101-A51(control "+" for the Pi-2 gene at loci RM-527 and SSR-140), 4 – Flagman (control "-" for the Pi-2 gene at loci RM-527 and SSR-140), 5 – 7698/Aisaule var. suberythroseros, 6 – Aru/7702 var. erythroceros, 7 – Aisaule/7689 var. zeravschanica, 8 – Aru/0327 var. vulgaris, 9 – Aru/0327 var. subpyrocarpa, 10 – Bakanasski/7667 var. subjanthoseros, 11 – Bakanasski/7667 var. vulgaris, 12 – Bakanasski/7701 var. vulgaris, 13 – Aisaule/7664 var. italica, 14 – Aisaule/7664 var. subvulgaris, 15 – Aisaule/04470 var. subvulgaris, 16 – Aisaule/04470 var. italica, 17 – Aisaule/Don 7712 var. sundensis, 18 – Aisaule/Don 7712 var. italica, 19 – Fatima/7695 var. italica (c): M – Molecular marker 100 bp; 1 – C101-Lac (control "+" for the Pi-1 gene at the Rm144 and RM224 loci), 2 – Flagman (control "-" for the Pi-1 gene at the Rm144 and RM224 loci), 3 – C101-A51 (control "+" for the Pi-2 gene at loci RM-527 and SSR-140), 4 – Flagman (control "-" for the Pi-2 gene at loci RM-527 and SSR-140), 5 – Fatima/7695 var. breviaristata, 6 – Bakanasski/7683 var. subjanthoseros, 7 – 7698/Aru var. erythroceros, 8 – 7698/Aru var. suberythroseros, 9 – Bakanasski/Don 7712, 10 – Bakanasski/Don 7712 var. Desvauxii, 11 – Aru/212-05 var. vulgaris, 12 – Aru/7668 var. vulgaris, 13 – Aisaule/04468 var. italica, 14 – Aru/04636 var. vulgaris, 15 – Bakanasski/7690 var. subjanthoseros, 16 – Bakanasski/7690 var. vulgaris, 17 – Bakanasski/04468 var. subvulgaris, 18 – Bakanasski/04468 var. vulgaris.
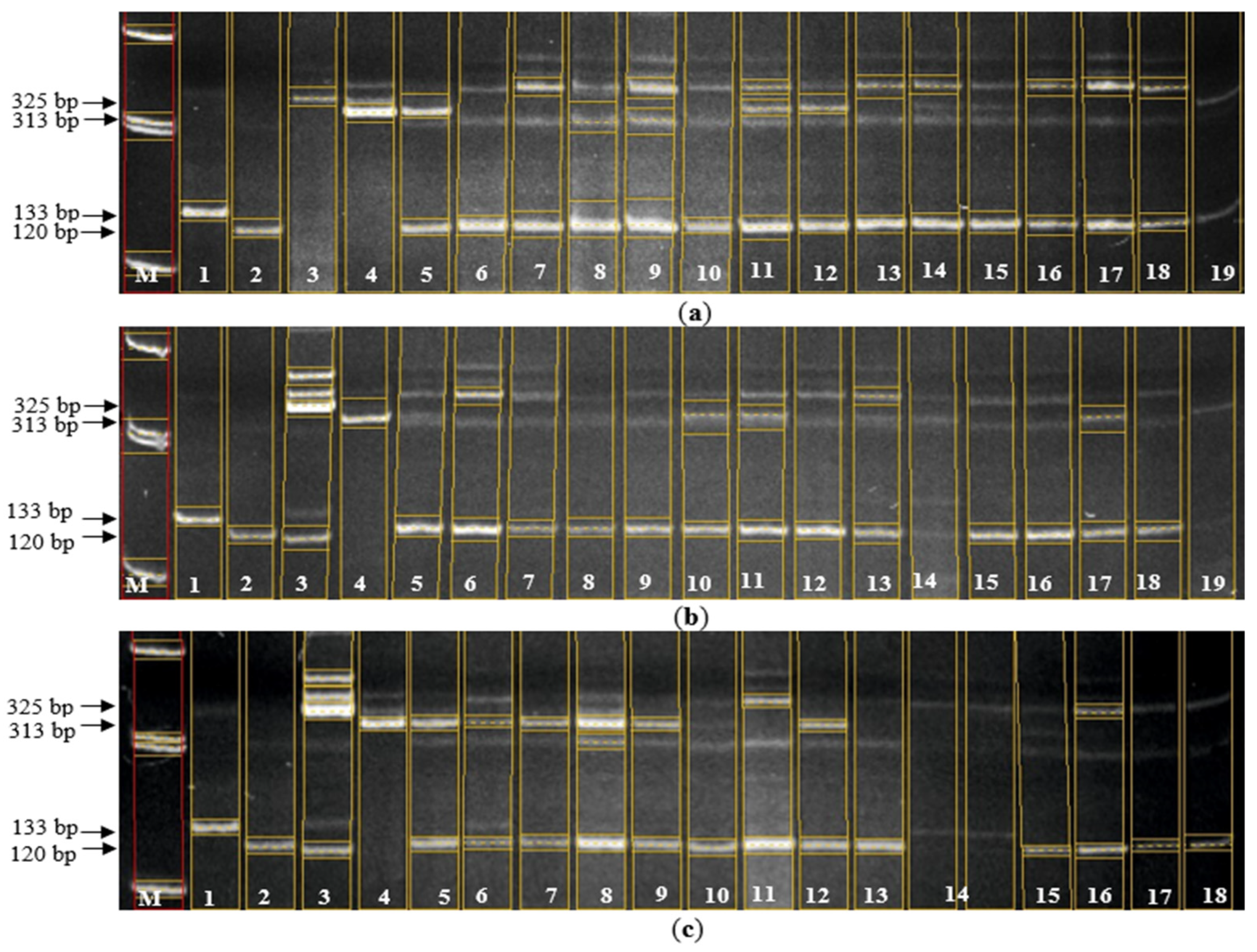
Figure 4.
Electropherogram of PCR products of hybrids for the presence of Pi-33 and Pi-ta genes. (а): M – Molecular marker 100 bp; 1 - C101-Lac (control "+" for the Pi-33 gene at the Rm310 and RM72 loci), 2 – Flagman (control "-" for the Pi-33 gene at the Rm310 and RM72 loci), 3 – IR36 (control "+" for the Pi-ta gene at the Pita loci), 4 – Flagman (control "-" for the Pi-ta gene at the Pita loci), 5 – Bakanasski/7668 var. subuzbekistanica, 6 – Bakanasski/04470 var. vulgaris, 7 – Bakanasski/7653 var. subuzbekistanica, 8 – Bakanasski/7653 var. subjanthoseros, 9 – Bakanasski/7684 var. subvulgaris, 10 – Bakanasski/7684 var. vulgaris, 11 – 7824/Aisaule var. italica, 12 – 7824/Aisaule var. subuzbekistanica, 13 – 7698/Bakanasski var. subvulgaris, 14 – 7698/Bakanasski var. italica, 15 – Aru/7701 var. vulgaris, 16 – Bakanasski/0327 var. vulgaris, 17 – Aisaule/7679 var. subvulgaris, 18 – 7698/Aisaule var. italica (в): M – Molecular marker 100 bp; 1 - C101-Lac (control "+" for the Pi-33 gene at the Rm310 and RM72 loci), 2 – Flagman (control "-" for the Pi-33 gene at the Rm310 and RM72 loci), 3 – IR36 (control "+" for the Pi-ta gene at the Pita loci), 4 – Flagman (control "-" for the Pi-ta gene at the Pita loci), 5 – 7698/Aisaule var. suberythroseros, 6 – Aru/7702 var. erythroceros, 7 – Aisaule/7689 var. zeravschanica, 8 – Aru/0327 var. vulgaris, 9 – Bakanasski/7667 var. vulgaris, 10 – Bakanasski/7701 var. vulgaris, 11 – Aisaule/7664 var. italica, 12 – Aisaule/7664 var. subvulgaris, 13 – Aisaule/04470 var. subvulgaris, 14 – Aisaule/04470 var. italica, 15 – Aisaule/Don 7712 var. sundensis, 16 – Aisaule/Don 7712 var. italica, 17 – Fatima/7695 var. breviaristata, 18 – Bakanasski/7683 var. subjanthoseros (с): M – Molecular marker 100 bp; 1 - C101-Lac (control "+" for the Pi-33 gene at the Rm310 and RM72 loci), 2 – Flagman (control "-" for the Pi-33 gene at the Rm310 and RM72 loci), 3 – IR36 (control "+" for the Pi-ta gene at the Pita loci), 4 – Flagman (control "-" for the Pi-ta gene at the Pita loci), 5 – Aru/212-05 var. vulgaris, 6 – Aru/7668 var. vulgaris, 7 – Aisaule/04468 var. italica, 8 – Aru/04636 var. vulgaris, 9 – Bakanasski/7690 var. subjanthoseros, 10 – Bakanasski/7690 var. vulgaris, 11 – Bakanasski/04468 var. subvulgaris, 12 – Bakanasski/04468 var. vulgaris, 13 – Aru/7666 var. suberythroseros, 14 – Fatima/7653 var. persica, 15 – Aru/Don 7712 var. Desvauxii semi-awned, 16 – Aru/7683 var. janthoceros, 17 – Aru/7683 var. vulgaris, 18 – Aisaule/7663 var. italica, 19 – Aru/0327 var. subpyrocarpa (d): M – Molecular marker 100 bp; 1 - C101-Lac (control "+" for the Pi-33 gene at the Rm310 and RM72 loci), 2 – Flagman (control "-" for the Pi-33 gene at the Rm310 and RM72 loci), 3 – IR36 (control "+" for the Pi-ta gene at the Pita loci), 4 – Flagman (control "-" for the Pi-ta gene at the Pita loci), 5 – Bakanasski/Don 7712, 6 – Fatima/7689 var. italica, 7 – Aru/04468 var. subvulgaris, 8 – Bakanasski/7667 var. subjanthoseros, 9 – Bakanasski/Don 7712 var. Desvauxii, 10 – Aru/Don 7712 var. vulgaris, 11 – 7698/Aru var. erythroceros, 12 – 7698/Aru var. suberythroseros, 13 – Aru/Don 7712 var. erythroceros, 14 – Aru/Don 7712 var. Desvauxii.
Figure 4.
Electropherogram of PCR products of hybrids for the presence of Pi-33 and Pi-ta genes. (а): M – Molecular marker 100 bp; 1 - C101-Lac (control "+" for the Pi-33 gene at the Rm310 and RM72 loci), 2 – Flagman (control "-" for the Pi-33 gene at the Rm310 and RM72 loci), 3 – IR36 (control "+" for the Pi-ta gene at the Pita loci), 4 – Flagman (control "-" for the Pi-ta gene at the Pita loci), 5 – Bakanasski/7668 var. subuzbekistanica, 6 – Bakanasski/04470 var. vulgaris, 7 – Bakanasski/7653 var. subuzbekistanica, 8 – Bakanasski/7653 var. subjanthoseros, 9 – Bakanasski/7684 var. subvulgaris, 10 – Bakanasski/7684 var. vulgaris, 11 – 7824/Aisaule var. italica, 12 – 7824/Aisaule var. subuzbekistanica, 13 – 7698/Bakanasski var. subvulgaris, 14 – 7698/Bakanasski var. italica, 15 – Aru/7701 var. vulgaris, 16 – Bakanasski/0327 var. vulgaris, 17 – Aisaule/7679 var. subvulgaris, 18 – 7698/Aisaule var. italica (в): M – Molecular marker 100 bp; 1 - C101-Lac (control "+" for the Pi-33 gene at the Rm310 and RM72 loci), 2 – Flagman (control "-" for the Pi-33 gene at the Rm310 and RM72 loci), 3 – IR36 (control "+" for the Pi-ta gene at the Pita loci), 4 – Flagman (control "-" for the Pi-ta gene at the Pita loci), 5 – 7698/Aisaule var. suberythroseros, 6 – Aru/7702 var. erythroceros, 7 – Aisaule/7689 var. zeravschanica, 8 – Aru/0327 var. vulgaris, 9 – Bakanasski/7667 var. vulgaris, 10 – Bakanasski/7701 var. vulgaris, 11 – Aisaule/7664 var. italica, 12 – Aisaule/7664 var. subvulgaris, 13 – Aisaule/04470 var. subvulgaris, 14 – Aisaule/04470 var. italica, 15 – Aisaule/Don 7712 var. sundensis, 16 – Aisaule/Don 7712 var. italica, 17 – Fatima/7695 var. breviaristata, 18 – Bakanasski/7683 var. subjanthoseros (с): M – Molecular marker 100 bp; 1 - C101-Lac (control "+" for the Pi-33 gene at the Rm310 and RM72 loci), 2 – Flagman (control "-" for the Pi-33 gene at the Rm310 and RM72 loci), 3 – IR36 (control "+" for the Pi-ta gene at the Pita loci), 4 – Flagman (control "-" for the Pi-ta gene at the Pita loci), 5 – Aru/212-05 var. vulgaris, 6 – Aru/7668 var. vulgaris, 7 – Aisaule/04468 var. italica, 8 – Aru/04636 var. vulgaris, 9 – Bakanasski/7690 var. subjanthoseros, 10 – Bakanasski/7690 var. vulgaris, 11 – Bakanasski/04468 var. subvulgaris, 12 – Bakanasski/04468 var. vulgaris, 13 – Aru/7666 var. suberythroseros, 14 – Fatima/7653 var. persica, 15 – Aru/Don 7712 var. Desvauxii semi-awned, 16 – Aru/7683 var. janthoceros, 17 – Aru/7683 var. vulgaris, 18 – Aisaule/7663 var. italica, 19 – Aru/0327 var. subpyrocarpa (d): M – Molecular marker 100 bp; 1 - C101-Lac (control "+" for the Pi-33 gene at the Rm310 and RM72 loci), 2 – Flagman (control "-" for the Pi-33 gene at the Rm310 and RM72 loci), 3 – IR36 (control "+" for the Pi-ta gene at the Pita loci), 4 – Flagman (control "-" for the Pi-ta gene at the Pita loci), 5 – Bakanasski/Don 7712, 6 – Fatima/7689 var. italica, 7 – Aru/04468 var. subvulgaris, 8 – Bakanasski/7667 var. subjanthoseros, 9 – Bakanasski/Don 7712 var. Desvauxii, 10 – Aru/Don 7712 var. vulgaris, 11 – 7698/Aru var. erythroceros, 12 – 7698/Aru var. suberythroseros, 13 – Aru/Don 7712 var. erythroceros, 14 – Aru/Don 7712 var. Desvauxii.
Figure 5.
Degree of leaf resistance to blast: (a) resistant, (b) susceptible, (c) moderately resistant.
Figure 5.
Degree of leaf resistance to blast: (a) resistant, (b) susceptible, (c) moderately resistant.
Table 1.
Phytopathological assessment of blast resistance of obtained pyramidal lines and local rice cultivars.
Table 1.
Phytopathological assessment of blast resistance of obtained pyramidal lines and local rice cultivars.
| Hybrid lines |
Blast resistance |
| Intensity of disease development, % |
Degree of resistance to disease |
Resistance genes |
Line’s reaction |
| Bakanasski |
55.6 |
susceptible |
Pi-2 |
S |
| Aisaule |
55.6 |
susceptible |
Pi-2 |
S |
| Aru |
66.7 |
susceptible |
Pi-2 |
S |
| Fatima |
44.4 |
moderately resistant |
- |
MR |
7698/Aisaule
var. suberythroseros
|
- |
- |
- |
This line didn't grow up |
Aru/7702
var. erythroceros
|
61.1 |
susceptible |
Pi-1, Pi-ta
|
S |
Aisaule/7689
var. zeravschanica
|
36.7 |
moderately resistant |
Pi-1, Pi-33, Pi-ta
|
MR
|
Bakanasski/7667
var. subjanthoseros
|
61.1 |
susceptible |
Pi-1, Pi-ta
|
S |
Bakanasski/7667
var. vulgaris
|
38.9 |
moderately resistant |
Pi-1, Pi-ta
|
MR |
Bakanasski/7701
var. vulgaris
|
72.2 |
susceptible |
Pi-1, Pi-ta
|
S |
Aisaule/7664
var. italica |
44.4 |
moderately resistant |
Pi-1, Pi-ta
|
MR |
| MR – moderately resistant, S – susceptible. |
Table 2.
Analyses of the productivity of pyramided rice lines by One-way ANOVA.
Table 2.
Analyses of the productivity of pyramided rice lines by One-way ANOVA.
| |
SS |
df |
MS |
F |
P-value |
F crit |
| Between |
367.12 |
6 |
61.19* |
27.07 |
0.00 |
3.25 |
| Within |
97.18 |
43 |
2.26 |
|
|
|
Table 3.
List of tested SSR primers for Pi genes of the blast resistance complex.
Table 3.
List of tested SSR primers for Pi genes of the blast resistance complex.
| Gene |
Localization on chromosome |
Name of a marker
|
Forward primer (5′-3′)
Reverse sequence (5′-3′) |
Pi-1
|
11 |
Rm 224 |
F - аtсgаtсgаtсttсасgаgg
R - tgctataaaaggcattcggg |
| Rm144 |
F - tgccctggcgcaaatttgatcc
R - gctagaggagatcagatggtagtgcatg |
| Pi-2 |
6 |
Rm 527 |
F - ggctcgatctagaaaatccg
R - ttgcacaggttgcgatagag |
| SSR 140 |
F - aaggtgtgaaacaagctagcaa
R - ttctaggggaggggtgtgaa |
| Pi-33 |
8 |
Rm 310 |
F - ссggсgаtаааасааtgаg
R - gсаtсggtссtаасtааggg |
| Rm 72 |
F - ссggсgаtаааасааtgаg
R - gсаtсggtссtаасtааggg |
| Pi-ta |
12 |
Pita |
F1 - gссgtggсttсtаtсtttасatg
R1 - аtссааgtgttаgggссаасаttс |
F2 - ttgасасtсtсаааggасtgggаt
R2 - tсааgtсаggttgааgаtgсаtсgа |









