Submitted:
06 May 2024
Posted:
08 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
Distribution of Corbicula in Argentina: Review and Sampling
Morphological Assignment of Lineages
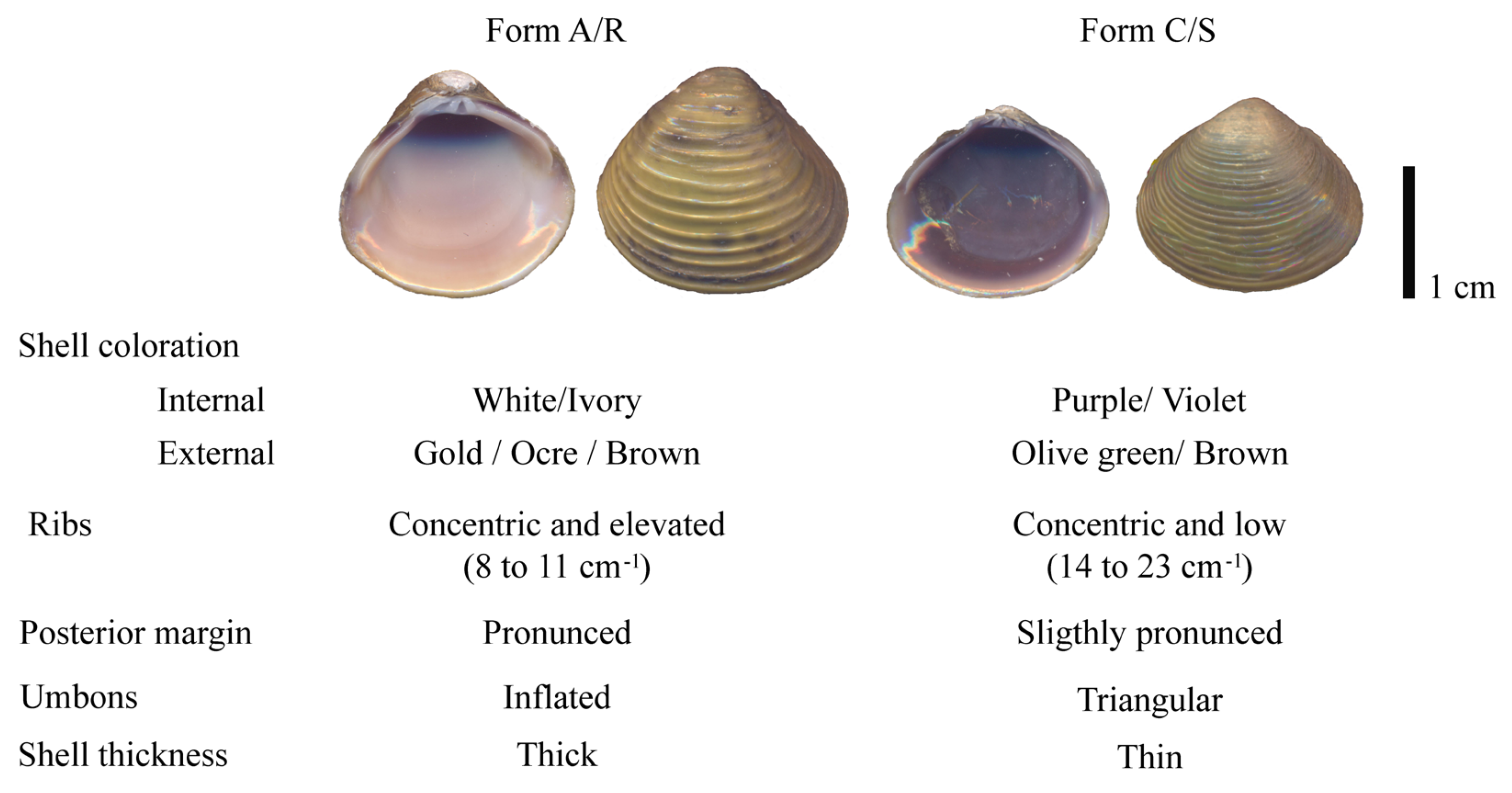
Shell Morphology
Internal Morphology
Population Selection for Molecular Analysis: Geographic Diversity across Argentina
DNA Extraction, PCR Amplification, and Sequencing
Data Analyses
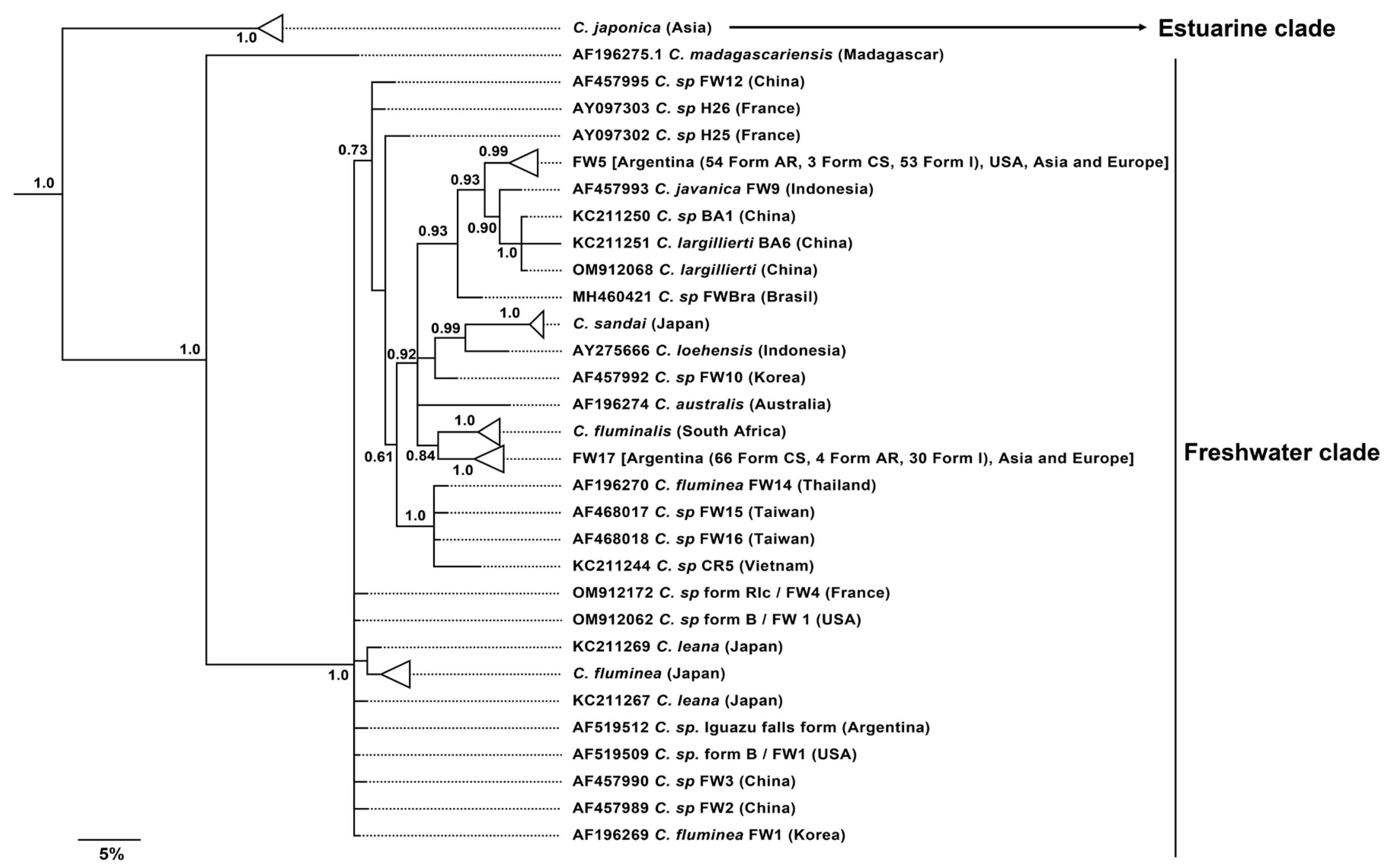
Phylogenetic and Phylogeographic Analyses
Morphological Clustering Analysis and Morphotype Reassignment
3. Results
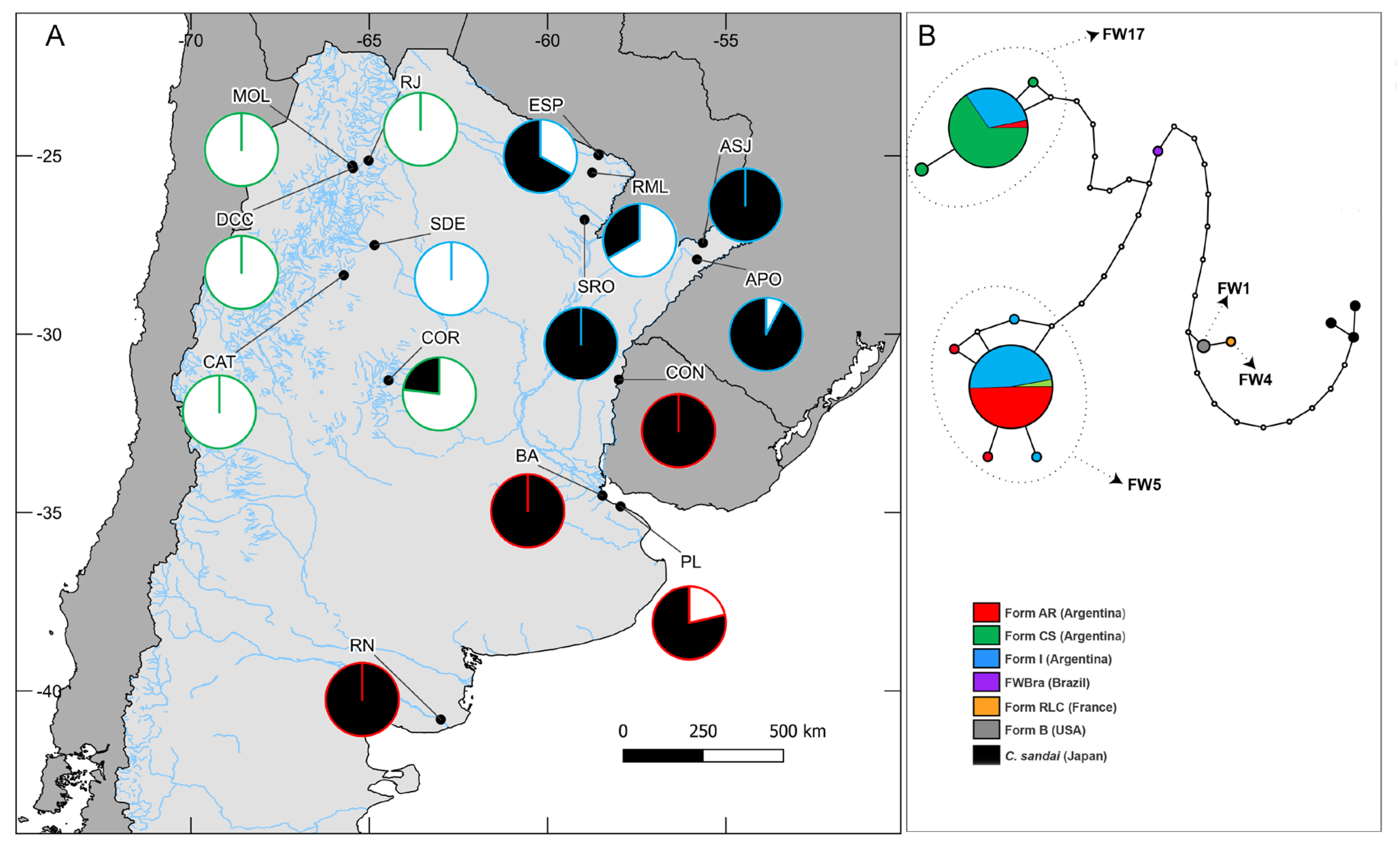
Distribution of Corbicula in Argentina: Review and Sampling
Phylogenetic and Phylogeographic Analyses
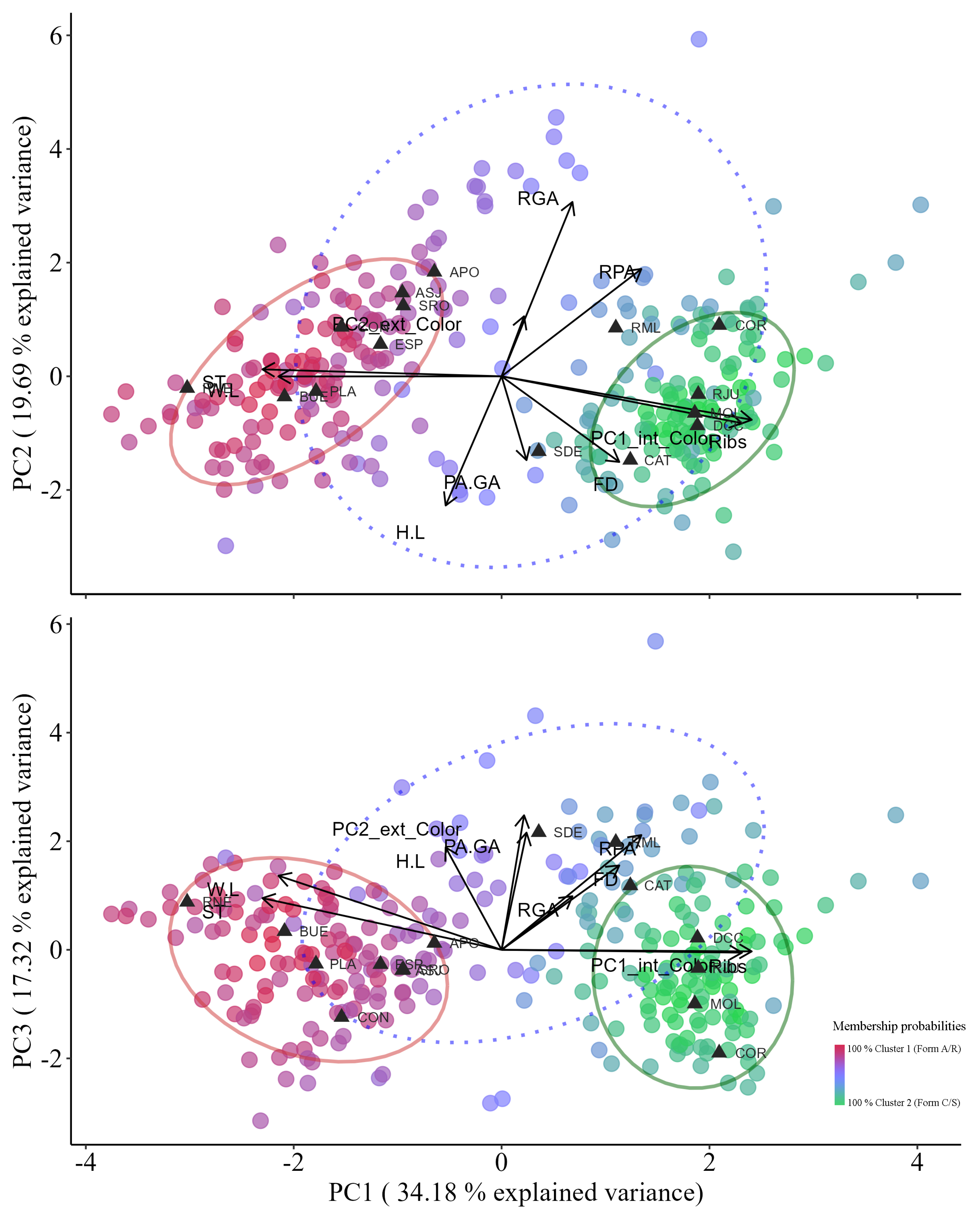
Morphological Clustering Analysis and Morphotype Reassignment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McMahon, R.F. The Occurence and Spread of the Introduced Asiatic Freshwater Clam, Corbicula fluminea (Muller) in North America: 1924-1982. Nautilus 1982, 96, 134–141. [Google Scholar]
- Araujo, R.; Moreno, D.; Ramos, M.A. The Asiatic Clam Corbicula fluminea (Müller, 1774) (Bivalvia: Corbiculidae) in Europe. Am. Malacol. Bull. 1993, 10, 39–49. [Google Scholar]
- Meijer, T.; Preece, R. A review of the occurrence of Corbicula in the Pleistocene of North-West Europe. Neth. J. Geosci. 2000, 79, 241–333. [Google Scholar] [CrossRef]
- Karatayev, A.Y.; Burlakova, L.E.; Padilla, D.K. Contrasting Distribution and Impacts of Two Freshwater Exotic Suspension Feeders, Dreissena polymorpha and Corbicula fluminea. In The Comparative Roles of Suspension-Feeders in Ecosystems. NATO Science Series IV: Earth and Environmental Series, vol 47; Dame, R.F., Olenin, S., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 239–262. [Google Scholar]
- Gama, M.; Crespo, D.; Dolbeth, M.; Anastácio, P.M. Ensemble forecasting of Corbicula fluminea worldwide distribution: Projections of the impact of climate change. Aquat. Conserv. Mar. Freshw. Ecosyst. 2017, 27, 675–684. [Google Scholar] [CrossRef]
- Pigneur, L.-M.; Marescaux, J.; Roland, K.; Etoundi, E.; Descy, J.-P.; Van Doninck, K. Phylogeny and androgenesis in the invasive Corbicula clams (Bivalvia, Corbiculidae) in Western Europe. BMC Evol. Biol. 2011, 11, 147. [Google Scholar] [CrossRef]
- Bespalaya, Y.V.; Kropotin, A.V.; Kondakov, A.V.; Aksenova, O.V.; Gofarov, M.Y.; Kim, S.K.; Lee, J.H.; Travina, O.V.; Vikhrev, I.V.; Vinarski, M.V.; et al. A taxonomic reassessment of native and invasive species of Corbicula clams (Bivalvia: Cyrenidae) from the Russian Far East and Korea. Zoöl. J. Linn. Soc. 2022, 197, 104–126. [Google Scholar] [CrossRef]
- Morton, B. Corbicula in Asia - an Updated Synthesis. Am. Malacol. Bull. 1986, Spetial Ed, 113–124. [Google Scholar]
- Pfenninger, M.; Reinhardt, F.; Streit, B. Evidence for cryptic hybridization between different evolutionary lineages of the invasive clam genus Corbicula (Veneroida, Bivalvia). J. Evol. Biol. 2002, 15, 818–829. [Google Scholar] [CrossRef]
- Marescaux, J.; Pigneur, L.-M.; Van Doninck, K. New records of Corbicula clams in French rivers. Aquat. Invasions 2010, 5, S35–S39. [Google Scholar] [CrossRef]
- Siripattrawan, S.; Park, J.-K.; Foighil, D. . Two lineages of the introduced Asian freshwater clam Corbicula occur in North America. J. Molluscan Stud. 2000, 66, 423–429. [Google Scholar] [CrossRef]
- Lee, T.; Siripattrawan, S.; Ituarte, C.F.; Foighil, D.Ó. Invasion of the Clonal Clams: Corbicula Lineages in the New World. Am. Malacol. Bull. 2005, 20, 113–122. [Google Scholar]
- Park, J.; Kim, W. Two Corbicula (Corbiculidae: Bivalvia) mitochondrial lineages are widely distributed in Asian freshwater environment. Mol. Phylogenetics Evol. 2003, 29, 529–539. [Google Scholar] [CrossRef]
- Pigneur, L.; Etoundi, E.; Aldridge, D.C.; Marescaux, J.; Yasuda, N.; Van Doninck, K. Genetic uniformity and long-distance clonal dispersal in the invasive androgenetic Corbicula clams. Mol. Ecol. 2014, 23, 5102–5116. [Google Scholar] [CrossRef]
- Capelle, J.J.; Hartog, E.; Bogaart, L.v.D.; Jansen, H.M.; Wijsman, J.W. Adaptation of gill-palp ratio by mussels after transplantation to culture plots with different seston conditions. Aquaculture 2021, 541, 736794. [Google Scholar] [CrossRef]
- Paolucci, E.M.; Sardiña, P.; Sylvester, F.; Perepelizin, P.V.; Zhan, A.; Ghabooli, S.; Cristescu, M.E.; Oliveira, M.D.; MacIsaac, H.J. Morphological and genetic variability in an alien invasive mussel across an environmental gradient in South America. Limnol. Oceanogr. 2014, 59, 400–412. [Google Scholar] [CrossRef]
- Hünicken, L.A.; Sylvester, F.; Paolucci, E.M. Physiological and morphological assessments suggest opposite structural allocation strategies between closely related invasive clams. Hydrobiologia 2022, 849, 2859–2875. [Google Scholar] [CrossRef]
- Modesto, V.; Ilarri, M.; Labecka, A.M.; Ferreira-Rodríguez, N.; Coughlan, N.E.; Liu, X.; Sousa, R. What we know and do not know about the invasive Asian clam Corbicula fluminea. Hydrobiologia 2023, 1–32. [Google Scholar] [CrossRef]
- Komaru, A.; Kawagishi, T.; Konishi, K. Cytological evidence of spontaneous androgenesis in the freshwater clam Corbicula leana Prime. Dev. Genes Evol. 1998, 208, 46–50. [Google Scholar] [CrossRef]
- Pigneur, L.-M.; Hedtke, S.M.; Etoundi, E.; Van Doninck, K. Androgenesis: a review through the study of the selfish shellfish Corbicula spp. Heredity 2012, 108, 581–591. [Google Scholar] [CrossRef]
- Hedtke, S.M.; Stanger-Hall, K.; Baker, R.J.; Hillis, D.M. All-Male asexuality: origin and maintenance of androgenesis in the asian clam corbicula. Evolution 2008, 62, 1119–1136. [Google Scholar] [CrossRef]
- Kropotin, A.V.; Bespalaya, Y.V.; Aksenova, O.V.; Kondakov, A.V.; Aksenov, A.S.; Khrebtova, I.S.; Palatov, D.M.; Travina, O.V.; Bolotov, I.N. Genetic and Morphological Characterization of the Invasive Corbicula Lineages in European Russia. Water 2023, 15, 3226. [Google Scholar] [CrossRef]
- Ludwig, S.; Darrigran, G.; Boeger, W.A. Opening the black box of the invasion of Corbicula clams (Bivalvia, Corbiculidae) in South America: a genetic and morphological evaluation. Hydrobiologia 2023, 851, 1203–1217. [Google Scholar] [CrossRef]
- Cazzaniga, N.J. Asiatic Clam,Corbicula fluminea, Reaching Patagonia (Argentina). J. Freshw. Ecol. 1997, 12, 629–630. [Google Scholar] [CrossRef]
- Cazzaniga, N.J.; Perez, C. Asiatic Clam, Corbicula fluminea, in Northwestern Patagonia (Argentina). J. Freshw. Ecol. 1999, 14, 551–552. [Google Scholar] [CrossRef]
- Martín, P.R.; Estebenet, A.L. Spread of the Asiatic Clam Corbicula fluminea in Southern Pampas and Northern Patagonia, Argentina. J. Freshw. Ecol. 2002, 17, 331–333. [Google Scholar] [CrossRef]
- Semenas, L.; Flores, V. Presence ofCorbicula flumineain the Upper Negro River Basin (Patagonia, Argentina). J. Freshw. Ecol. 2005, 20, 615–616. [Google Scholar] [CrossRef]
- Martin, P.R.; Tiecher, M.J. Hallazgo de La Almeja Invasora Corbicula fluminea En El Río Sauce Grande (Provincia de Buenos Aires, Argentina). BioScriba 2009, 2, 115–120. [Google Scholar]
- Reyna, P.B.; Morán, A.G.; Tatián, M. Taxonomy, distribution and population structure of invasive Corbiculidae (Mollusca, Bivalvia) in the Suquía River basin, Córdoba, Argentina. Iheringia, Série Zool. 2013, 103, 77–84. [Google Scholar] [CrossRef]
- Torre, L.; Reyna, P. Bivalvia, Veneroidea, Corbiculidae, Corbicula largillierti (Philippi, 1844): New distribution record in the Del Valle Central basin, Catamarca Province, Argentina. Check List. 2013, 9, 165. [Google Scholar] [CrossRef]
- Molina, L.M.; Pereyra, P.J.; Carrizo, N.G.M.; Abrameto, M.A. Here come the clam: Southernmost record worldwide of the Asian clam Corbicula fluminea (Patagonia, Argentina). Russ. J. Biol. Invasions 2015, 6, 129–134. [Google Scholar] [CrossRef]
- Trovant, B.; Signorelli, J.H.; Battini, N. Invasive pest spreads beyond the last frontier: Corbicula clam in the Chubut River, Patagonia. Limnology 2023, 24, 1–8. [Google Scholar] [CrossRef]
- Ituarte, C.F. Corbicula and Neocorbicula (Bivalvia: Corbiculidae) in the Paraná, Uruguay, and Río de La Plata Basins. Nautilus 1994, 107, 129–135. [Google Scholar]
- Mansur, M.C.D.; Pereira, D. Bivalves límnicos da bacia do rio dos Sinos, Rio Grande do Sul, Brasil (Bivalvia, Unionoida, Veneroida e Mytiloida). Rev. Bras. de Zoöl. 2006, 23, 1123–1147. [Google Scholar] [CrossRef]
- Coughlan, N.E.; Cunningham, E.M.; Cuthbert, R.N.; Joyce, P.W.S.; Anastácio, P.; Banha, F.; Bonel, N.; Bradbeer, S.J.; Briski, E.; Butitta, V.L.; et al. Biometric conversion factors as a unifying platform for comparative assessment of invasive freshwater bivalves. J. Appl. Ecol. 2021, 58, 1945–1956. [Google Scholar] [CrossRef]
- Dutertre, M.; Barillé, L.; Beninger, P.G.; Rosa, P.; Gruet, Y. Variations in the pallial organ sizes of the invasive oyster, Crassostrea gigas, along an extreme turbidity gradient. Estuarine, Coast. Shelf Sci. 2009, 85, 431–436. [Google Scholar] [CrossRef]
- Ivanova, N.V; DeWard, J.; Hebert, P.D.N. An Inexpensive, Automation-Friendly Protocol for Recovering High-Quality DNA. Mol. Ecol. Notes 2006, 6, 998–1002. [Google Scholar] [CrossRef]
- Ivanova, N.V.; Fazekas, A.J.; Hebert, P.D.N. Semi-automated, Membrane-Based Protocol for DNA Isolation from Plants. Plant Mol. Biol. Rep. 2008, 26, 186–198. [Google Scholar] [CrossRef]
- Whitlock, R.; Hipperson, H.; Mannarelli, M.; Burke, T. A High Throughput Protocol for Extracting High-Purity Genomic DNA from Plants and Animals. Mol. Ecol. Resour. 2008, 8, 736–741. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.Y. A Pattern Matching Algorithm for Double-Type Characters. Appl. Mech. Mater. 2014, 571-572, 461–464. [Google Scholar] [CrossRef]
- Hart, M.W.; Sunday, J. Things fall apart: biological species form unconnected parsimony networks. Biol. Lett. 2007, 3, 509–512. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, A.M.; Cabezas, M.P.; I Tavares, A.; Xavier, R.; Branco, M. tcsBU: a tool to extend TCS network layout and visualization. Bioinformatics 2016, 32, 627–628. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Librado, P.; Sanchez-Delbarrio, J.; Messeguer, X.; Rozas, R. DnaSP 5. 10. 00 2009.
- Legendre, P.; Legendre, L. Numerical Ecology, 3rd ed.; Developments in Environmental Modelling; Elsevier: Amsterdam, The Netherlands; Oxford, UK, 2012; Volume 24, ISBN 978-0-444-53868-0. [Google Scholar]
- Maechler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M.; Studer, M.; Gonzalez, J. Package ‘Cluster’ 2023.
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses, R Package v. 1.0. 7 2020.
- Reshaid, Y.; Cao, L.; Brea, F.; Blanche, M.O.; Torres, S.; Darrigran, G. Variation in the distribution of Corbicula species (Mollusca: Bivalvia: Corbiculidae) after 25 years of its introduction in the Río de la Plata, Argentina. Zoöl. 2017, 34, 1–6. [Google Scholar] [CrossRef]
- Peñarrubia, L.; Araguas, R.-M.; Vidal, O.; Pla, C.; Viñas, J.; Sanz, N. Genetic characterization of the Asian clam species complex (Corbicula) invasion in the Iberian Peninsula. Hydrobiologia 2016, 784, 349–365. [Google Scholar] [CrossRef]
- Bespalaya, Y.V.; Bolotov, I.N.; Aksenova, O.V.; Kondakov, A.V.; Gofarov, M.Y.; Laenko, T.M.; Sokolova, S.E.; Shevchenko, A.R.; Travina, O.V. Aliens are moving to the Arctic frontiers: an integrative approach reveals selective expansion of androgenic hybrid Corbicula lineages towards the North of Russia. Biol. Invasions 2018, 20, 2227–2243. [Google Scholar] [CrossRef]
- Renard, E.; Bachmann, V.; Cariou, M.L.; Moreteau, J.C. Morphological and molecular differentiation of invasive freshwater species of the genus Corbicula (Bivalvia,Corbiculidea) suggest the presence of three taxa in French rivers. Mol. Ecol. 2000, 9, 2009–2016. [Google Scholar] [CrossRef] [PubMed]
- Sousa, R.; Freire, R.; Rufino, M.; Méndez, J.; Gaspar, M.; Antunes, C.; Guilhermino, L. Genetic and shell morphological variability of the invasive bivalve Corbicula fluminea (Müller, 1774) in two Portuguese estuaries. Estuarine, Coast. Shelf Sci. 2007, 74, 166–174. [Google Scholar] [CrossRef]
- Pereira, D.; Dreher Mansur, M.C.D.; Duarte, L.D.S.; de Oliveira, A.S.; Pimpão, D.M.; Callil, C.T.; Ituarte, C.; Parada, E.; Peredo, S.; Darrigran, G.; et al. Bivalve distribution in hydrographic regions in South America: historical overview and conservation. Hydrobiologia 2014, 735, 15–44. [Google Scholar] [CrossRef]
- Ituarte, C.F. Growth Dynamics in a Natural Population of Corbicula fluminea (Bivalvia Sphaeriacea) at Punta Atalaya, Rio de La Plata, Argentina. Stud. Neotrop. Fauna Environ. 1985, 20, 217–225. [Google Scholar] [CrossRef]
- Bódis, E.; Nosek, J.; Oertel, N.; Tóth, B.; Fehér, Z. A Comparative Study of Two Corbicula Morphs (Bivalvia, Corbiculidae) Inhabiting River Danube. Int. Rev. Hydrobiol. 2011, 96, 257–273. [Google Scholar] [CrossRef]
- Szarmach, D.; Wiśniewski, K.; Kobak, J.; Kakareko, T.; Labecka, A.M.; Sousa, R.; Poznańska-Kakareko, M. Differences in substratum preferences and behaviour within the invasive Corbicula species complex. Freshw. Biol. 2023, 68, 1489–1502. [Google Scholar] [CrossRef]
- Ituarte, C.F. Aspectos Biológicos de Las Poblaciones de Corbicula largillierti Phillippi (Mollusca Pelecypoda) En El Río de La Plata. Rev. del Mus. La Plata (Nueva Ser. 1984, Tomo XIII, 231–247.
- Darrigran, G.A.; Maroñas, M.E. Crecimiento de Las Poblaciones Naturales de Corbicula fluminea (Müller, 1774) y C. largillierti (Phillippi, 1844) (Bivalvia: Sphaeriacea) En El Litoral de Punta Blanca, Estuario Del Río de La Plata, República Argentina. Comun. la Soc. Malacológica del Uruguay 1989, VII, 139–157.
- Mansur, M.C.D.; Vanin, A.S.; Bergonci, P.E.A.; Oliveira, A.S. de Dinâmica Reprodutiva de Corbicula fluminea e Corbicula largillierti. In Moluscos límnicos invasores no Brasil: biologia, prevençâo e controle; Mansur, M.C.D., dos Santos, C.P., Pereira, D., Padula Paz, I.C., Zurita, M.L.L., Raya Rodriguez, M.T., Nehrke, M.V., Bergonci, P.E.A., Eds.; Redes Editora: Porto Alegre, 2012; pp. 119–124. [Google Scholar]
- Marescaux, J.; Falisse, E.; Lorquet, J.; Van Doninck, K.; Beisel, J.-N.; Descy, J.-P. Assessing filtration rates of exotic bivalves: dependence on algae concentration and seasonal factors. Hydrobiologia 2016, 777, 67–78. [Google Scholar] [CrossRef]
| ID | Region; Province | Water body; Basin | Lat. Lon. | N | Haplotypic group | Form |
| DCC | Northwest; Salta | Cabra Corral reservoir; Río de la Plata basin | -25,34;$$$-65,45 | 15 | FW17 | C/S |
| RJU | Northwest; Salta | Juramento river; Río de la Plata basin | -25,13;$$$-65,01 | 11 | FW17 | C/S |
| MOL | Northwest; Salta | Moldes irrigation ditch; Río de la Plata basin | -25,27;$$$-65,47 | 15 | FW17 | C/S |
| SDE | Northwest; Sgo. del Estero | Dulce river; Mar Chiquita basin | -27,50;$$$-64,85 | 15 | FW17 | C/S and Corbicula sp. |
| CAT | Northwest; Catamarca | del Valle river; Del Valle basin | -28,34;$$$-65,71 | 15 | FW17 | C/S |
| COR | Center; Córdoba | Cosquín river; Mar Chiquita basin | -31,29;$$$-64,46 | 13 | FW17(10); FW5(3) | C/S |
| PLA | Estuary; Buenos Aires | Río de la Plata estuary | -34,82;$$$-57,95 | 14 | FW5(11); FW17(3) | A/R |
| BUE | Estuary; Buenos Aires | Río de la Plata estuary | -34,52;$$$-58,45 | 14 | FW5 | A/R |
| RNE | Patagonia; Río Negro | Negro river | -40,80;$$$-62,99 | 15 | FW5 | A/R |
| CON | East plain river; Entre Ríos | Ayuí Grande stream; Río de la Plata basin | -31,27;$$$-58,00 | 15 | FW5 | A/R |
| APO | Northest; Misiones | Chirimay stream; Río de la Plata basin | -27,90;$$$-55,81 | 14 | FW5 | Corbicula sp. |
| ASJ | Northest; Misiones | San Juan stream; Río de la Plata Basin | -27,44;$$$-55,64 | 13 | FW5(12); FW17(1) | Corbicula sp. |
| SRO | Northest; Chaco | de Oro river; Río de la Plata basin | -26,78;$$$-58,96 | 14 | FW5 | Corbicula sp. |
| ESP | Northest; Formosa | Porteño river; Río de la Plata basin | -24,97;$$$-58,56 | 12 | FW5(8); FW17(4) | Corbicula sp. |
| RML | Northest; Formosa | Monte Lindo river; Río de la Plata basin | -25,47;$$$-58,75 | 15 | FW5(5); FW17(10) | Corbicula sp. |
| Haplogroup (Argentina) | ||
| FW5 | FW17 | |
| n | 111 | 99 |
| π | 0.00013 | 0.00012 |
| Form A/R | 55 | 3 |
| Form C/S | 3 | 66 |
| Form I | 53 | 30 |
| p-distance | 2.28% | |
| Variable | Principal Component | ||
| 1 | 2 | 3 | |
| PC1_int_Color | 0.89 | -0.21 | -0.01 |
| PC2_ext_Color | 0.08 | 0.30 | 0.65 |
| H:L | -0.20 | -0.64 | 0.50 |
| W:L | -0.79 | 0.00 | 0.36 |
| ST | -0.85 | 0.03 | 0.25 |
| RGA | 0.25 | 0.86 | 0.26 |
| Ribs | 0.86 | -0.23 | -0.01 |
| RPA | 0.50 | 0.53 | 0.56 |
| PA:GA | 0.09 | -0.41 | 0.57 |
| FD | 0.42 | -0.42 | 0.41 |
| Standard deviation | 1.85 | 1.40 | 1.32 |
| Proportion of Variance | 0.34 | 0.20 | 0.17 |
| Cumulative Proportion | 0.34 | 0.54 | 0.71 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





