Submitted:
28 February 2024
Posted:
29 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
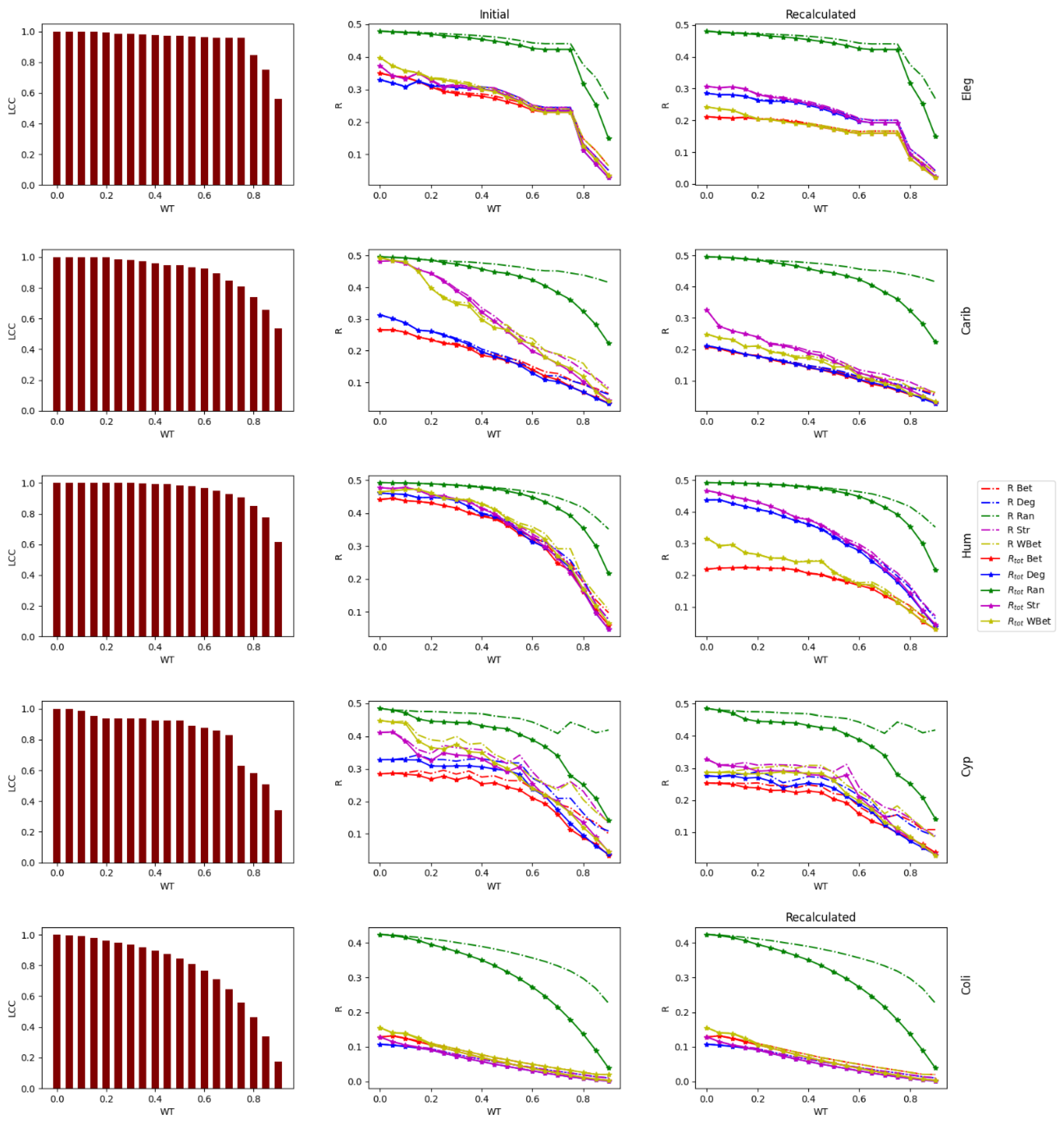
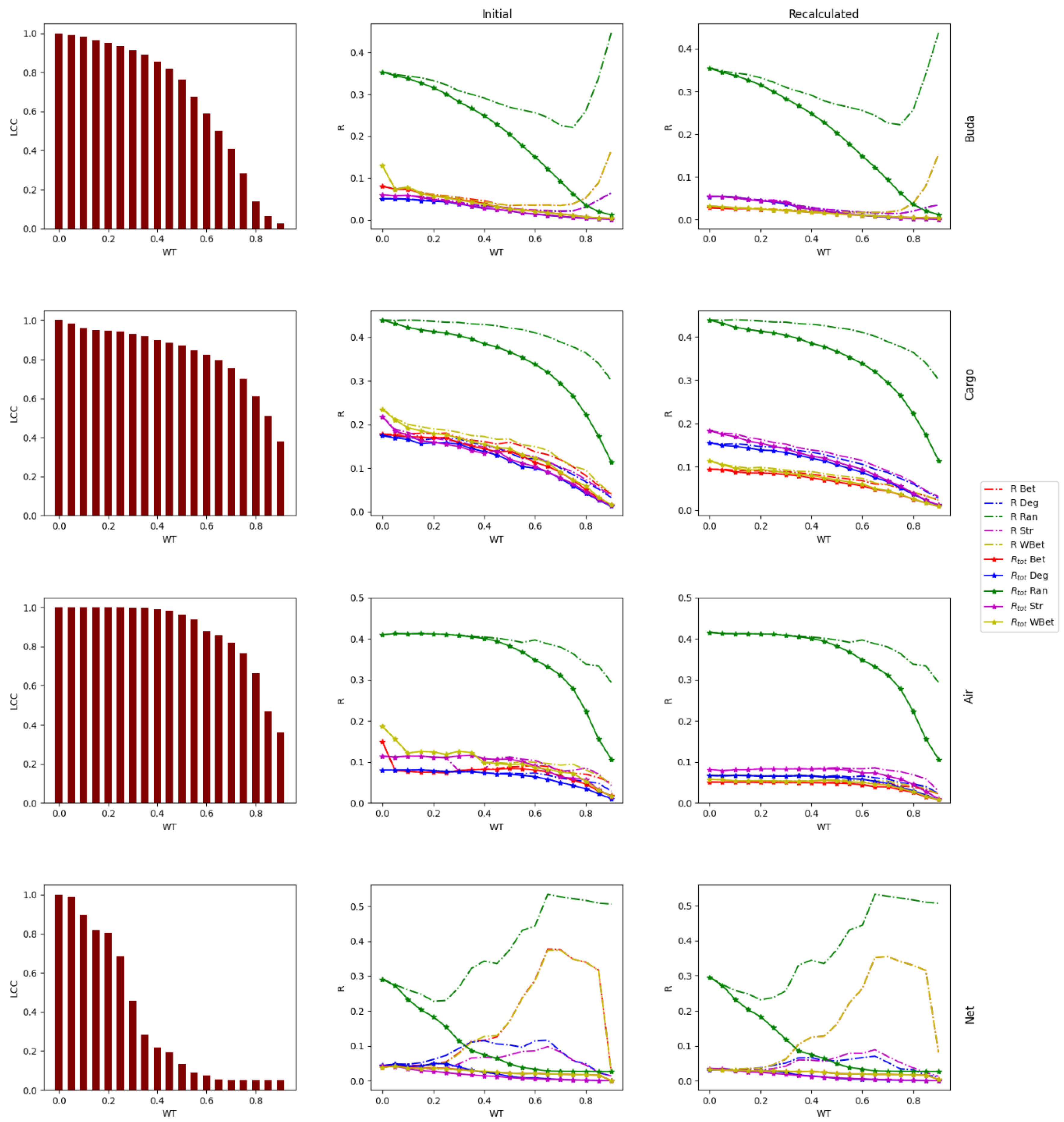
2.1. Real-World Networks
2.2. Attack strategies
- Degree (Deg): Nodes having the highest degree (hubs) are removed first [22,24,25,26,27]. The degree of a node is the number of links connected to it. The degree of node i is given bywhere indicates the presence of a link between nodes i and j and is 0 otherwise. is the number of nodes in the network.
-
Strength (Str): A node’s strength is the sum of the weights of the links connected to that node. It is a weighted version of the degree centrality [28], and it is also called weighted degree.Mathematically, the strength of node i is:where indicates the presence of a link between nodes i and j and is 0 otherwise. is the weight of the link between i and j. In this attack strategy, nodes with the highest strength are removed first.
- Betweenness (Bet): Betweenness of a node is the number of shortest paths (between all the pairs of nodes) passing through it [24,25,26]. This binary metric defines the shortest path between two nodes as the minimum number of links needed to travel from one node to another. Mathematically, the betweenness of node i is:where is the number of shortest paths between nodes s and t passing through the node i. is the total number of shortest paths between nodes s and t. Based on this global metric, this attack strategy first removes nodes with the highest betweenness.
-
Weighted Betweenness (WBet): Weighted betweenness of a node is defined as the number of weighted shortest paths passing through that node [29].Weighted Betweenness of node i is:where is the number of weighted shortest paths between nodes s and t passing through the node i. is the total number of weighted shortest paths between nodes s and t.
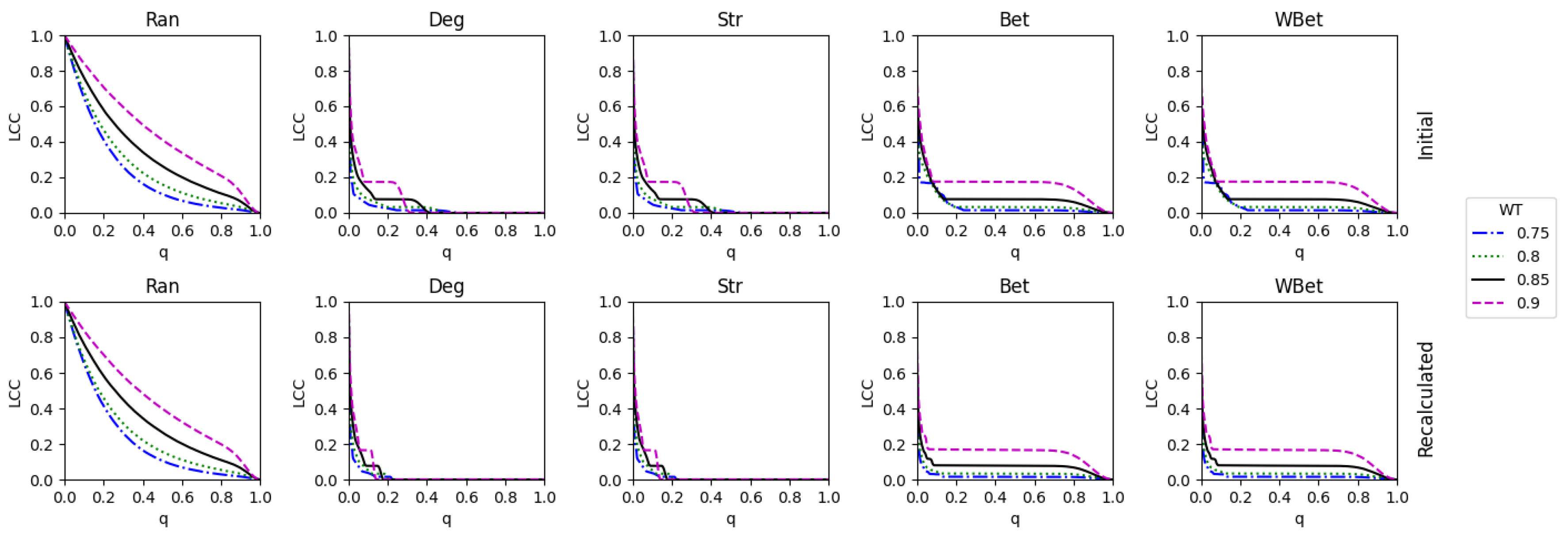
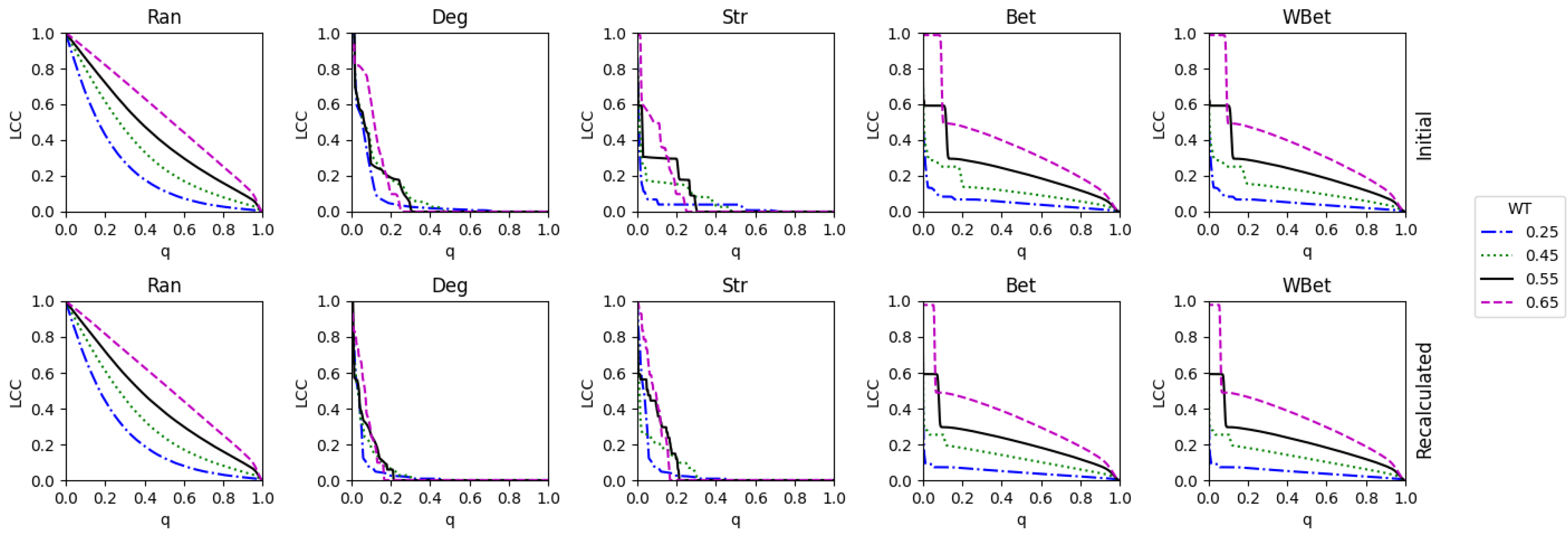
2.3. Weight thresholding
| Algorithm 1: Methodology of WT analysis. | |
| Procedure Weight Thresholding (G, N, L) | |
| 1: | WT = {0.0, 0.05, 0.1,…………., 0.85, 0.9} |
| 2: | for each WT |
| 3: | for i =1 to m |
| 4: | link_set= {links in the decreasing order of their weight} |
| 5: | strong_linkset = {WT fraction of strong links from link_set} |
| 6: | G’= G - strong_linkset |
| 7: | Initial attack (G’, N, L’) |
| 8: | Recalculated attack (G’, N, L’) |
| Procedure Initial attack (G’, N, L’) | |
| 1: | Find Initial LCC |
| 2: | for i =1 to n |
| 3: | node_set = { nodes of G’ in the decreasing order of centrality measure } |
| 4: | while (LCC !=1) |
| 5: | Remove a node x from the G’ (in the order of node_set) |
| 6: | Find LCC of new network |
| 7: | node_set = node_set - x |
| Procedure Recalculated attack (G’, N, L’) | |
| 1: | Find Initial LCC |
| 2: | for i=1 to n |
| 3: | while (LCC !=1) |
| 4: | Calculate centrality meaures |
| 5: | node_set = { nodes of G’ in the decreasing order of centrality measure } |
| 6: | Remove a node x from the G’ (in the order of node_set) |
| 7: | Find LCC of new network |
| 8: | node_set = node_set - x |
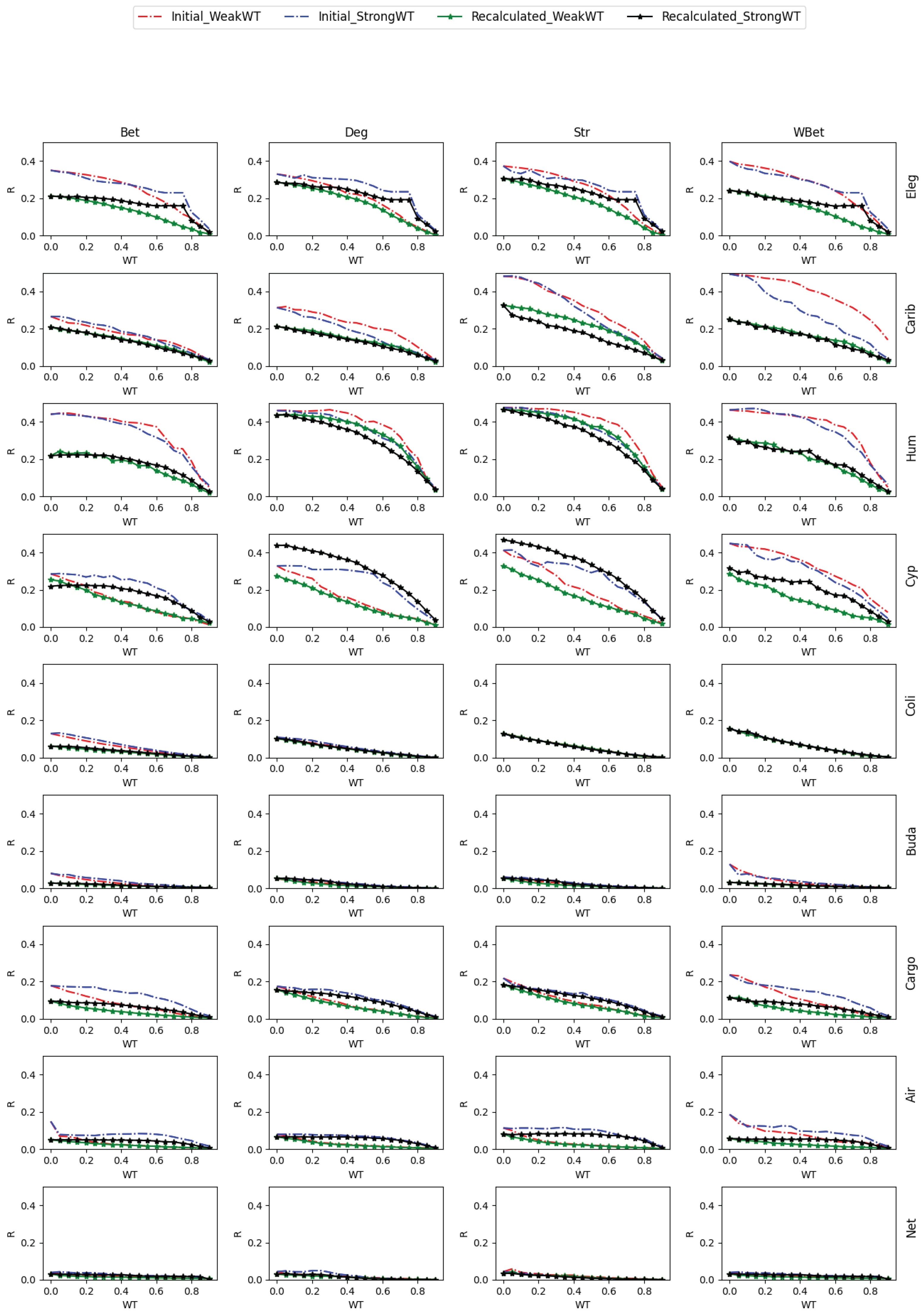
2.4. Network robustness indicator
- 1)
- One way is to normalize the LCC after node removal by the initial LCC value (before node attack) of the network after WT. In this case, we are considering each thresholded network as an independent network, and we do not account for the LCC decrease directly caused by the WT procedure.
- 2)
- A second way is to normalize the LCC after node removal by the initial LCC at WT=0, i.e., we normalize using the LCC of the original network. In this second case, we consider the LCC decrease triggered by the link removal of the WT procedure. This normalization is intended to analyze the joint effect of the weight thresholding and node attack to directly decrease the LCC. This is the total LCC decrease.
3. Results and Discussion
3.1. Robustness against WT
3.2. Robustness to WT and node attack
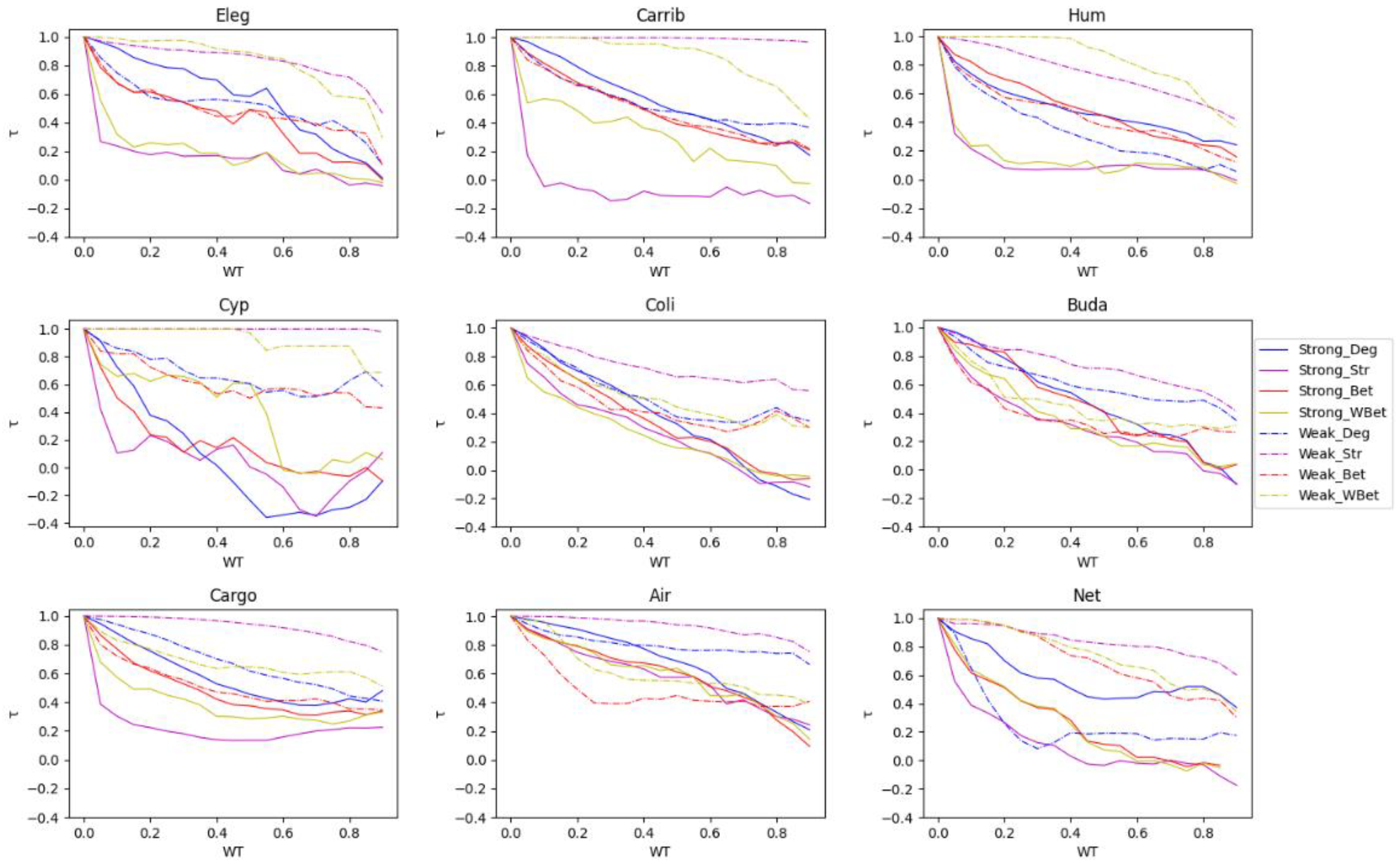
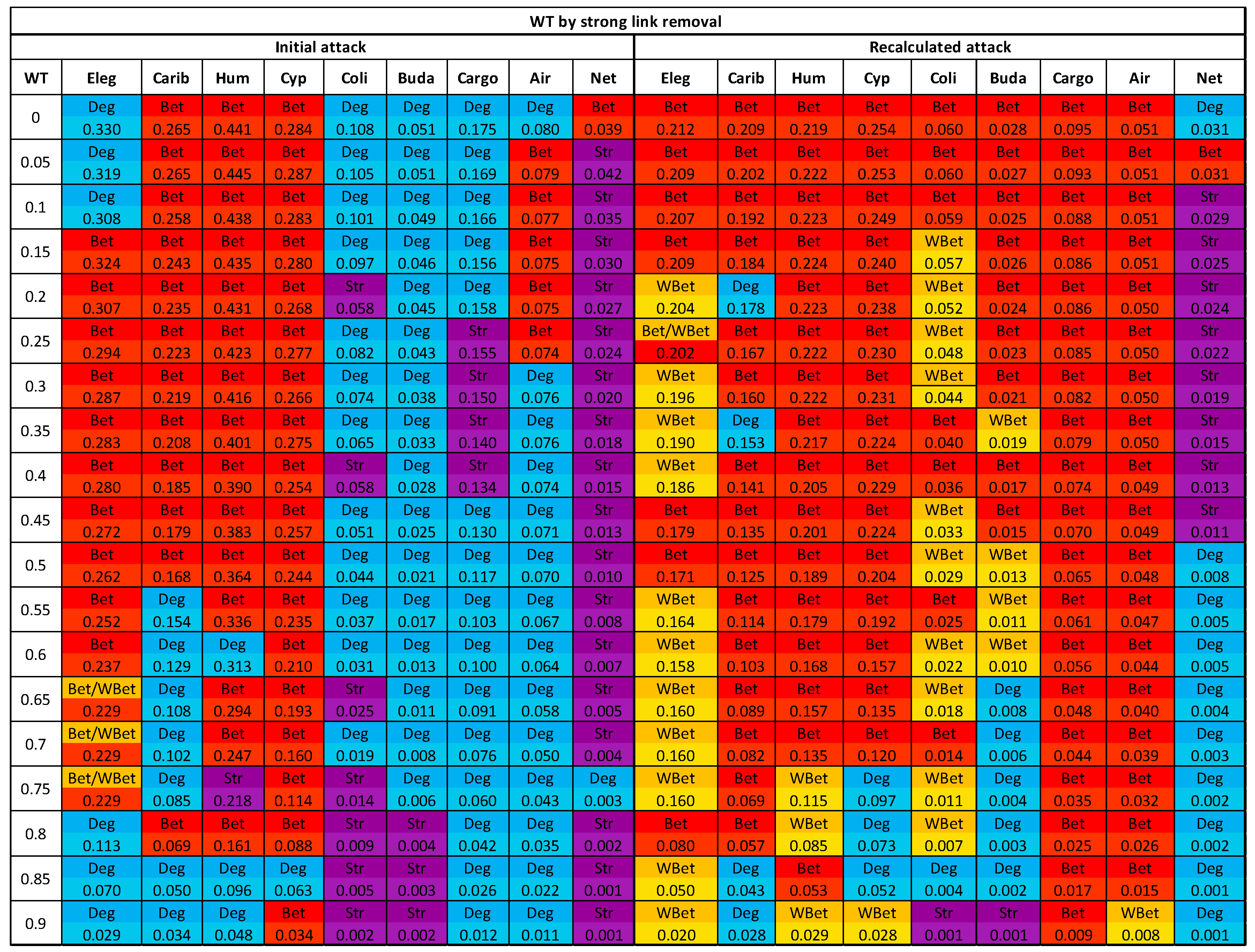
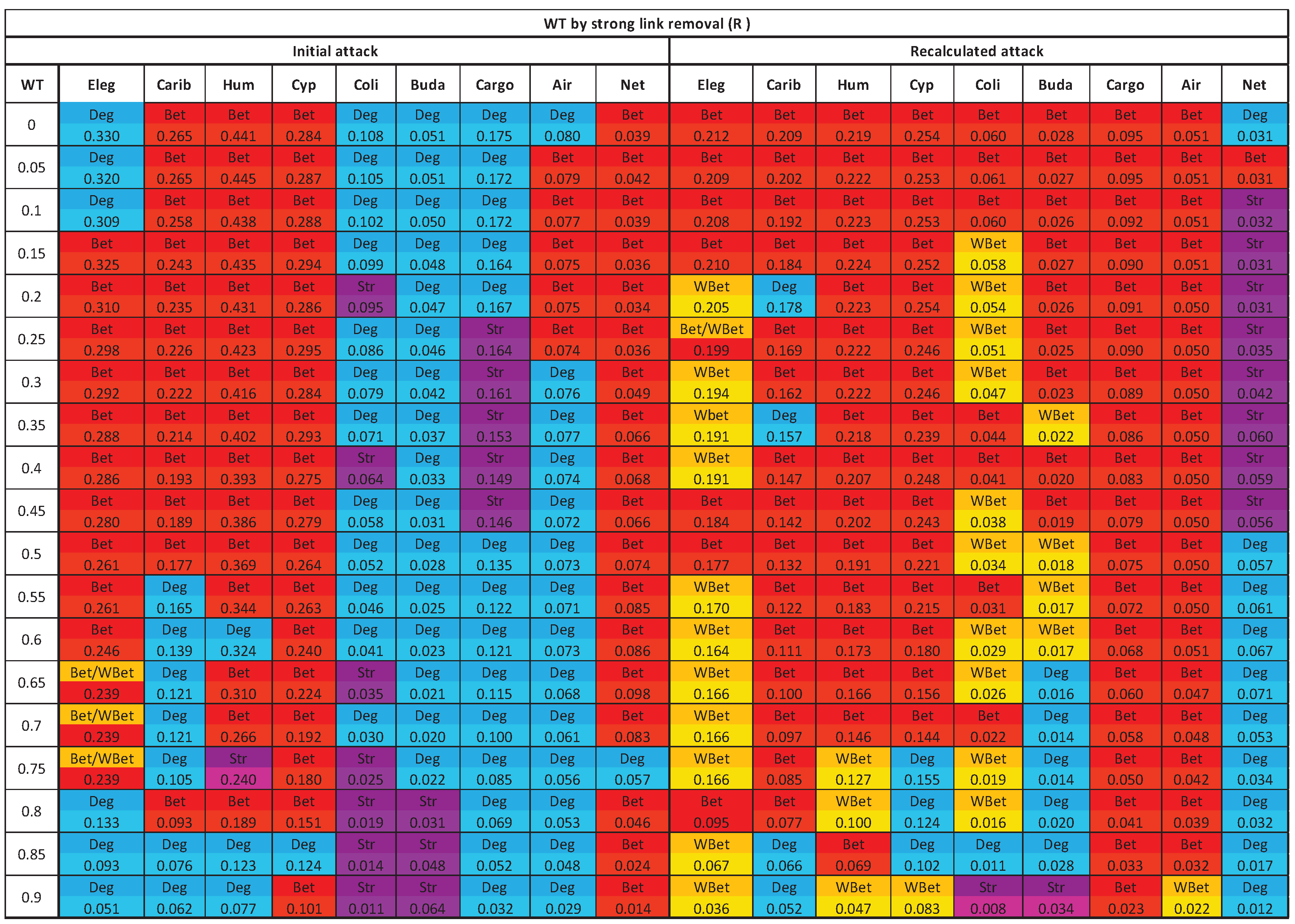
3.3. The efficacy of the node attack strategies
3.4. Comparing strong and weak WT procedures
4. Conclusion
Acknowledgments
References
- X. Yan, L.. G. S. Jeub, A. Flammini, F. Radicchi and. S. Fortunato, “Weight thresholding on complex networks,” Physical Review E, vol. 98, no. 4, 2018.
- J. M. John, M. Bellingeri, D. S. Lekha, D. Cassi and R. Alfieri, “Effect of Weight Thresholding on the Robustness of Real-World Complex Networks to Central Node Attacks,” Mathematics, vol. 11, 2023.
- A. Namaki, A. H. Shirazi, R. Raei and G. Jafari, “Network analysis of a financial market based on genuine correlation and threshold method,” Physica A: Statistical Mechanics and its Applications, vol. 390, pp. 3835--3841, 2011.
- M.-E. Lynall, D. S. Bassett, R. Kerwin, P. J. McKenna, M. Kitzbichler, U. Muller and E. Bullmore, “Functional connectivity and brain networks in schizophrenia,” Journal of Neuroscience, vol. 30, pp. 9477--9487, 2010.
- S. Allesina, A. Bodini and C. Bondavalli, “Secondary extinctions in ecological networks: bottlenecks unveiled,” Ecological Modelling, vol. 194, pp. 150--161, 2006.
- K. A. Garrison, D. Scheinost, E. S. Finn, X. Shen and R. T. Constable, “The (in) stability of functional brain network measures across thresholds,” Neuroimage, vol. 118, pp. 651--661, 2015.
- T. Kavzoglu and P. M. Mather, “Assessing artificial neural network pruning algorithms,” in Proceedings of the 24th annual conference and exhibition of the remote sensing society, Greenwich, UK, 1998.
- P. Hongwu, G. Deniz, H. Shaoyi, G. Tong, J. Weiwen, K. Orner and D. Caiwen, “Towards Sparsification of Graph Neural Networks,” in IEEE 40th International Conference on Computer Design (ICCD), Olympic Valley, CA, USA, 2022.
- D.. J. Watts and. S. H. Strogatz, “Collective dynamics of ‘small-world’ networks,” Nature, vol. 393, p. 440–2, 1998.
- V. Latora, V. Nicosia and G. Russo, Complex Networks: Principles, Methods and Applications., 2017.
- A. Allard, M. A. Serrano,. G. Garcia-Perez and M. Boguna, “The geometric nature of weights in real complex networks.,” Nature communications, vol. 8, pp. 1-8, 2017.
- V. Colizza, R. Pastor-Satorras and A. Vespignani, “Reaction-diffusion processes and metapopulation models in heterogeneous,” Nature Physics, vol. 3, p. 276–282, 2007.
- A. M. Serrano,. M. Boguna and. F. Sagues, “Uncovering the hidden geometry behind metabolic networks,” Molecular BioSystems, vol. 8, p. 843–850, 2012.
- M.. E. J. Newman, “Finding community structure in networks using the eigenvectors of matrices,” Phys. Rev., vol. 74, p. 036104, 2006.
- A. Avena-Koenigsberger, J. Goni, R. F. Betzel,. M. P. v. d. Heuvel, A. Griffa,. P. Hagmann, J.-P. Thiran and. O. Sporns, “Using Pareto optimality to explore the topology and dynamics of the human connectome,” Philos Trans R Soc Lond B Biol Sci., vol. 369, 2014.
- P. Hagmann, L. Cammoun,. X. Gigandet,. R. Meuli,. C. J. Honey, V. J. Wedeen and O. Sporns, “Mapping the structural core of human cerebral cortex,” PLoS biology, vol. 6, p. 1479–1493, 2008.
- M. Bellingeri and A. Bodini, “Food web's backbones and energy delivery in ecosystems,” Oikos, vol. 125, pp. 586-594, 2016.
- S. Opitz, Trophic Interactions in Caribbean Coral Reefs, ICLARM, 1996.
- J.. J. Heymans, R. E. Ulanowicz and C. Bondavalli, “Network analysis of the South Florida Everglades graminoid marshes and comparison with nearby cypress ecosystems,” Ecological Modelling, vol. 149, pp. 5-23, 2002.
- M. Bellingeri and S. Vincenzi, “Robustness of empirical food webs with varying consumer's sensitivities to loss of resources,” Journal of Theoretical Biology, vol. 333, pp. 18-26, 2013.
- B. Szalkai, C. Kerepesi, B. Varga and V. Grolmusz, “The Budapest Reference Connectome Server v2.0,” Neuroscience Letters, vol. 595, pp. 60-62, 2015.
- R. Albert, H. Jeong and. A.-L. Barabasi, “Error and attack tolerance of complex networks,” Nature volume, vol. 406, p. 378–382, 2000.
- R. Albert and A.-L. Barabasi, “Statistical mechanics of complex networks,” Review of Modern Physics, vol. 74, no. 1, pp. 47-97, 2002.
- P. Holme, B. J. Kim, C. N. Yoon and S. K. Han, “Attack vulnerability of complex networks,” Phys. Rev. E, vol. 65, no. 5, p. 056109, 2002.
- M. Bellingeri,. D. Cassi and S. Vincenzi, “Efficiency of attack strategies on complex model and real-world networks,” Physica A: Statistical Mechanics and its Applications, vol. 414, pp. 174-180, 2014.
- S. Iyer, T. Killingback,. B. Sundaram and Z. Wang, “Attack robustness and centrality of complex networks,” PLoS One, vol. 8, pp. 1-17, 2013.
- R. Cohen,. K. Erez,. D. Ben-Avraham and S. Havlin, “Breakdown of the internet under intentional attack,” Physical Review Letters, vol. 86, p. 3682–3685, 2001.
- M. Bellingeri and D. Cassi, “Robustness of weighted networks,” Physica A: Statistical Mechanics and its Applications, vol. 489, pp. 47-55, 2018.
- Q. Nguyen, N. K. K. Nguyen, D. Cassi and M. Bellingeri, “New betweenness centrality node attack strategies for real-world complex weighted networks,” Complexity, vol. 2021, 2021.
- M. Bellingeri, D. Bevacqua, F. Sartori, M. Turchetto, F. Scotognella, R. Alfieri, N. K. K. Nguyen, T. T. Le, Q. Nguyen and D. Cassi, “Considering weights in real social networks: A review,” Frontiers in Physics, vol. 11, 2023.
- M. Bellingeri, D. Bevacqua, F. a. Scotognella, R. Alfieri and D. Cassi, “A comparative analysis of link removal strategies in real complex weighted networks,” Scientific reports, vol. 10, pp. {1--15, 2020.
- R. K. Pan and J. Saramaki, “The strength of strong ties in scientific collaboration networks,” Europhysics Letters, vol. 97, no. 1, p. 18007, 2012.
- F. Jordán, “Keystone Species and Food Webs,” Philosophical transactions of the Royal Society of London. Series B, Biological sciences, vol. 364, pp. 1733-41, 2009.
- T. Nie, Z. Guo, K. Zhao and Z.-M. Lu, “New attack strategies for complex networks,” Physica A: Statistical Mechanics and its Applications, vol. 424, pp. 248-253, 2015.
- M. G. Kendall, “The treatment of ties in ranking problems,” Biometrika, vol. 33, pp. 239--251, 1945.
- L. Lü, D. Chen, X.-L. Ren, Q.-M. Zhang, Y.-C. Zhang and T. Zhou, “Vital nodes identification in complex networks,” Physics Reports, vol. 650, pp. 1-63, 2016.
| Networks | Key | Ref. | Type | Node | Link | Weight | N | L | <k> | <w> | LCC |
| C. Elegans | Eleg | [9,10] | Biological | Neurons | Neurons connection | Number of Connections | 297 | 2344 | 15.8 | 3.761 | 297 |
| Cargoship | Cargo | [11] | Transport | Ports | Route | Shipping journeys | 834 | 4348 | 10.4 | 97.709 | 821 |
| USairport | Air | [12] | Transport | Airports | Route | Passengers | 500 | 2979 | 11.9 | 152320.2 | 500 |
| E. Coli | Coli | [11,13] | Biological | Metabolites | Common reaction | Number of Common reactions | 1100 | 3636 | 6.61 | 1.364 | 1100 |
| Netscience | Net | [14] | Social | authors | Coauthorship | Number of Common papers | 1461 | 2741 | 3.75 | 0.434 | 379 |
| Human12a | Hum | [15,16] | Biological | Brain regions | Connection between regions | Connection density | 501 | 6038 | 24.1 | 0.01 | 501 |
| Caribbean | Carib | [17,18] | Ecological Food web | Species | Trophic relation | Amount of biomass | 249 | 3503 | 28.13 | 0.067 | 249 |
| CypDry | Cyp | [19,20] | Ecological Food web | Species | Trophic relation | Amount of biomass | 66 | 503 | 15.24 | 0.358 | 65 |
| Budapest | Buda | [21] | Biological | Brain regions | Neural connection | Amount of track flow | 480 | 1000 | 4.167 | 5.024 | 467 |
| Abbreviation | Full name |
|---|---|
| WT | Weight thresholding |
| LCC | Size of largest connected component |
| N | Number of nodes |
| L | Number of links |
| <w> | Average weight |
| <k> | Average degree |
| Ran | Random node attack |
| Deg | Degree node attack |
| Str | Strength node attack |
| Bet | Betweenness node attack |
| WBet | Weighted Betweenness node attack |
| G | Weighted network |
| G' | Thresholded network |
| L’ | Number of links in G' |
| q | Fraction of nodes removed |
| R | Robustness |
| Rtot | Total Robustness |
| Initial_Weak WT | WT by weak link removal with initial node attack strategy |
| Initial_Strong WT | WT by strong link removal with initial node attack strategy |
| Recalculated_Weak WT | WT by weak link removal with recalculated node attack strategy |
| Recalculated_Strong WT | WT by strong link removal with recalculated node attack strategy |
 |
 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





