Submitted:
31 January 2024
Posted:
31 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
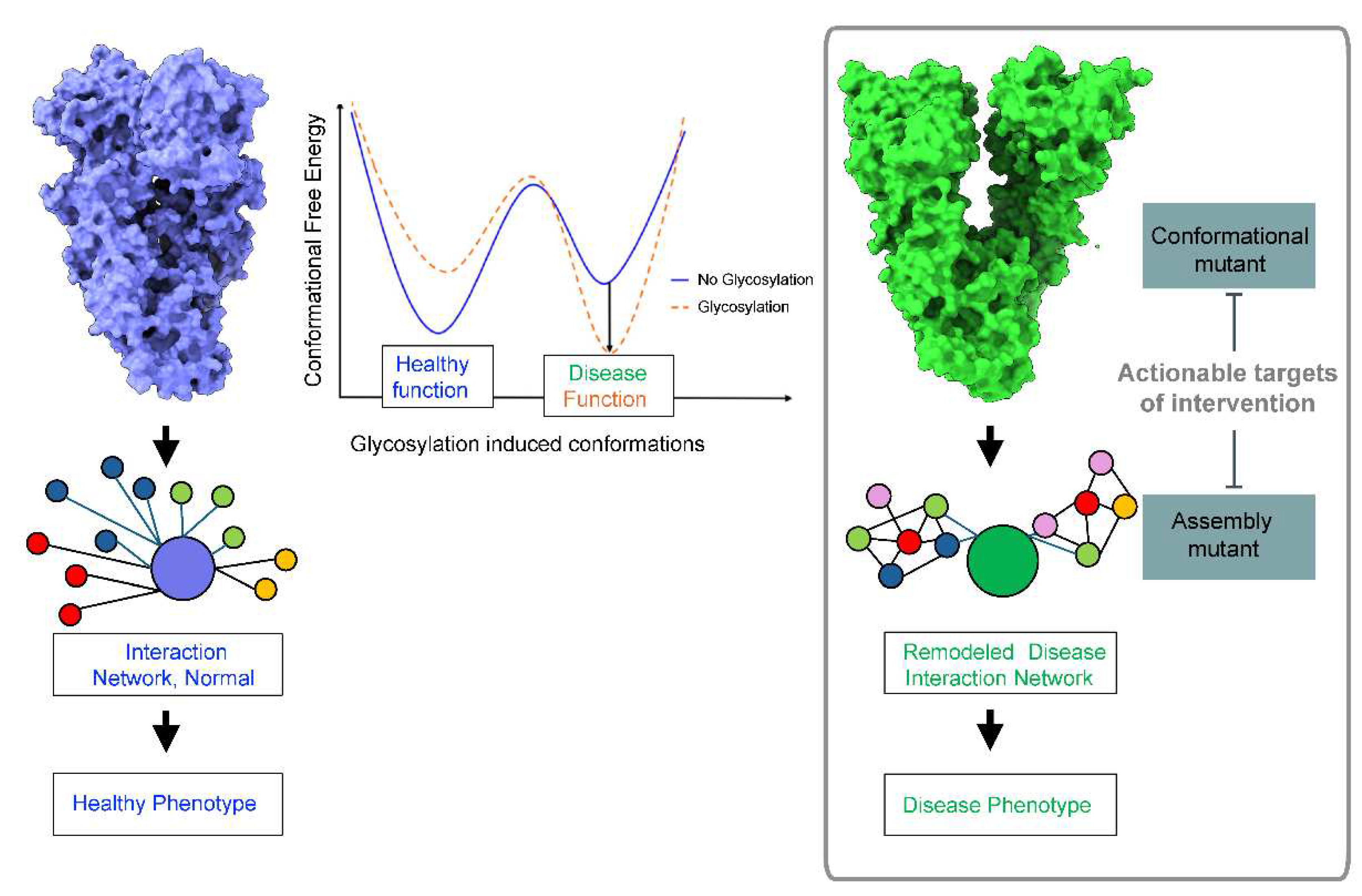
2. Glycosylation and Protein Conformation
3. N-glycans’ Effect on Pathologic Protein Conformations in Disease
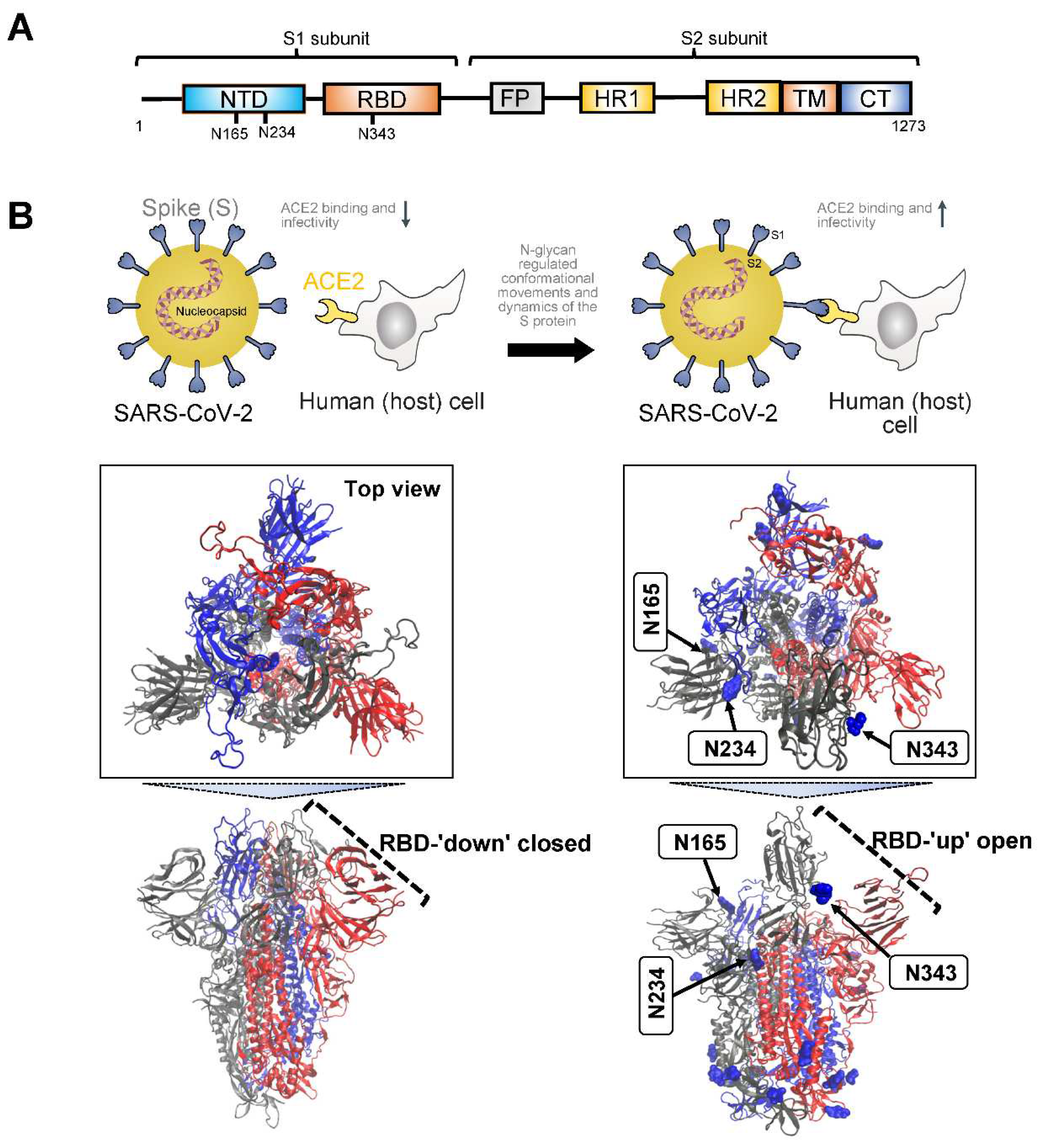
3.1. Severe Acute Respiratory Syndrome SARS Proteins
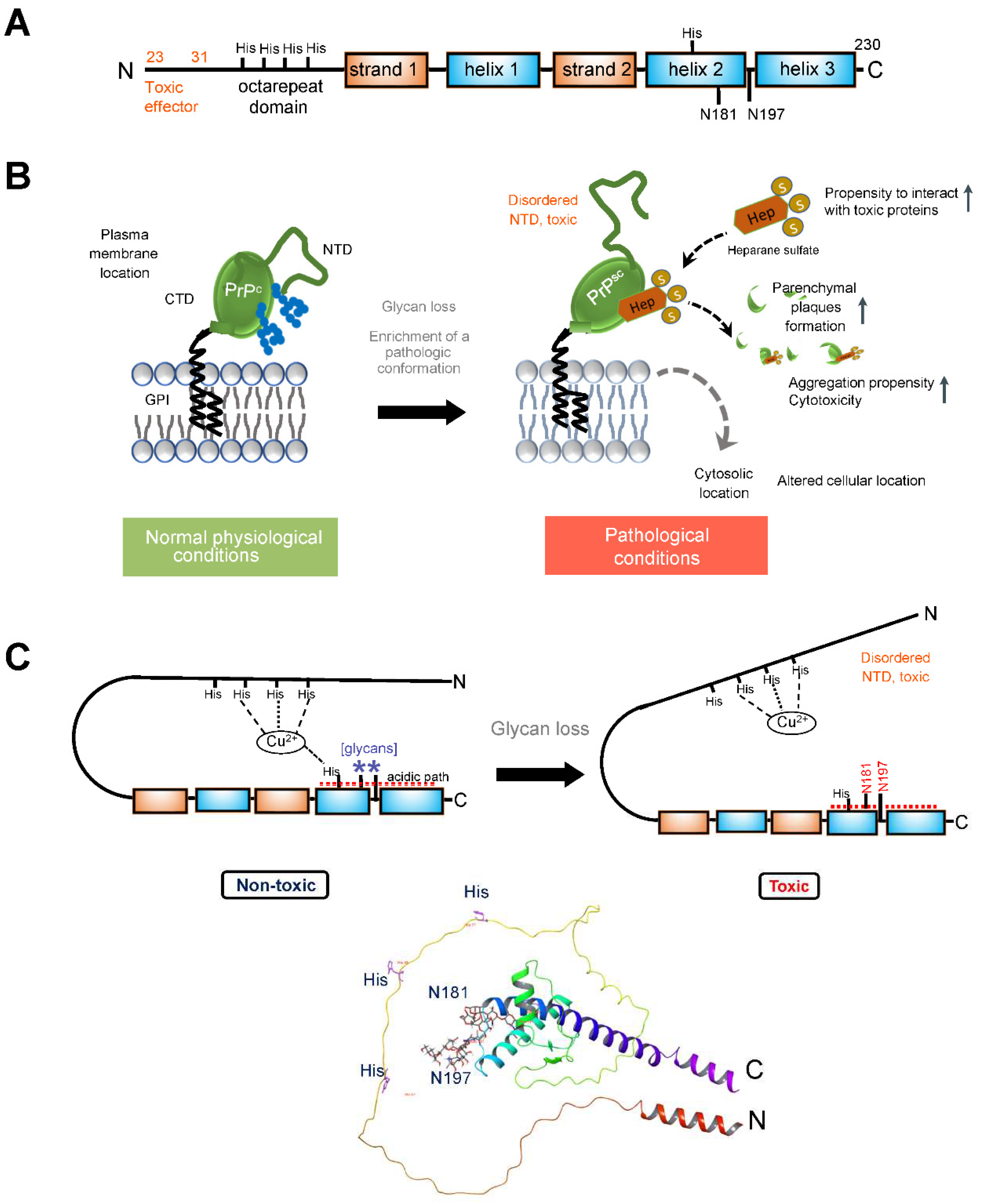
3.2. Prion Protein
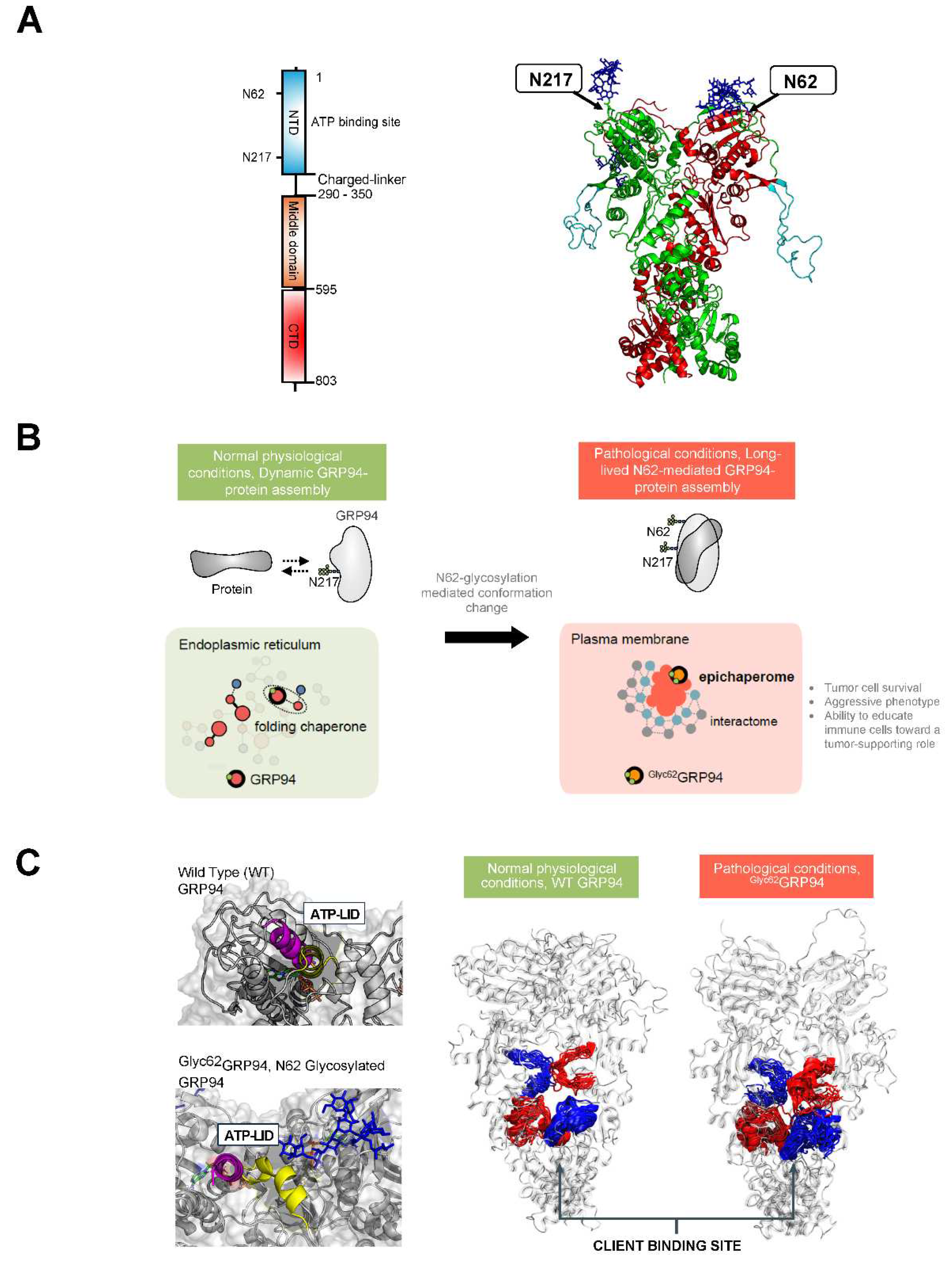
3.3. Glucose Regulated Protein 94 (GRP94)
4. Therapeutic Implications - Targeting Conformational and Assembly Mutants in Disease
4.1. Targeting The viral Protein Conformations
4.2. Correcting the Prion Protein Conformation
4.3. Targeting Pathologic GRP94 Conformers and Assemblies
5. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Moremen, K.W.; Tiemeyer, M.; Nairn, A.V. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol 2012, 13, 448-462. [CrossRef]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat Rev Nephrol 2019, 15, 346-366. [CrossRef]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global view of human protein glycosylation pathways and functions. Nat Rev Mol Cell Biol 2020, 21, 729-749. [CrossRef]
- Varki, A. Biological roles of glycans. Glycobiology 2017, 27, 3-49. [CrossRef]
- Rini, J.M.; Moremen, K.W.; Davis, B.G.; Esko, J.D. Glycosyltransferases and Glycan-Processing Enzymes. In Essentials of Glycobiology, 4th ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Mohnen, D., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor (NY), 2022; pp. 67-78.
- Corfield, A.P.; Berry, M. Glycan variation and evolution in the eukaryotes. Trends Biochem Sci 2015, 40, 351-359. [CrossRef]
- Lebrilla, C.B.; Liu, J.; Widmalm, G.; Prestegard, J.H. Oligosaccharides and Polysaccharides. In Essentials of Glycobiology, 4th ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Mohnen, D., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor (NY), 2022; pp. 33-42.
- Seeberger, P.H. Monosaccharide Diversity. In Essentials of Glycobiology, 4th ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Mohnen, D., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor (NY), 2022; pp. 21-32.
- Rudd, P.M.; Karlsson, N.G.; Khoo, K.H.; Thaysen-Andersen, M.; Wells, L.; Packer, N.H. Glycomics and Glycoproteomics. In Essentials of Glycobiology, 4th ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Mohnen, D., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor (NY), 2022; pp. 689-704.
- Chao, Q.; Ding, Y.; Chen, Z.H.; Xiang, M.H.; Wang, N.; Gao, X.D. Recent Progress in Chemo-Enzymatic Methods for the Synthesis of N-Glycans. Front Chem 2020, 8, 513. [CrossRef]
- de Haas, P.; Hendriks, W.; Lefeber, D.J.; Cambi, A. Biological and Technical Challenges in Unraveling the Role of N-Glycans in Immune Receptor Regulation. Front Chem 2020, 8, 55. [CrossRef]
- Fang, P.; Ji, Y.; Oellerich, T.; Urlaub, H.; Pan, K.T. Strategies for Proteome-Wide Quantification of Glycosylation Macro- and Micro-Heterogeneity. Int J Mol Sci 2022, 23. [CrossRef]
- Schmaltz, R.M.; Hanson, S.R.; Wong, C.H. Enzymes in the synthesis of glycoconjugates. Chem Rev 2011, 111, 4259-4307. [CrossRef]
- Zacchi, L.F.; Schulz, B.L. N-glycoprotein macroheterogeneity: biological implications and proteomic characterization. Glycoconj J 2016, 33, 359-376. [CrossRef]
- Springer, S.A.; Gagneux, P. Glycan evolution in response to collaboration, conflict, and constraint. J Biol Chem 2013, 288, 6904-6911. [CrossRef]
- Gong, Y.; Qin, S.; Dai, L.; Tian, Z. The glycosylation in SARS-CoV-2 and its receptor ACE2. Signal Transduct Target Ther 2021, 6, 396. [CrossRef]
- Otaki, M.; Hirane, N.; Natsume-Kitatani, Y.; Nogami Itoh, M.; Shindo, M.; Kurebayashi, Y.; Nishimura, S.I. Mouse tissue glycome atlas 2022 highlights inter-organ variation in major N-glycan profiles. Sci Rep 2022, 12, 17804. [CrossRef]
- Lumibao, J.C.; Tremblay, J.R.; Hsu, J.; Engle, D.D. Altered glycosylation in pancreatic cancer and beyond. J Exp Med 2022, 219. [CrossRef]
- Saleh, F.M.; Chandra, P.K.; Lin, D.; Robinson, J.E.; Izadpanah, R.; Mondal, D.; Bollensdorff, C.; Alt, E.U.; Zhu, Q.; Marasco, W.A.; et al. A New Humanized Mouse Model Mimics Humans in Lacking alpha-Gal Epitopes and Secreting Anti-Gal Antibodies. J Immunol 2020, 204, 1998-2005. [CrossRef]
- Riley, N.M.; Hebert, A.S.; Westphall, M.S.; Coon, J.J. Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis. Nat Commun 2019, 10, 1311. [CrossRef]
- Yang, X.; Wang, Z.; Guo, L.; Zhu, Z.; Zhang, Y. Proteome-Wide Analysis of N-Glycosylation Stoichiometry Using SWATH Technology. J Proteome Res 2017, 16, 3830-3840. [CrossRef]
- Bagdonaite, I.; Malaker, S.A.; Polasky, D.A.; Riley, N.M.; Schjoldager, K.; Vakhrushev, S.Y.; Halim, A.; Aoki-Kinoshita, K.F.; Nesvizhskii, A.I.; Bertozzi, C.R.; et al. Glycoproteomics. Nature Reviews Methods Primers 2022, 2, 48. [CrossRef]
- Hu, Y.; Pan, J.; Shah, P.; Ao, M.; Thomas, S.N.; Liu, Y.; Chen, L.; Schnaubelt, M.; Clark, D.J.; Rodriguez, H.; et al. Integrated Proteomic and Glycoproteomic Characterization of Human High-Grade Serous Ovarian Carcinoma. Cell Rep 2020, 33, 108276. [CrossRef]
- Campbell, M.P.; Peterson, R.; Mariethoz, J.; Gasteiger, E.; Akune, Y.; Aoki-Kinoshita, K.F.; Lisacek, F.; Packer, N.H. UniCarbKB: building a knowledge platform for glycoproteomics. Nucleic Acids Res 2014, 42, D215-221. [CrossRef]
- Li, X.; Xu, Z.; Hong, X.; Zhang, Y.; Zou, X. Databases and Bioinformatic Tools for Glycobiology and Glycoproteomics. Int J Mol Sci 2020, 21. [CrossRef]
- Toukach, P.V.; Egorova, K.S. Carbohydrate structure database merged from bacterial, archaeal, plant and fungal parts. Nucleic Acids Res 2016, 44, D1229-1236. [CrossRef]
- Scherbinina, S.I.; Toukach, P.V. Three-Dimensional Structures of Carbohydrates and Where to Find Them. Int J Mol Sci 2020, 21. [CrossRef]
- Bohm, M.; Bohne-Lang, A.; Frank, M.; Loss, A.; Rojas-Macias, M.A.; Lutteke, T. Glycosciences.DB: an annotated data collection linking glycomics and proteomics data (2018 update). Nucleic Acids Res 2019, 47, D1195-D1201. [CrossRef]
- Aoki-Kinoshita, K.F.; Kanehisa, M. Glycomic Analysis Using KEGG GLYCAN. In Glycoinformatics, Lütteke, T., Frank, M., Eds.; Springer New York: New York, NY, 2015; pp. 97-107.
- Maeda, M.; Fujita, N.; Suzuki, Y.; Sawaki, H.; Shikanai, T.; Narimatsu, H. JCGGDB: Japan Consortium for Glycobiology and Glycotechnology Database. In Glycoinformatics, Lütteke, T., Frank, M., Eds.; Springer New York: New York, NY, 2015; pp. 161-179.
- Toukach, P.V.; Egorova, K.S. Source files of the Carbohydrate Structure Database: the way to sophisticated analysis of natural glycans. Scientific Data 2022, 9, 131. [CrossRef]
- Sun, S.; Hu, Y.; Ao, M.; Shah, P.; Chen, J.; Yang, W.; Jia, X.; Tian, Y.; Thomas, S.; Zhang, H. N-GlycositeAtlas: a database resource for mass spectrometry-based human N-linked glycoprotein and glycosylation site mapping. Clin Proteomics 2019, 16, 35. [CrossRef]
- Abrahams, J.L.; Taherzadeh, G.; Jarvas, G.; Guttman, A.; Zhou, Y.; Campbell, M.P. Recent advances in glycoinformatic platforms for glycomics and glycoproteomics. Curr Opin Struct Biol 2020, 62, 56-69. [CrossRef]
- Egorova, K.S.; Toukach, P.V. Glycoinformatics: Bridging Isolated Islands in the Sea of Data. Angew Chem Int Ed Engl 2018, 57, 14986-14990. [CrossRef]
- Breitling, J.; Aebi, M. N-linked protein glycosylation in the endoplasmic reticulum. Cold Spring Harb Perspect Biol 2013, 5, a013359. [CrossRef]
- Hanson, S.R.; Culyba, E.K.; Hsu, T.L.; Wong, C.H.; Kelly, J.W.; Powers, E.T. The core trisaccharide of an N-linked glycoprotein intrinsically accelerates folding and enhances stability. Proc Natl Acad Sci U S A 2009, 106, 3131-3136. [CrossRef]
- Xu, C.; Ng, D.T. Glycosylation-directed quality control of protein folding. Nat Rev Mol Cell Biol 2015, 16, 742-752. [CrossRef]
- Ohtsubo, K.; Marth, J.D. Glycosylation in cellular mechanisms of health and disease. Cell 2006, 126, 855-867. [CrossRef]
- Wolfert, M.A.; Boons, G.J. Adaptive immune activation: glycosylation does matter. Nat Chem Biol 2013, 9, 776-784. [CrossRef]
- Dzobo, K.; Dandara, C. The Extracellular Matrix: Its Composition, Function, Remodeling, and Role in Tumorigenesis. Biomimetics (Basel) 2023, 8. [CrossRef]
- Pinho, S.S.; Alves, I.; Gaifem, J.; Rabinovich, G.A. Immune regulatory networks coordinated by glycans and glycan-binding proteins in autoimmunity and infection. Cellular & Molecular Immunology 2023, 20, 1101-1113. [CrossRef]
- Stowell, S.R.; Ju, T.; Cummings, R.D. Protein glycosylation in cancer. Annu Rev Pathol 2015, 10, 473-510. [CrossRef]
- Taniguchi, N.; Kizuka, Y. Glycans and cancer: role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv Cancer Res 2015, 126, 11-51. [CrossRef]
- Chandler, K.B.; Costello, C.E.; Rahimi, N. Glycosylation in the Tumor Microenvironment: Implications for Tumor Angiogenesis and Metastasis. Cells 2019, 8. [CrossRef]
- Costa, A.F.; Campos, D.; Reis, C.A.; Gomes, C. Targeting Glycosylation: A New Road for Cancer Drug Discovery. Trends Cancer 2020, 6, 757-766. [CrossRef]
- Mereiter, S.; Balmana, M.; Campos, D.; Gomes, J.; Reis, C.A. Glycosylation in the Era of Cancer-Targeted Therapy: Where Are We Heading? Cancer Cell 2019, 36, 6-16. [CrossRef]
- Thomas, D.; Rathinavel, A.K.; Radhakrishnan, P. Altered glycosylation in cancer: A promising target for biomarkers and therapeutics. Biochim Biophys Acta Rev Cancer 2021, 1875, 188464. [CrossRef]
- Haukedal, H.; Freude, K.K. Implications of Glycosylation in Alzheimer's Disease. Front Neurosci 2020, 14, 625348. [CrossRef]
- Pradeep, P.; Kang, H.; Lee, B. Glycosylation and behavioral symptoms in neurological disorders. Transl Psychiatry 2023, 13, 154. [CrossRef]
- Zhao, J.; Lang, M. New insight into protein glycosylation in the development of Alzheimer's disease. Cell Death Discov 2023, 9, 314. [CrossRef]
- Conroy, L.R.; Hawkinson, T.R.; Young, L.E.A.; Gentry, M.S.; Sun, R.C. Emerging roles of N-linked glycosylation in brain physiology and disorders. Trends Endocrinol Metab 2021, 32, 980-993. [CrossRef]
- Paprocka, J.; Jezela-Stanek, A.; Tylki-Szymanska, A.; Grunewald, S. Congenital Disorders of Glycosylation from a Neurological Perspective. Brain Sci 2021, 11. [CrossRef]
- Groux-Degroote, S.; Cavdarli, S.; Uchimura, K.; Allain, F.; Delannoy, P. Glycosylation changes in inflammatory diseases. Adv Protein Chem Struct Biol 2020, 119, 111-156. [CrossRef]
- Kissel, T.; Toes, R.E.M.; Huizinga, T.W.J.; Wuhrer, M. Glycobiology of rheumatic diseases. Nat Rev Rheumatol 2023, 19, 28-43. [CrossRef]
- Loke, I.; Kolarich, D.; Packer, N.H.; Thaysen-Andersen, M. Emerging roles of protein mannosylation in inflammation and infection. Mol Aspects Med 2016, 51, 31-55. [CrossRef]
- Ugonotti, J.; Chatterjee, S.; Thaysen-Andersen, M. Structural and functional diversity of neutrophil glycosylation in innate immunity and related disorders. Mol Aspects Med 2021, 79, 100882. [CrossRef]
- Lin, B.; Qing, X.; Liao, J.; Zhuo, K. Role of Protein Glycosylation in Host-Pathogen Interaction. Cells 2020, 9. [CrossRef]
- Wang, S.H.; Wu, T.J.; Lee, C.W.; Yu, J. Dissecting the conformation of glycans and their interactions with proteins. J Biomed Sci 2020, 27, 93. [CrossRef]
- Varki, A.; Freeze, H.H. Glycans in Acquired Human Diseases. In Essentials of Glycobiology, Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor Laboratory Press Copyright © 2009, The Consortium of Glycobiology Editors, La Jolla, California.: Cold Spring Harbor (NY), 2009.
- Zhou, J.Y.; Cobb, B.A. Glycans in Immunologic Health and Disease. Annual Review of Immunology 2021, 39, 511-536. [CrossRef]
- Barb, A.W. Fc gamma receptor compositional heterogeneity: Considerations for immunotherapy development. J Biol Chem 2021, 296, 100057. [CrossRef]
- Saporiti, S.; Parravicini, C.; Pergola, C.; Guerrini, U.; Rossi, M.; Centola, F.; Eberini, I. IgG1 conformational behavior: elucidation of the N-glycosylation role via molecular dynamics. Biophys J 2021, 120, 5355-5370. [CrossRef]
- Subedi, G.P.; Hanson, Q.M.; Barb, A.W. Restricted motion of the conserved immunoglobulin G1 N-glycan is essential for efficient FcgammaRIIIa binding. Structure 2014, 22, 1478-1488. [CrossRef]
- Sun, Y.; Izadi, S.; Callahan, M.; Deperalta, G.; Wecksler, A.T. Antibody-receptor interactions mediate antibody-dependent cellular cytotoxicity. J Biol Chem 2021, 297, 100826. [CrossRef]
- Chen, B.; Liu, W.; Li, Y.; Ma, B.; Shang, S.; Tan, Z. Impact of N-Linked Glycosylation on Therapeutic Proteins. Molecules 2022, 27. [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer 2015, 15, 540-555. [CrossRef]
- Buffone, A.; Weaver, V.M. Don't sugarcoat it: How glycocalyx composition influences cancer progression. J Cell Biol 2020, 219. [CrossRef]
- Lee, H.S.; Qi, Y.; Im, W. Effects of N-glycosylation on protein conformation and dynamics: Protein Data Bank analysis and molecular dynamics simulation study. Sci Rep 2015, 5, 8926. [CrossRef]
- Mule, S.N.; Rosa-Fernandes, L.; Coutinho, J.V.P.; Gomes, V.M.; Macedo-da-Silva, J.; Santiago, V.F.; Quina, D.; de Oliveira, G.S.; Thaysen-Andersen, M.; Larsen, M.R.; et al. Systems-wide analysis of glycoprotein conformational changes by limited deglycosylation assay. J Proteomics 2021, 248, 104355. [CrossRef]
- Papaleo, E.; Saladino, G.; Lambrughi, M.; Lindorff-Larsen, K.; Gervasio, F.L.; Nussinov, R. The Role of Protein Loops and Linkers in Conformational Dynamics and Allostery. Chemical Reviews 2016, 116, 6391-6423. [CrossRef]
- Cascella, M.; Rajnik, M.; Aleem, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation, and Treatment of Coronavirus (COVID-19). In StatPearls; Treasure Island (FL), 2023.
- Casalino, L.; Gaieb, Z.; Goldsmith, J.A.; Hjorth, C.K.; Dommer, A.C.; Harbison, A.M.; Fogarty, C.A.; Barros, E.P.; Taylor, B.C.; McLellan, J.S.; et al. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Cent Sci 2020, 6, 1722-1734. [CrossRef]
- Malaquias, M.A.S.; Gadotti, A.C.; Motta-Junior, J.D.S.; Martins, A.P.C.; Azevedo, M.L.V.; Benevides, A.P.K.; Cezar-Neto, P.; Panini do Carmo, L.A.; Zeni, R.C.; Raboni, S.M.; et al. The role of the lectin pathway of the complement system in SARS-CoV-2 lung injury. Transl Res 2021, 231, 55-63. [CrossRef]
- Jiang, S.; Hillyer, C.; Du, L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses: (Trends in Immunology 41, 355-359; 2020). Trends Immunol 2020, 41, 545. [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260-1263. [CrossRef]
- Du, L.; He, Y.; Zhou, Y.; Liu, S.; Zheng, B.J.; Jiang, S. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat Rev Microbiol 2009, 7, 226-236. [CrossRef]
- Song, W.; Gui, M.; Wang, X.; Xiang, Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog 2018, 14, e1007236. [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271-280.e278. [CrossRef]
- Benton, D.J.; Wrobel, A.G.; Xu, P.; Roustan, C.; Martin, S.R.; Rosenthal, P.B.; Skehel, J.J.; Gamblin, S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 2020, 588, 327-330. [CrossRef]
- Lu, M.; Uchil, P.D.; Li, W.; Zheng, D.; Terry, D.S.; Gorman, J.; Shi, W.; Zhang, B.; Zhou, T.; Ding, S.; et al. Real-Time Conformational Dynamics of SARS-CoV-2 Spikes on Virus Particles. Cell Host Microbe 2020, 28, 880-891 e888. [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 183, 1735. [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215-220. [CrossRef]
- Tian, Y.; Parsons, L.M.; Jankowska, E.; Cipollo, J.F. Site-Specific Glycosylation Patterns of the SARS-CoV-2 Spike Protein Derived From Recombinant Protein and Viral WA1 and D614G Strains. Front Chem 2021, 9, 767448. [CrossRef]
- Cai, Y.; Zhang, J.; Xiao, T.; Peng, H.; Sterling, S.M.; Walsh, R.M., Jr.; Rawson, S.; Rits-Volloch, S.; Chen, B. Distinct conformational states of SARS-CoV-2 spike protein. Science 2020, 369, 1586-1592. [CrossRef]
- Fan, X.; Cao, D.; Kong, L.; Zhang, X. Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein. Nat Commun 2020, 11, 3618. [CrossRef]
- Harbison, A.M.; Fogarty, C.A.; Phung, T.K.; Satheesan, A.; Schulz, B.L.; Fadda, E. Fine-tuning the spike: role of the nature and topology of the glycan shield in the structure and dynamics of the SARS-CoV-2 S. Chem Sci 2022, 13, 386-395. [CrossRef]
- Ray, D.; Le, L.; Andricioaei, I. Distant residues modulate conformational opening in SARS-CoV-2 spike protein. Proc Natl Acad Sci U S A 2021, 118. [CrossRef]
- Zimmerman, M.I.; Porter, J.R.; Ward, M.D.; Singh, S.; Vithani, N.; Meller, A.; Mallimadugula, U.L.; Kuhn, C.E.; Borowsky, J.H.; Wiewiora, R.P.; et al. SARS-CoV-2 simulations go exascale to predict dramatic spike opening and cryptic pockets across the proteome. Nat Chem 2021, 13, 651-659. [CrossRef]
- Fallon, L.; Belfon, K.A.A.; Raguette, L.; Wang, Y.; Stepanenko, D.; Cuomo, A.; Guerra, J.; Budhan, S.; Varghese, S.; Corbo, C.P.; et al. Free Energy Landscapes from SARS-CoV-2 Spike Glycoprotein Simulations Suggest that RBD Opening Can Be Modulated via Interactions in an Allosteric Pocket. Journal of the American Chemical Society 2021, 143, 11349-11360. [CrossRef]
- Sztain, T.; Ahn, S.H.; Bogetti, A.T.; Casalino, L.; Goldsmith, J.A.; Seitz, E.; McCool, R.S.; Kearns, F.L.; Acosta-Reyes, F.; Maji, S.; et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat Chem 2021, 13, 963-968. [CrossRef]
- Zhang, B.W.; Jasnow, D.; Zuckerman, D.M. The “weighted ensemble” path sampling method is statistically exact for a broad class of stochastic processes and binning procedures. The Journal of Chemical Physics 2010, 132, 054107. [CrossRef]
- Pang, Y.T.; Acharya, A.; Lynch, D.L.; Pavlova, A.; Gumbart, J.C. SARS-CoV-2 spike opening dynamics and energetics reveal the individual roles of glycans and their collective impact. Commun Biol 2022, 5, 1170. [CrossRef]
- Dodero-Rojas, E.; Onuchic, J.N.; Whitford, P.C. Sterically confined rearrangements of SARS-CoV-2 Spike protein control cell invasion. Elife 2021, 10. [CrossRef]
- Geschwind, M.D. Prion Diseases. Continuum (Minneap Minn) 2015, 21, 1612-1638. [CrossRef]
- Collinge, J. Mammalian prions and their wider relevance in neurodegenerative diseases. Nature 2016, 539, 217-226. [CrossRef]
- Johnson, R.T. Prion diseases. The Lancet Neurology 2005, 4, 635-642. [CrossRef]
- Westergard, L.; Christensen, H.M.; Harris, D.A. The cellular prion protein (PrPC): Its physiological function and role in disease. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 2007, 1772, 629-644. [CrossRef]
- Ermonval, M.; Mouillet-Richard, S.; Codogno, P.; Kellermann, O.; Botti, J. Evolving views in prion glycosylation: functional and pathological implications. Biochimie 2003, 85, 33-45. [CrossRef]
- Xiao, X.; Yuan, J.; Haik, S.; Cali, I.; Zhan, Y.; Moudjou, M.; Li, B.; Laplanche, J.L.; Laude, H.; Langeveld, J.; et al. Glycoform-selective prion formation in sporadic and familial forms of prion disease. PLoS One 2013, 8, e58786. [CrossRef]
- Mallucci, G.; Dickinson, A.; Linehan, J.; Klohn, P.C.; Brandner, S.; Collinge, J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science 2003, 302, 871-874. [CrossRef]
- Aguilar-Calvo, P.; Callender, J.A.; Sigurdson, C.J. Short and sweet: How glycans impact prion conversion, cofactor interactions, and cross-species transmission. PLOS Pathogens 2021, 17, e1009123. [CrossRef]
- Aguilar-Calvo, P.; Xiao, X.; Bett, C.; Erana, H.; Soldau, K.; Castilla, J.; Nilsson, K.P.; Surewicz, W.K.; Sigurdson, C.J. Post-translational modifications in PrP expand the conformational diversity of prions in vivo. Sci Rep 2017, 7, 43295. [CrossRef]
- Angers, R.C.; Kang, H.E.; Napier, D.; Browning, S.; Seward, T.; Mathiason, C.; Balachandran, A.; McKenzie, D.; Castilla, J.; Soto, C.; et al. Prion strain mutation determined by prion protein conformational compatibility and primary structure. Science 2010, 328, 1154-1158. [CrossRef]
- Nakic, N.; Tran, T.H.; Novokmet, M.; Andreoletti, O.; Lauc, G.; Legname, G. Site-specific analysis of N-glycans from different sheep prion strains. PLoS Pathog 2021, 17, e1009232. [CrossRef]
- Sevillano, A.M.; Aguilar-Calvo, P.; Kurt, T.D.; Lawrence, J.A.; Soldau, K.; Nam, T.H.; Schumann, T.; Pizzo, D.P.; Nystrom, S.; Choudhury, B.; et al. Prion protein glycans reduce intracerebral fibril formation and spongiosis in prion disease. J Clin Invest 2020, 130, 1350-1362. [CrossRef]
- Corsaro, A.; Thellung, S.; Villa, V.; Nizzari, M.; Florio, T. Role of prion protein aggregation in neurotoxicity. Int J Mol Sci 2012, 13, 8648-8669. [CrossRef]
- Poggiolini, I.; Saverioni, D.; Parchi, P. Prion protein misfolding, strains, and neurotoxicity: an update from studies on Mammalian prions. Int J Cell Biol 2013, 2013, 910314. [CrossRef]
- Salamat, M.K.; Dron, M.; Chapuis, J.; Langevin, C.; Laude, H. Prion propagation in cells expressing PrP glycosylation mutants. J Virol 2011, 85, 3077-3085. [CrossRef]
- Cancellotti, E.; Mahal, S.P.; Somerville, R.; Diack, A.; Brown, D.; Piccardo, P.; Weissmann, C.; Manson, J.C. Post-translational changes to PrP alter transmissible spongiform encephalopathy strain properties. EMBO J 2013, 32, 756-769. [CrossRef]
- Yi, C.W.; Wang, L.Q.; Huang, J.J.; Pan, K.; Chen, J.; Liang, Y. Glycosylation Significantly Inhibits the Aggregation of Human Prion Protein and Decreases Its Cytotoxicity. Sci Rep 2018, 8, 12603. [CrossRef]
- Schilling, K.M.; Jorwal, P.; Ubilla-Rodriguez, N.C.; Assafa, T.E.; Gatdula, J.R.P.; Vultaggio, J.S.; Harris, D.A.; Millhauser, G.L. N-glycosylation is a potent regulator of prion protein neurotoxicity. J Biol Chem 2023, 299, 105101. [CrossRef]
- DeArmond, S.J.; Sanchez, H.; Yehiely, F.; Qiu, Y.; Ninchak-Casey, A.; Daggett, V.; Camerino, A.P.; Cayetano, J.; Rogers, M.; Groth, D.; et al. Selective neuronal targeting in prion disease. Neuron 1997, 19, 1337-1348. [CrossRef]
- Giachin, G.; Biljan, I.; Ilc, G.; Plavec, J.; Legname, G. Probing early misfolding events in prion protein mutants by NMR spectroscopy. Molecules 2013, 18, 9451-9476. [CrossRef]
- Riek, R.; Hornemann, S.; Wider, G.; Glockshuber, R.; Wuthrich, K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23-231). FEBS Lett 1997, 413, 282-288. [CrossRef]
- Zuegg, J.; Gready, J.E. Molecular dynamics simulation of human prion protein including both N-linked oligosaccharides and the GPI anchor. Glycobiology 2000, 10, 959-974. [CrossRef]
- Schilling, K.M.; Tao, L.; Wu, B.; Kiblen, J.T.M.; Ubilla-Rodriguez, N.C.; Pushie, M.J.; Britt, R.D.; Roseman, G.P.; Harris, D.A.; Millhauser, G.L. Both N-Terminal and C-Terminal Histidine Residues of the Prion Protein Are Essential for Copper Coordination and Neuroprotective Self-Regulation. Journal of Molecular Biology 2020, 432, 4408-4425. [CrossRef]
- Evans, Eric G.B.; Pushie, M.J.; Markham, Kate A.; Lee, H.-W.; Millhauser, Glenn L. Interaction between Prion Protein's Copper-Bound Octarepeat Domain and a Charged C-Terminal Pocket Suggests a Mechanism for N-Terminal Regulation. Structure 2016, 24, 1057-1067. [CrossRef]
- Spevacek, Ann R.; Evans, Eric G.B.; Miller, Jillian L.; Meyer, Heidi C.; Pelton, Jeffrey G.; Millhauser, Glenn L. Zinc Drives a Tertiary Fold in the Prion Protein with Familial Disease Mutation Sites at the Interface. Structure 2013, 21, 236-246. [CrossRef]
- Aguilar-Calvo, P.; Sevillano, A.M.; Bapat, J.; Soldau, K.; Sandoval, D.R.; Altmeppen, H.C.; Linsenmeier, L.; Pizzo, D.P.; Geschwind, M.D.; Sanchez, H.; et al. Shortening heparan sulfate chains prolongs survival and reduces parenchymal plaques in prion disease caused by mobile, ADAM10-cleaved prions. Acta Neuropathol 2020, 139, 527-546. [CrossRef]
- Marzec, M.; Eletto, D.; Argon, Y. GRP94: An HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochim Biophys Acta 2012, 1823, 774-787. [CrossRef]
- Ansa-Addo, E.A.; Thaxton, J.; Hong, F.; Wu, B.X.; Zhang, Y.; Fugle, C.W.; Metelli, A.; Riesenberg, B.; Williams, K.; Gewirth, D.T.; et al. Clients and Oncogenic Roles of Molecular Chaperone gp96/grp94. Curr Top Med Chem 2016, 16, 2765-2778. [CrossRef]
- Eletto, D.; Dersh, D.; Argon, Y. GRP94 in ER quality control and stress responses. Semin Cell Dev Biol 2010, 21, 479-485. [CrossRef]
- Lee, A.S. Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nat Rev Cancer 2014, 14, 263-276. [CrossRef]
- Wiersma, V.R.; Michalak, M.; Abdullah, T.M.; Bremer, E.; Eggleton, P. Mechanisms of Translocation of ER Chaperones to the Cell Surface and Immunomodulatory Roles in Cancer and Autoimmunity. Front Oncol 2015, 5, 7. [CrossRef]
- Altmeyer, A.; Maki, R.G.; Feldweg, A.M.; Heike, M.; Protopopov, V.P.; Masur, S.K.; Srivastava, P.K. Tumor-specific cell surface expression of the-KDEL containing, endoplasmic reticular heat shock protein gp96. Int J Cancer 1996, 69, 340-349. [CrossRef]
- Booth, C.; Koch, G.L. Perturbation of cellular calcium induces secretion of luminal ER proteins. Cell 1989, 59, 729-737. [CrossRef]
- Martins, M.; Custodio, R.; Camejo, A.; Almeida, M.T.; Cabanes, D.; Sousa, S. Listeria monocytogenes triggers the cell surface expression of Gp96 protein and interacts with its N terminus to support cellular infection. J Biol Chem 2012, 287, 43083-43093. [CrossRef]
- Rothan, H.A.; Zhong, Y.; Sanborn, M.A.; Teoh, T.C.; Ruan, J.; Yusof, R.; Hang, J.; Henderson, M.J.; Fang, S. Small molecule grp94 inhibitors block dengue and Zika virus replication. Antiviral Res 2019, 171, 104590. [CrossRef]
- Sumitomo, T.; Nakata, M.; Nagase, S.; Takahara, Y.; Honda-Ogawa, M.; Mori, Y.; Akamatsu, Y.; Yamaguchi, M.; Okamoto, S.; Kawabata, S. GP96 Drives Exacerbation of Secondary Bacterial Pneumonia following Influenza A Virus Infection. mBio 2021, 12, e0326920. [CrossRef]
- Liu, B.; Dai, J.; Zheng, H.; Stoilova, D.; Sun, S.; Li, Z. Cell surface expression of an endoplasmic reticulum resident heat shock protein gp96 triggers MyD88-dependent systemic autoimmune diseases. Proc Natl Acad Sci U S A 2003, 100, 15824-15829. [CrossRef]
- Li, X.; Sun, L.; Hou, J.; Gui, M.; Ying, J.; Zhao, H.; Lv, N.; Meng, S. Cell membrane gp96 facilitates HER2 dimerization and serves as a novel target in breast cancer. Int J Cancer 2015, 137, 512-524. [CrossRef]
- Patel, P.D.; Yan, P.; Seidler, P.M.; Patel, H.J.; Sun, W.; Yang, C.; Que, N.S.; Taldone, T.; Finotti, P.; Stephani, R.A.; et al. Paralog-selective Hsp90 inhibitors define tumor-specific regulation of HER2. Nat Chem Biol 2013, 9, 677-684. [CrossRef]
- Yan, P.; Patel, H.J.; Sharma, S.; Corben, A.; Wang, T.; Panchal, P.; Yang, C.; Sun, W.; Araujo, T.L.; Rodina, A.; et al. Molecular Stressors Engender Protein Connectivity Dysfunction through Aberrant N-Glycosylation of a Chaperone. Cell Rep 2020, 31, 107840. [CrossRef]
- Chavany, C.; Mimnaugh, E.; Miller, P.; Bitton, R.; Nguyen, P.; Trepel, J.; Whitesell, L.; Schnur, R.; Moyer, J.; Neckers, L. p185erbB2 binds to GRP94 in vivo. Dissociation of the p185erbB2/GRP94 heterocomplex by benzoquinone ansamycins precedes depletion of p185erbB2. J Biol Chem 1996, 271, 4974-4977. [CrossRef]
- Chaumonnot, K.; Masson, S.; Sikner, H.; Bouchard, A.; Baverel, V.; Bellaye, P.S.; Collin, B.; Garrido, C.; Kohli, E. The HSP GRP94 interacts with macrophage intracellular complement C3 and impacts M2 profile during ER stress. Cell Death Dis 2021, 12, 114. [CrossRef]
- Ratna, A.; Lim, A.; Li, Z.; Argemi, J.; Bataller, R.; Chiosis, G.; Mandrekar, P. Myeloid Endoplasmic Reticulum Resident Chaperone GP96 Facilitates Inflammation and Steatosis in Alcohol-Associated Liver Disease. Hepatol Commun 2021, 5, 1165-1182. [CrossRef]
- Pagetta, A.; Tramentozzi, E.; Tibaldi, E.; Cendron, L.; Zanotti, G.; Brunati, A.M.; Vitadello, M.; Gorza, L.; Finotti, P. Structural insights into complexes of glucose-regulated Protein94 (Grp94) with human immunoglobulin G. relevance for Grp94-IgG complexes that form in vivo in pathological conditions. PLoS One 2014, 9, e86198. [CrossRef]
- Cherepanova, N.A.; Venev, S.V.; Leszyk, J.D.; Shaffer, S.A.; Gilmore, R. Quantitative glycoproteomics reveals new classes of STT3A- and STT3B-dependent N-glycosylation sites. J Cell Biol 2019, 218, 2782-2796. [CrossRef]
- Wearsch, P.A.; Nicchitta, C.V. Purification and partial molecular characterization of GRP94, an ER resident chaperone. Protein Expr Purif 1996, 7, 114-121. [CrossRef]
- Wen, P.; Chen, J.; Zuo, C.; Gao, X.; Fujita, M.; Yang, G. Proteome and Glycoproteome Analyses Reveal the Protein N-Linked Glycosylation Specificity of STT3A and STT3B. Cells 2022, 11. [CrossRef]
- Gottschalk, N.; Kimmig, R.; Lang, S.; Singh, M.; Brandau, S. Anti-epidermal growth factor receptor (EGFR) antibodies overcome resistance of ovarian cancer cells to targeted therapy and natural cytotoxicity. Int J Mol Sci 2012, 13, 12000-12016. [CrossRef]
- Sun, S.; Zhang, H. Large-Scale Measurement of Absolute Protein Glycosylation Stoichiometry. Anal Chem 2015, 87, 6479-6482. [CrossRef]
- Castelli, M.; Yan, P.; Rodina, A.; Digwal, C.S.; Panchal, P.; Chiosis, G.; Moroni, E.; Colombo, G. How aberrant N-glycosylation can alter protein functionality and ligand binding: An atomistic view. Structure 2023, 31, 987-1004.e1008. [CrossRef]
- Bouchard, A.; Sikner, H.; Baverel, V.; Garnier, A.R.; Monterrat, M.; Moreau, M.; Limagne, E.; Garrido, C.; Kohli, E.; Collin, B.; et al. The GRP94 Inhibitor PU-WS13 Decreases M2-like Macrophages in Murine TNBC Tumors: A Pharmaco-Imaging Study with (99m)Tc-Tilmanocept SPECT. Cells 2021, 10. [CrossRef]
- Rodina, A.; Xu, C.; Digwal, C.S.; Joshi, S.; Patel, Y.; Santhaseela, A.R.; Bay, S.; Merugu, S.; Alam, A.; Yan, P.; et al. Systems-level analyses of protein-protein interaction network dysfunctions via epichaperomics identify cancer-specific mechanisms of stress adaptation. Nature Communications 2023, 14, 3742. [CrossRef]
- Chiosis, G.; Digwal, C.S.; Trepel, J.B.; Neckers, L. Structural and functional complexity of HSP90 in cellular homeostasis and disease. Nature Reviews Molecular Cell Biology 2023, 24, 797-815. [CrossRef]
- Rodina, A.; Wang, T.; Yan, P.; Gomes, E.D.; Dunphy, M.P.; Pillarsetty, N.; Koren, J.; Gerecitano, J.F.; Taldone, T.; Zong, H.; et al. The epichaperome is an integrated chaperome network that facilitates tumour survival. Nature 2016, 538, 397-401. [CrossRef]
- Barouch, D.H. Covid-19 Vaccines - Immunity, Variants, Boosters. N Engl J Med 2022, 387, 1011-1020. [CrossRef]
- Corti, D.; Purcell, L.A.; Snell, G.; Veesler, D. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell 2021, 184, 4593-4595. [CrossRef]
- Raybould, M.I.J.; Kovaltsuk, A.; Marks, C.; Deane, C.M. CoV-AbDab: the coronavirus antibody database. Bioinformatics 2021, 37, 734-735. [CrossRef]
- Voss, W.N.; Hou, Y.J.; Johnson, N.V.; Delidakis, G.; Kim, J.E.; Javanmardi, K.; Horton, A.P.; Bartzoka, F.; Paresi, C.J.; Tanno, Y.; et al. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes. Science 2021, 372, 1108-1112. [CrossRef]
- Serapian, S.A.; Marchetti, F.; Triveri, A.; Morra, G.; Meli, M.; Moroni, E.; Sautto, G.A.; Rasola, A.; Colombo, G. The Answer Lies in the Energy: How Simple Atomistic Molecular Dynamics Simulations May Hold the Key to Epitope Prediction on the Fully Glycosylated SARS-CoV-2 Spike Protein. The Journal of Physical Chemistry Letters 2020, 11, 8084-8093. [CrossRef]
- Triveri, A.; Serapian, S.A.; Marchetti, F.; Doria, F.; Pavoni, S.; Cinquini, F.; Moroni, E.; Rasola, A.; Frigerio, F.; Colombo, G. SARS-CoV-2 Spike Protein Mutations and Escape from Antibodies: A Computational Model of Epitope Loss in Variants of Concern. Journal of Chemical Information and Modeling 2021, 61, 4687-4700. [CrossRef]
- Triveri, A.; Casali, E.; Frasnetti, E.; Doria, F.; Frigerio, F.; Cinquini, F.; Pavoni, S.; Moroni, E.; Marchetti, F.; Serapian, S.A.; et al. Conformational Behavior of SARS-Cov-2 Spike Protein Variants: Evolutionary Jumps in Sequence Reverberate in Structural Dynamic Differences. Journal of Chemical Theory and Computation 2023, 19, 2120-2134. [CrossRef]
- Mead, S.; Whitfield, J.; Poulter, M.; Shah, P.; Uphill, J.; Campbell, T.; Al-Dujaily, H.; Hummerich, H.; Beck, J.; Mein, C.A.; et al. A novel protective prion protein variant that colocalizes with kuru exposure. N Engl J Med 2009, 361, 2056-2065. [CrossRef]
- Asante, E.A.; Smidak, M.; Grimshaw, A.; Houghton, R.; Tomlinson, A.; Jeelani, A.; Jakubcova, T.; Hamdan, S.; Richard-Londt, A.; Linehan, J.M.; et al. A naturally occurring variant of the human prion protein completely prevents prion disease. Nature 2015, 522, 478-481. [CrossRef]
- Zheng, Z.; Zhang, M.; Wang, Y.; Ma, R.; Guo, C.; Feng, L.; Wu, J.; Yao, H.; Lin, D. Structural basis for the complete resistance of the human prion protein mutant G127V to prion disease. Sci Rep 2018, 8, 13211. [CrossRef]
- Kuwata, K. Logical design of medical chaperone for prion diseases. Curr Top Med Chem 2013, 13, 2432-2440. [CrossRef]
- Yamaguchi, K.; Kamatari, Y.O.; Ono, F.; Shibata, H.; Fuse, T.; Elhelaly, A.E.; Fukuoka, M.; Kimura, T.; Hosokawa-Muto, J.; Ishikawa, T.; et al. A designer molecular chaperone against transmissible spongiform encephalopathy slows disease progression in mice and macaques. Nat Biomed Eng 2019, 3, 206-219. [CrossRef]
- Lysek, D.A.; Schorn, C.; Nivon, L.G.; Esteve-Moya, V.; Christen, B.; Calzolai, L.; von Schroetter, C.; Fiorito, F.; Herrmann, T.; Guntert, P.; et al. Prion protein NMR structures of cats, dogs, pigs, and sheep. Proc Natl Acad Sci U S A 2005, 102, 640-645. [CrossRef]
- Duan, X.; Iwanowycz, S.; Ngoi, S.; Hill, M.; Zhao, Q.; Liu, B. Molecular Chaperone GRP94/GP96 in Cancers: Oncogenesis and Therapeutic Target. Front Oncol 2021, 11, 629846. [CrossRef]
- Wu, B.X.; Hong, F.; Zhang, Y.; Ansa-Addo, E.; Li, Z. GRP94/gp96 in Cancer: Biology, Structure, Immunology, and Drug Development. Adv Cancer Res 2016, 129, 165-190. [CrossRef]
- Gewirth, D.T. Paralog Specific Hsp90 Inhibitors - A Brief History and a Bright Future. Curr Top Med Chem 2016, 16, 2779-2791. [CrossRef]
- Patel, H.J.; Patel, P.D.; Ochiana, S.O.; Yan, P.; Sun, W.; Patel, M.R.; Shah, S.K.; Tramentozzi, E.; Brooks, J.; Bolaender, A.; et al. Structure-activity relationship in a purine-scaffold compound series with selectivity for the endoplasmic reticulum Hsp90 paralog Grp94. J Med Chem 2015, 58, 3922-3943. [CrossRef]
- Shrestha, L.; Patel, H.J.; Chiosis, G. Chemical Tools to Investigate Mechanisms Associated with HSP90 and HSP70 in Disease. Cell Chem Biol 2016, 23, 158-172. [CrossRef]
- Sousa, S.; Brion, R.; Lintunen, M.; Kronqvist, P.; Sandholm, J.; Monkkonen, J.; Kellokumpu-Lehtinen, P.L.; Lauttia, S.; Tynninen, O.; Joensuu, H.; et al. Human breast cancer cells educate macrophages toward the M2 activation status. Breast Cancer Res 2015, 17, 101. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




