1. Introduction
Microbiota refers to the collection of microbes residing in and on the body of a host, with the highest density being in the gastrointestinal tract. Gut microbiota has been detected in nearly all metazoans both vertebrates and invertebrates and include bacteria, archaea, fungi, viruses and protozoa having mostly commensal or mutualistic behaviours [1] [2]. The genetic complement of these gut microorganisms is referred to as a second genome or a supporting organ that continuously interacts with the host [3] [4] [5]. The roles of gut microbiota are increasingly being documented and include contributions to health and disease states, as well as, to the physiology and homeostasis of the associated hosts [6]. A large portion of our current understanding comes from studies focusing on human microbiota, with comparatively less data being available on other animals. Given that resident microbial communities contribute to the well-being and health status of their hosts, it is crucial to explore these roles in food production animals most especially ruminants such as dairy cattle [7] [8] [9]. Production animals contribute almost 40% of the total agricultural output in developed countries and 20% in developing countries [10]. The agricultural industry ensures a constant food supply and food security for the population and is considered to be the backbone of the economy for developing countries [11]. For instance, the dairy cattle industry produces the largest proportion (82.7%) of global milk and is a rapidly growing sector with a projected increase of 177 million tonnes by 2025 to meet the mounting consumer demand on dairy products [12].
Studies that have researched the microbiome of production animals and their effect on ruminant hosts indicate that their microbiome composition and abundance is primarily shaped by the diet [8] [13]. Ruminants regurgitate and remasticate forage from their rumen to reduce the size of the pre-ingested bolus. This effectively increases the digestibility of the fibrous ingesta and facilitates passage to the small intestine. The rumen microbiome breaks down complex structural polysaccharides enabling nutrient absorption and consequently, indigestible plant biomass is converted to energy and further to milk or meat production for human consumption [14] [15].
Pre-existing studies have largely concentrated on the microbiome of the rumen, whereas similar investigations on the large intestinal gut (lower gut) and its effect on the host’s health and welfare are minimal. In order to uncover the diversity and composition of the cattle microbiome, a global approach of the whole gastrointestinal tract is necessary. Similar to the rumen microbiome, the specific composition of the lower gut microbiome can also influence feed and milk production efficiency and quality in cattle as well as the production of methane, a significant greenhouse gas contributor [9] [16] [17]. The lower gut microbiome is also involved in establishing a functional mucosal immune system in ruminants, which directly influences the host’s gut health and is the first line of defence against pathogens and disease [18]. Importantly, cows have been shown to be reservoirs for several zoonotic pathogens (Escherichia coli, Mycobacterium tuberculosis, Campylobacter, Cryptosporidium and Bacillus anthracis) and a healthy, diverse microbiome greatly contributes to the defense against them [19] [20].
To our knowledge, few studies exist taking a holistic approach investigating the role of microbiome of livestock in health and disease, especially in a geographically confined region. Herein, we describe the establishment of a ruminant gut microbiome biobank in Cyprus.
Why establish such a ruminant microbiome biobank in Cyprus? Cyprus is the third largest island in the Mediterranean with a unique climate, which influences its environment, biodiversity, and farm husbandry practices. All these constitute factors of varying effects on the ruminant gut microbiome diversity and composition. The biobank will provide an unprecedented opportunity for detailed investigation of the relationship between these distinctive characteristics and the ruminant microbiome in an island setting. The biobank is aimed at investigating the composition of the associated resident microbiome and its role in health and disease in dairy cattle and other ruminants, specifically in Cyprus. This will be the first agricultural biobank in Cyprus. Importantly, gathering this data will allow for understanding how microbial communities can enhance disease resistance, improve productivity, and mitigate the impact of infectious agents prevalent in livestock. We envision that the availability of samples will extend beyond the proposed investigations and will forge strong regional and international ties and networks.
Objectives
Secondary objectives
Determine the presence of zoonotic organisms in faecal, milk and environmental samples
Determine transmission dynamics of maternal microbiome to the offspring (calves)
Characterize antimicrobial resistant bacteria at the level of the dairy cattle farm
Investigate the interplay of gut microbiome and milk production
Explore associations between environmental factors (including geographical location) and husbandry practices on the composition and abundance of the gut microbiome.
Investigate links between dairy cattle gut microbiome profiles and pathogens (biomarkers)
Compare microbiomes of cattle raised under various farming practices
2. Materials and Methods
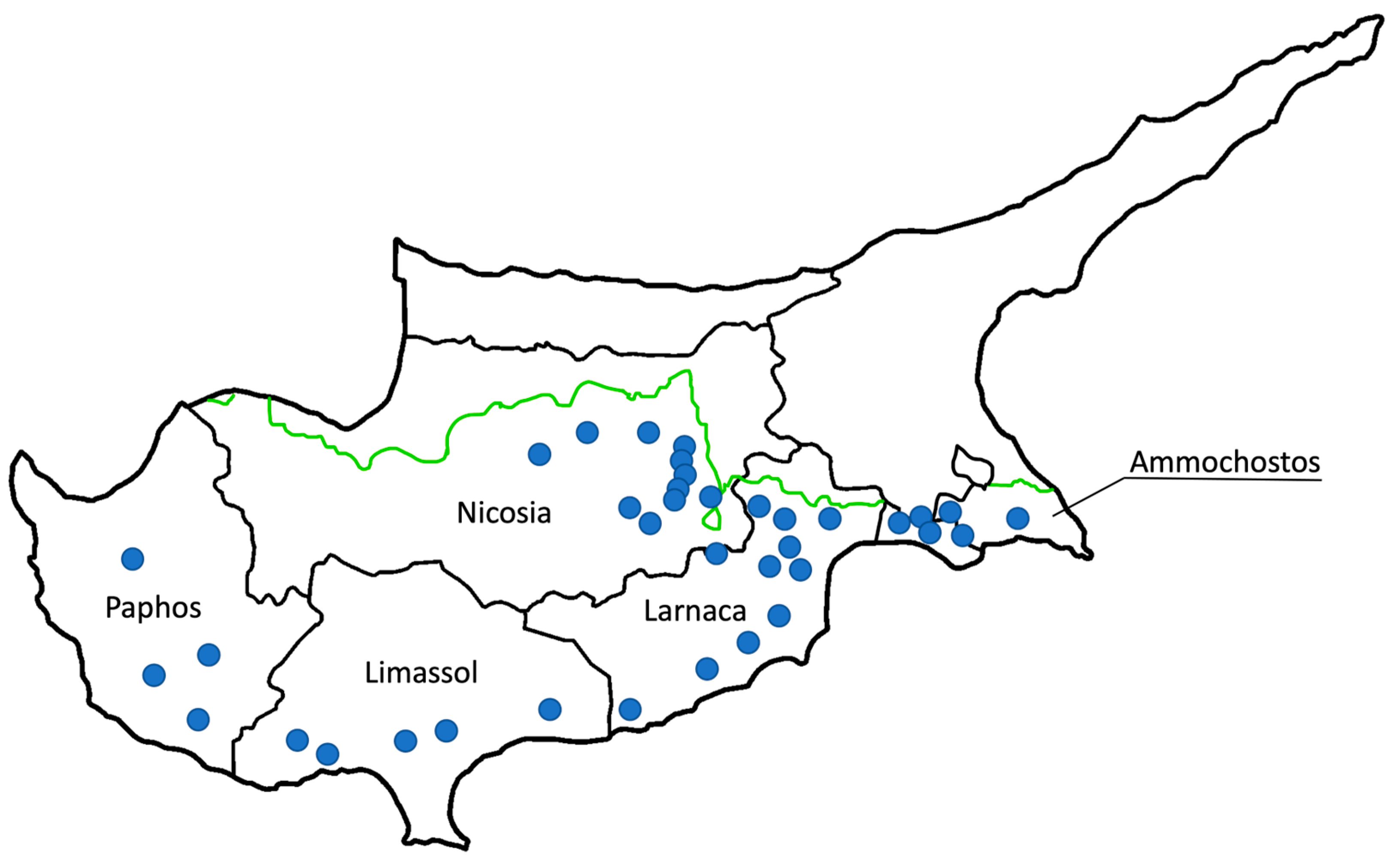
The study will focus on the Nicosia, Limassol, Larnaca, Ammochostos, and Paphos districts of Cyprus. Cattle farms in Cyprus tend to be of both dairy and beef production consisting mostly of the Holstein breed. All the bovine farms and animals are registered in the official database of the Cyprus Veterinary Services. Based on 2022 data, there is a total of 358 farms in these districts. Specifically, Ammochostos has 33 farms, Larnaca 131, Limassol 41, Nicosia 117, and Paphos has 36.
Farms housing more than 100 cows of the Holstein breed have been selected. The percentage of farms in each district with the selected criteria was calculated (
Table 1). Initially, a total of 37 farms will be sampled. The representative number of farms selected from each district were proportional to the total number of farms in that area. Within the districts, the selected farms are spread across different regions and altitudes to represent all the microclimates of the island. Farms in the districts of Nicosia and Larnaca are in closer proximity to each other compared to other districts as they have the highest aggregation of farms (
Figure 1). Thirty seven farms from five districts of Cyprus were selected. The ability to measure milk production and keep records to enable future follow-ups were also considered.
Sheets containing information on the study and its importance will be provided and consent forms will be obtained by the farmers participating in the study. Participants will complete questionnaires on diet, treatments (including antibiotic use), housing arrangements, hygiene practices and other husbandry aspects.
The project was submitted for bioethical approval to the Cyprus National Bioethics Committee, which deemed that no approval was necessary because the collection of samples is considered part of the regular practices of the State. Approval from the Government Veterinary Services has been obtained and no further licenses or documents are required.
Each farm participating in the study has signed a consent form voluntarily agreeing to take part in the project and authorising the researcher to collect, process and use their data. The farmers can stop their participation at any time during the study once they have informed the researcher. All results, data and information arising from this research project will be treated with confidentiality and the identity of each farm will not be disclosed in any publication or to a third party not involved in the project.
Data and sample collection
A trained researcher will provide structured questionnaires on herd health management and husbandry to be completed by the farm owners and veterinarians during the farm visits. More specifically, the questionnaire will include sections and information on the herd, dry cow management, calving, first management of new-born calves, colostrum management, calf rearing, disease prevention and sanitation procedures, sick calf management, deceased animal disposal, milking, milking machine maintenance and mastitis management. The questionnaire is a translated and modified version of a questionnaire already implemented in countries including the Netherlands, France, Belgium and the United Kingdom [21] [22]. The questionnaire has been translated to Greek using an authorised translation service. Google Forms are used for the completion of the questionnaires and the results will be automatically exported to a Microsoft Excel spreadsheet. Data collection on weather and climate variations in the regions of the farms during the period of sample collection will also be recorded.
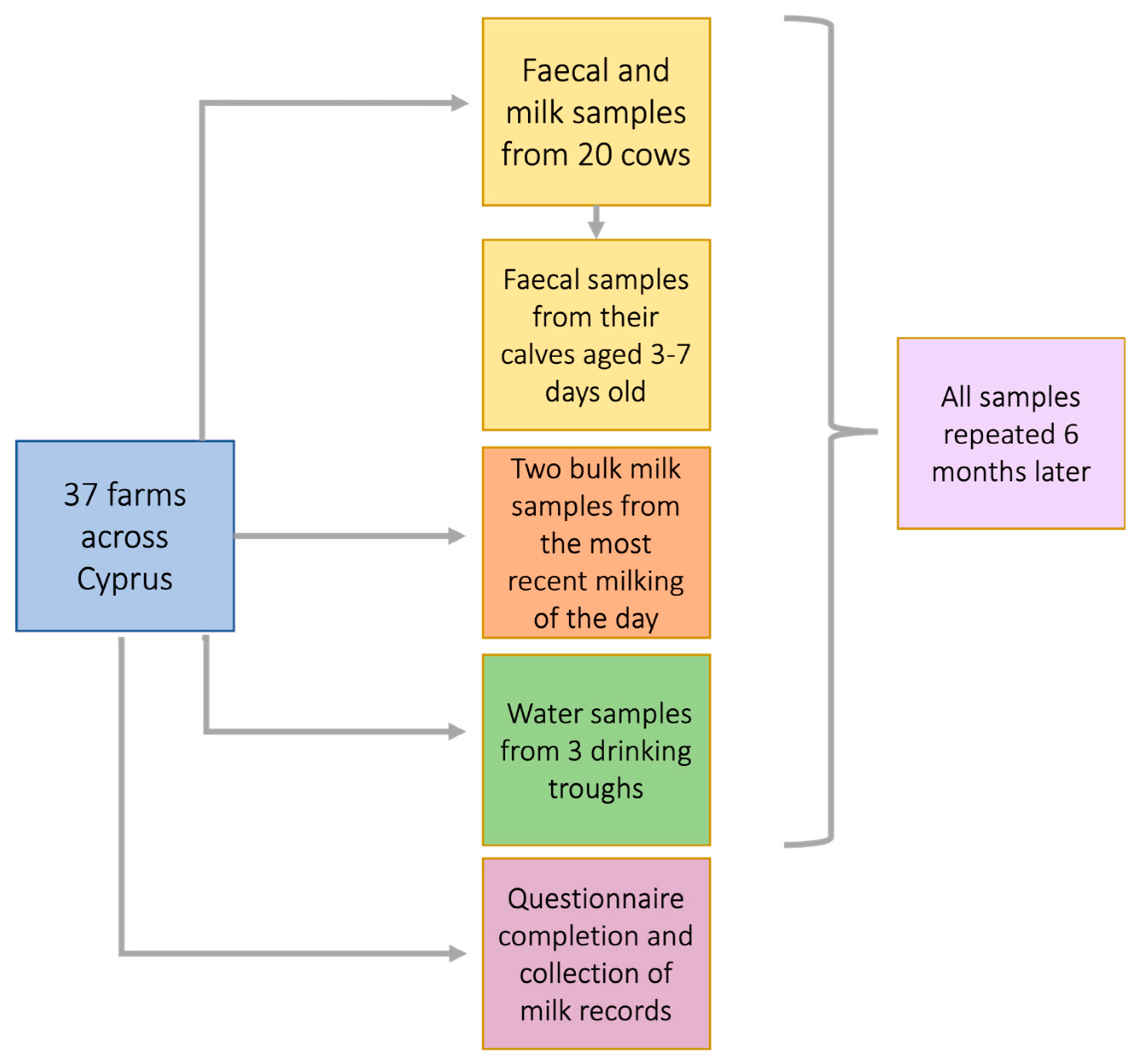
Samples will be collected as follows: at least 20 individual milk and 40 fecal samples per farm will be collected at two time points in a two-year interval in addition to bulk milk, and water samples as shown in
Figure 2. Faecal samples will be collected in individual sampling pots from at least 20 cows that have given birth within 3 - 7 days and their corresponding calves by performing a rectal procedure by authorized trained samplers. The rectal procedure is regularly undertaken in farm animal practice for various applications. Milk samples will be obtained by hand milking both teats from each cow to collect approximately 10 – 15 ml of milk. These will come from the same animals from which faecal samples will be obtained. Bulk milk, defined as the collective milk of the cows on a farm that have been milked in the milking parlour will also be sampled. Milking is usually performed twice a day (morning and afternoon). Two bulk milk samples (approximately 10 – 15 ml) will be collected after the most recent milking session of the day. Milk production records will be obtained at both visits from the cows whose faecal samples were taken. Environmental samples will include water from the cows’ drinking troughs and the water reservoir of the farm, if available (
Figure 2). All samples collected will be subsequently stored in polystyrene boxes with ice packs and immediately transferred to -80
0C.
Biorepository development and sample banking
The faecal, individual cow milk, bulk milk and environmental samples will be stored. Each sample will be labelled according to the sample type and given a code to represent the specific cow and farm from which the sample originated from. Samples will then be grouped and stored according to the farm from which they were collected and each farm will be assigned a code. All samples will be stored at -800C.
DNA from all samples will be extracted using a microbiome purification kit. DNA will be sent to an external provider for amplicon sequencing. Sanger sequencing will also be performed for selected pathogens.
The microbiome data will be processed as follows: (a) Pre-processing and filtering: De-multiplexing and adapter/barcoding sequencing trimming will be carried out using UMI tools in R and FastQC, respectively. The latter will also be used for quality control and filtering of low quality reads from the fastq files. Chimera checking and elimination will be carried out using the relevant QIIME2 tools. (b) Clustering and classification: The filtered sequencing data will be clustered and classified using the de novo and/or open-reference Operational Taxonomic Unit (OTU) mode in QIIME2 using a threshold of 97%. The representative OTU sequence matching will be based on the NCBI 16S taxonomy marker database. The coverage check for each sample will be performed using rarefaction analysis based on alpha-diversity measures like richness and Shannon entropy as a function of sequences number. (c) Primary analysis: Downstream analysis will be performed at the phylum, order, class, family and genus levels. Relative abundance and compositions will be calculated in R using the phyloseq and microbiomeR packages. Alpha and beta diversity will be calculated. Next, pairwise comparisons between samples grouped at different levels – by farm, by region and between faecal samples including those with and without select pathogens will be performed. Sample comparisons will be undertaken between samples from the same groups collected at different time points. (d) Meta-analysis and knowledge extraction: A network based approach will be implemented to obtain and analyse the microbial concurrence and anti-concurrence of specific taxa in faecal samples. Moreover, a network of farms and regions will be constructed based on their physical distance and observed microbial beta-diversity distance in order to identify clusters with distinct microbial signatures. Finally, functional analysis of faecal microecology will be carried out using LEfSE in order to pinpoint importance and abundance of specific taxa.
All samples will be screened for specific eukaryotic and prokaryotic pathogens using established in-house protocols [20], [34]. Transmission dynamics will be assessed by considering microbiota of cows, their offspring and the environment. Select fecal samples per farm will be used for antimicrobial resistance detection using standard state protocols.
Statistical analyses will also be performed to evaluate associations between microbiome (or specific microbial taxa) and metadata gathered with the questionnaire.
4. Discussion
Research on the lower gut microbiome in cattle is still limited and this is particularly evident in the Mediterranean region, where such investigations are scarce. This is an oversight considering the more recent discoveries of their important roles in health, disease, zoonoses and animal productivity. The aim of the GUTBIOME CY study is to address this research gap and understand the complex interactions between host and lower gut microbes on immune defense, milk production efficiency and quality, and the effect of the environment. These data can then be used to present primary targets for microbiome manipulation towards a sustainable and environmentally friendly agriculture.
The ruminant microbiome biobank establishment will open various avenues for research. The composition of the gut microbiome in cattle has been shown to influence both milk production and its quality, through feed utilisation, which involves dietary energy derived from feed [16] [17] [9]. Due to global increases in food demand particularly for milk and animal protein, the efficiency of animal food production requires improvement. Through the GUTBIOME CY study, a greater understanding of the existing microbes and their mechanisms in fibre degradation could be achieved, leading to the identification of biomarkers of feed efficiency to help develop methods of microbiome manipulation and optimise milk production phenotypes [23]. Ultimately, these strategies would improve animal productivity, which is of significant economic and welfare interest [24].
Previous studies worldwide revealed that the microbiome of the lower gut is involved in establishing a functional mucosal immune system in cattle. This directly influences the host’s gut health and helps fight infections. Antibiotics are often the choice of treatment to fight these infections but their overuse on farms is a major concern. Animals consume more than twice as many antibiotics than humans thereby increasing antibiotic resistance bacteria in agricultural environments [25]. Recent investigations have revealed that resistant bacteria in farm animals can also reach consumers through the food chain [26]. This is of particular importance in Cyprus which is usually in the top five countries for antibiotic use in farm animals in the EU [27]. As such, by exploring the composition of gut microbes through the GUTBIOME CY study, alternative avenues to guide antibiotic stewardship efforts and preserve their efficacy can be developed and implemented.
The GUTBIOME CY study involves collection of metadata and as such it has an epidemiological component looking into herd health management and husbandry practices on the farms and how these might relate to the microbiome composition individually or in synergy. Questionnaires completed with farmers on diet, treatments (antibiotic use), housing arrangements, hygiene practices and other husbandry aspects will be analysed for any notable associations to the GIT microbiome and possible causal-effect relationships. By investigating these factors in relation to the gut microbiome, new and improved farm management practices can be introduced. In addition, the welfare of the farm animals can be improved by curtailing infections and guiding breeding programs to influence the health of future offspring.
Through the GUTBIOME CY study, the authors will create a biobank of faecal, milk and environmental samples from farms across the whole island. Accessibility to this large biobank of samples will foster additional transdisciplinary investigations of host microbiome associations and their direct environment as well as promote collaborative research and discussions on the topic within the microbiome community at large.
Author Contributions
Conceptualization, A.D.T. and S.M.; methodology, D.E.M., M.L., E.G. and A.D.T.; validation, D.I., E.G. and A.D.T.; formal analysis, D.E.M.; investigation, D.E.M.; resources, M.L., C.P., S.M., and A.D.T..; data curation, E.G..; writing—original draft preparation, D.E.M.; writing—review and editing, E.G. and A.D.T..; visualization, A.D.T.; supervision, D.I., S.M., E.G. and A.D.T.; project administration, A.D.T.. All authors have read and agreed to the published version of the manuscript.
Funding
Please add: “This research received no external funding”.
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
Not Applicable.
Acknowledgments
We would like to thank the state veterinereans in Cyprus for the organization of the ssampling in each district. .
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Y. a. P. O. Fan, "Gut microbiota in human metabolic health and disease.," vol. 19, 2021. [CrossRef]
- W. a. H. K. Lee, "Gut microbiota–generated metabolites in animal health and disease," vol. 10, 2014. [CrossRef]
- J. Qin, R. Li, J. Raes, M. Arumugam, K. Solvsten, C. Manichanh, T. Nielsen, N. Pons, F. Levenez, T. Yamada, D. R. Mende, J. Li, J. Xu, S. Li and D. Li, "A human gut microbial gene catalogue established by metagenomic sequencing," vol. 464, 2010. [CrossRef]
- X. W. a. L. L. Baoli Zhu, "Human gut microbiome: the second genome of human body," vol. 1, no. 8, 2010.
- C. L. Lemm, "The second genome," vol. 3, no. 8, 2018. [CrossRef]
- T. L. a. E. E. Danping Zheng, "Interaction between microbiota and immunity in health and disease," vol. 30, 2020. [CrossRef]
- N. Malmuthuge, P. Griebel and L. L. Guan, "The Gut Microbiome and Its Potential Role in the Development and Function of Newborn Calf Gastrointestinal Tract," Frontiers in Veterinary Science, vol. 2, no. 36, 2015. [CrossRef]
- H. Shen, Z. Xu, Z. Shen and Z. Lu, "The Regulation of Ruminal Short-Chain Fatty Acids on the Functions of Rumen Barriers," Frontiers in Physiology, vol. 10, p. 1305, 2019. [CrossRef]
- J. Wallace, G. Sasson, P. Garnsworthy, I. Tapio, E. Gregson, P. Bani, P. Huhtanen, A. Bayat and F. Strozzi, "A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions," Science Advances, vol. 5, 2019. [CrossRef]
- Food and Agricultural Organization of the United Nations, "Animal Production," [Online]. Available: https://www.fao.org/animal-production/en. [Accessed 1 10 2023].
- Organisation for Economic Co-operation and Development, "Agriculture," [Online]. Available: https://www.oecd.org/gov/pcsd/25507214.pdf. [Accessed 1 10 2023].
- Food and Agricultural Organization of the United Nations, "The Global Dairy Sector: Facts," [Online]. Available: https://www.fil-idf.org/wp-content/uploads/2016/12/FAO-Global-Facts-1.pdf. [Accessed 1 10 2023].
- G. Henderson, F. Cox, S. Ganesh, A. Jonker, W. Young, G. R. C. Collaborators and P. H. Janssen, "Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range," Nature, vol. 5, p. 14567, 2015. [CrossRef]
- S. Paudyal, "Using rumination time to manage health and reproduction in dairy cattle: a review," vol. 41, no. 1, 2021. [CrossRef]
- K. C. Swanson, "Small Intestinal Anatomy, Physiology, and Digestion in Ruminants," 2019. [CrossRef]
- S. K. B. Shabat, G. Sasson, A. Doron-Faigenboim, T. Durman, S. Yaacoby, M. E. B. Miller, B. A. White, N. Shterzer and I. Mizrahi, "Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants," The ISME journal, vol. 10, p. 2958–2972, 2016. [CrossRef]
- C. Matthews, F. Crispie, E. Lewis, M. Reid, P. W. O’Toole and P. D. Cotter, "The rumen microbiome: a crucial consideration when optimising milk and meat production and nitrogen utilisation efficiency," Gut microbes, vol. 10, no. 2, pp. 115-132, 2019. [CrossRef]
- E. O’Hara, A. Neves, Y. Song and L. L. Guan, "The Role of the Gut Microbiome in Cattle Production and Health: Driver or Passenger?," Annual Reviews of Animal Biosciences, vol. 8, pp. 199-220, 2020. [CrossRef]
- C. Delling and A. Daugschies, "Literature Review: Coinfection in Young Ruminant Livestock—Cryptosporidium spp. and Its Companions," Pathogens, vol. 11, no. 1, p. 103, 2022. [CrossRef]
- P. Pinto, C. A. Ribeiro, S. Hoque, O. Hammouma, H. Leruste, S. Détriché, E. Canniere, Y. Daandels, M. Dellevoet, J. Roemen, A. B. Bourgeois, M. Kváč, J. Follet and A. D. Tsaousis, "Cross-Border Investigations on the Prevalence and Transmission Dynamics of Cryptosporidium Species in Dairy Cattle Farms in Western Mainland Europe," Microorganisms , vol. 9, no. 11, 2021. [CrossRef]
- M. Roblin, E. Canniere, A. Barbier, Y. Daandels, M. Dellevoet-Groenewegen, P. Pinto, A. Tsaousis, H. Leruste, J. Brainard, P. R. Hunter and J. Follet, "Study of the economic impact of cryptosporidiosis in calves after implementing good practices to manage the disease on dairy farms in Belgium, France, and the Netherlands," vol. 4, no. 100149, 2023. [CrossRef]
- "Interreg 2 Seas Mers Zeeën H4DC," [Online]. Available: https://h4dc-interreg2seas.eu. [Accessed 23 November 2021].
- B. Clemmons, B. Voy and P. Myer, "Altering the Gut Microbiome of Cattle: Considerations of Host-Microbiome Interactions for Persistent Microbiome Manipulation," Microbial Ecology, vol. 77, p. 523–536, 2019. [CrossRef]
- L. Wang, G. Zhang, Y. Li and Y. Zhang, "Effects of High Forage/Concentrate Diet on Volatile Fatty Acid Production and the Microorganisms Involved in VFA Production in Cow Rumen," Animals, vol. 10, p. 223, 2020. [CrossRef]
- F. Aarestrup, "Sustainable farming: Get pigs off antibiotics," Nature, vol. 486, no. 7404, pp. 465-466, 2012. [CrossRef]
- C. L. Ventola, "The Antibiotic Resistance Crisis Part 1: Causes and Threats," Pharmacy and Therapeutics, vol. 40, no. 4, pp. 277-283, 2015.
- "Save our antibiotics," [Online]. Available: https://www.saveourantibiotics.org/media/1738/farm-antibiotic-use-in-cyprus.pdf. [Accessed 2 August 2022].
- E. O’Hara, A. L. Neves, Y. Song and L. L. Guan, "The Role of the Gut Microbiome in Cattle Production and Health: Driver or Passenger?," Annual Reviews of Animal Biosciences, vol. 8, pp. 199-220, 2020. [CrossRef]
- M. L. Hutton, S. J. Mileto, M. L. James, C. Evans, R. M. Shah, A. B. Ghodke, K. E. Hillyer, S. S. Metcalfe, J.-W. Liu, T. Walsh, D. Lyras, E. A. Palombo and D. J. Beale, "Cryptosporidiosis modulates gut microbiome metabolism and the immune response in an infected host," Research square, 2020.
- K. Tiseo, L. Huber, M. Gilbert, T. Robinson and T. Van Boeckel, "Global Trends in Antimicrobial Use in Food Animals from 2017 to 2030," Antibiotics, vol. 9, p. 918, 2020. [CrossRef]
- S. Ji, T. Jiang, H. Yan, C. Guo, J. Liu, H. Su, G. Alugongo, H. Shi, Y. Wang, Z. Cao and S. Li, "Ecological Restoration of Antibiotic-Disturbed Gastrointestinal Microbiota in Foregut and Hindgut of Cows," Frontiers in Cellular and Infection Microbiology, vol. 8, p. 79, 2018. [CrossRef]
- S. Ghosh and T. M. LaPara, "The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria," The ISME Journal, vol. 1, pp. 191-203, 2007. [CrossRef]
- "Disarm Project," [Online]. Available: https://disarmproject.eu. [Accessed 18 April 2021].
- S. Hoque, D. E. Mavrides, P. Pinto, C. Azevedo-Ribeiro, M. Kváč, S. Malas, E. Gentekaki and A. Tsaousis, "High prevalence of Cryptosporidium parvum in Cypriot dairy cow farms," Frontiers in Veterinary Science, Under preparation.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).