Submitted:
29 October 2023
Posted:
30 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Identification and Sequence Analysis of GS and GOGAT Gene Family Members in Pecan
| Gene Name | Gene ID | Exon No. | AA | CDS (bp) | MW (kDa) | pI | Instability Index | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| CiGS1.1a | CiPaw.03G073300.1 | 12 | 356 | 1071 | 39.31 | 6.68 | 39.29 | -0.48 | Cytoplasm |
| CiGS1.1b | CiPaw.04G048000.1 | 12 | 356 | 1071 | 39.02 | 5.49 | 36.56 | -0.40 | Cytoplasm |
| CiGS1.1c | CiPaw.05G168500.1 | 13 | 356 | 1071 | 39.21 | 5.79 | 39.24 | -0.42 | Chloroplast Cytoplasm |
| CiGS1.2 | CiPaw.16G097000.1 | 13 | 356 | 1071 | 39.28 | 5.82 | 39.61 | -0.46 | Chloroplast Cytoplasm |
| CiGS2a | CiPaw.01G157900.1 | 14 | 432 | 1299 | 47.59 | 8.07 | 45.71 | -0.48 | Chloroplast Mitochondrion |
| CiGS2b | CiPaw.02G090300.1 | 14 | 432 | 1299 | 47.78 | 6.48 | 44.12 | -0.50 | Chloroplas Mitochondrion |
| CiNADH-GOGATa | CiPaw.05G252100.1 | 23 | 2222 | 6669 | 244.32 | 6.75 | 36.48 | -0.30 | Chloroplast |
| CiNADH-GOGATb | CiPaw.06G009900.1 | 23 | 2305 | 6918 | 254.99 | 6.46 | 36.66 | -0.28 | Chloroplast |
| CiFd-GOGATa | CiPaw.09G047200.1 | 33 | 1490 | 4473 | 162.42 | 5.94 | 34.55 | -0.15 | Chloroplast |
| CiFd-GOGATb | CiPaw.10G039900.1 | 33 | 1637 | 4914 | 178.25 | 6.23 | 37.12 | -0.15 | Chloroplast |
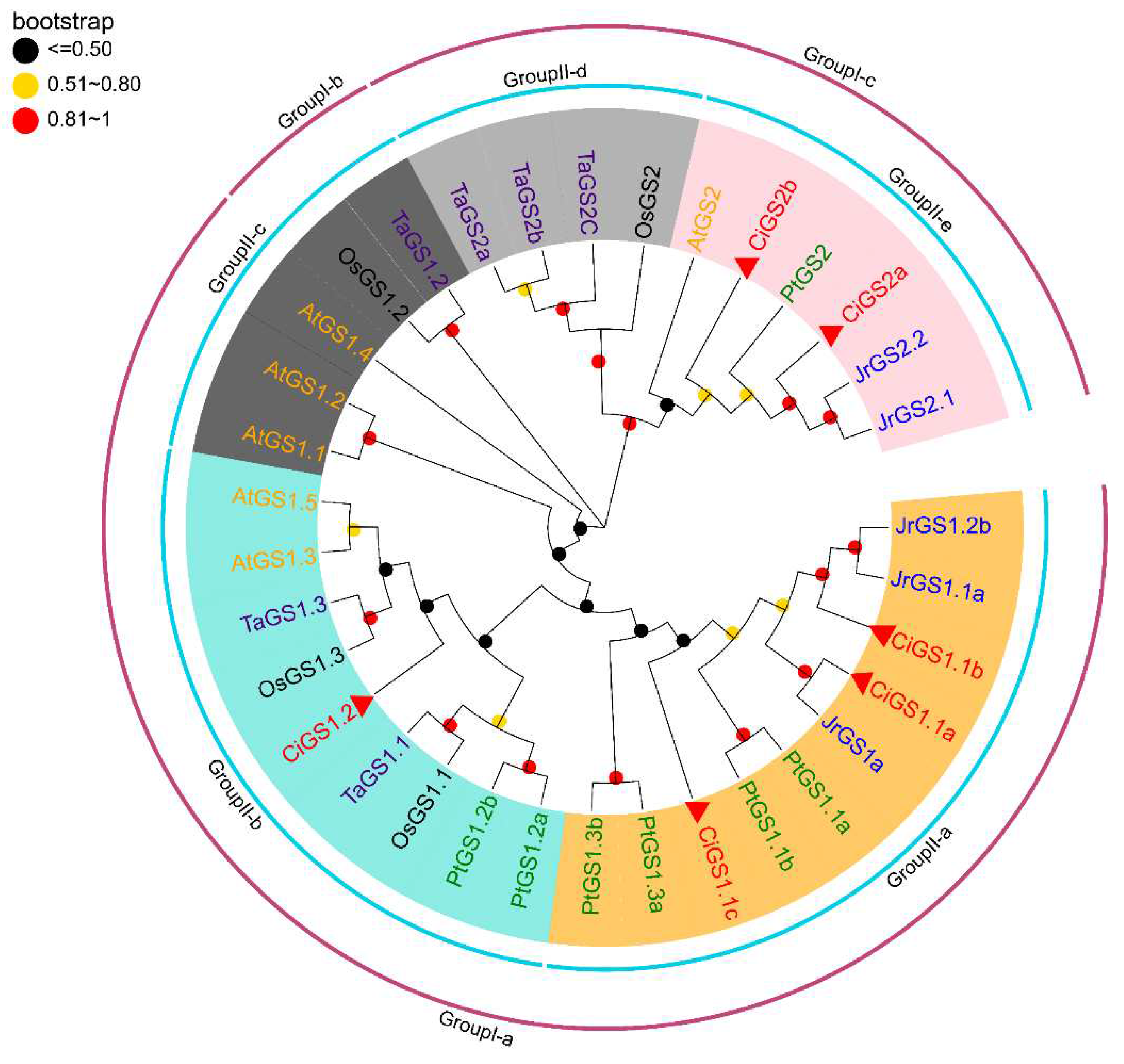
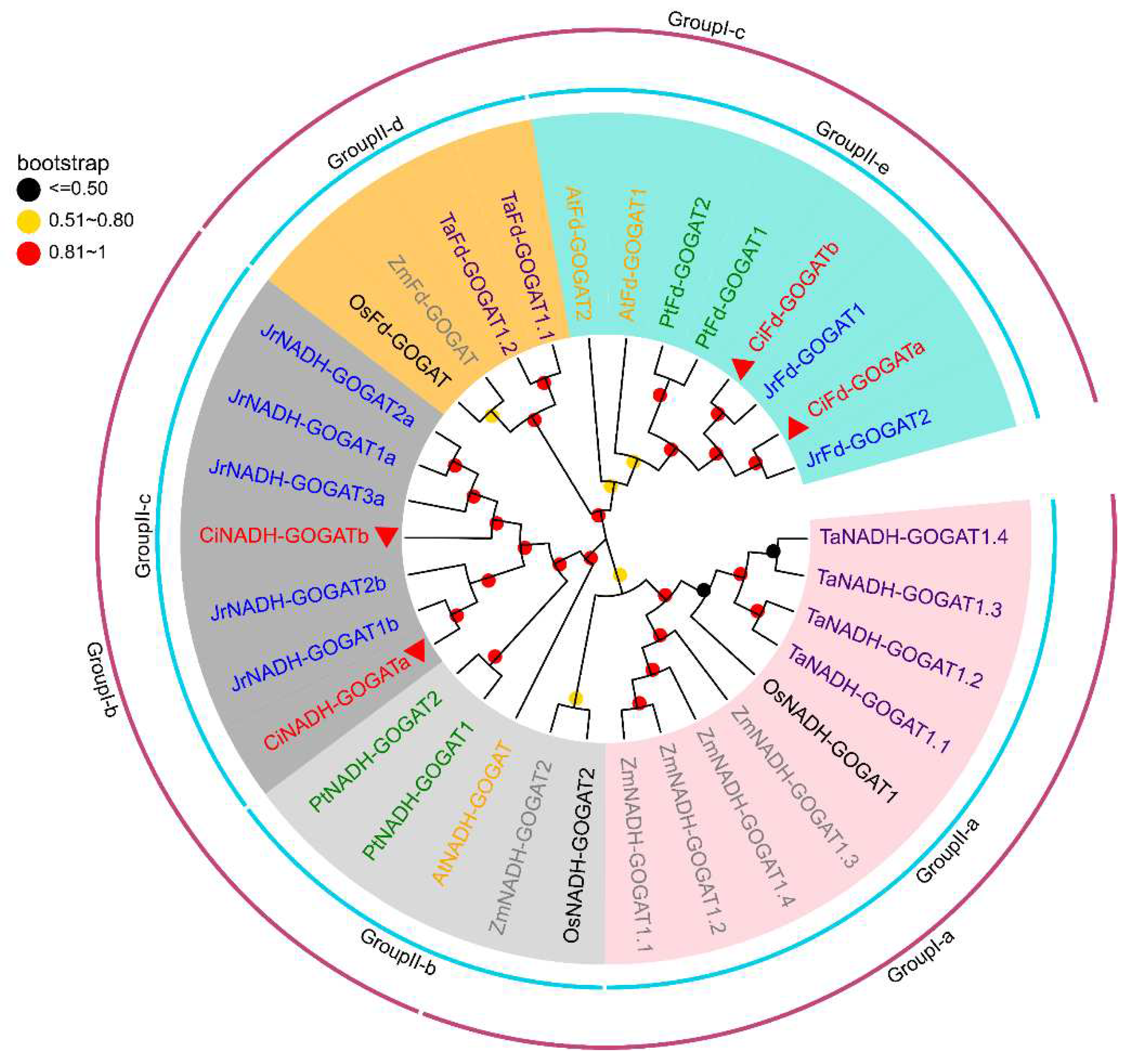
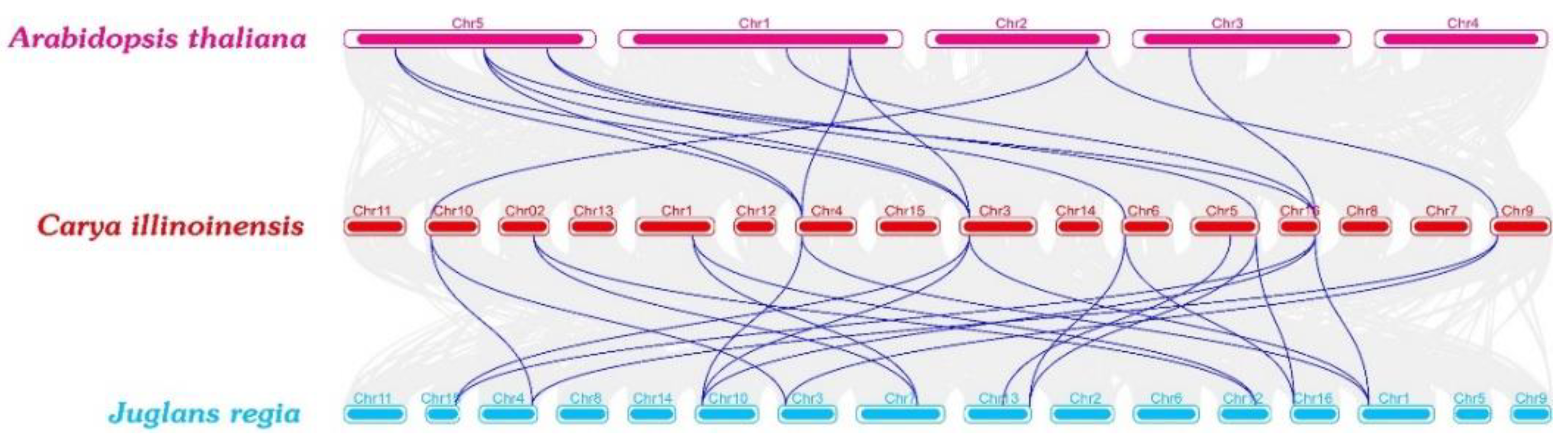
2.2. Phylogenetic Analysis of the GS and GOGAT in Different Species
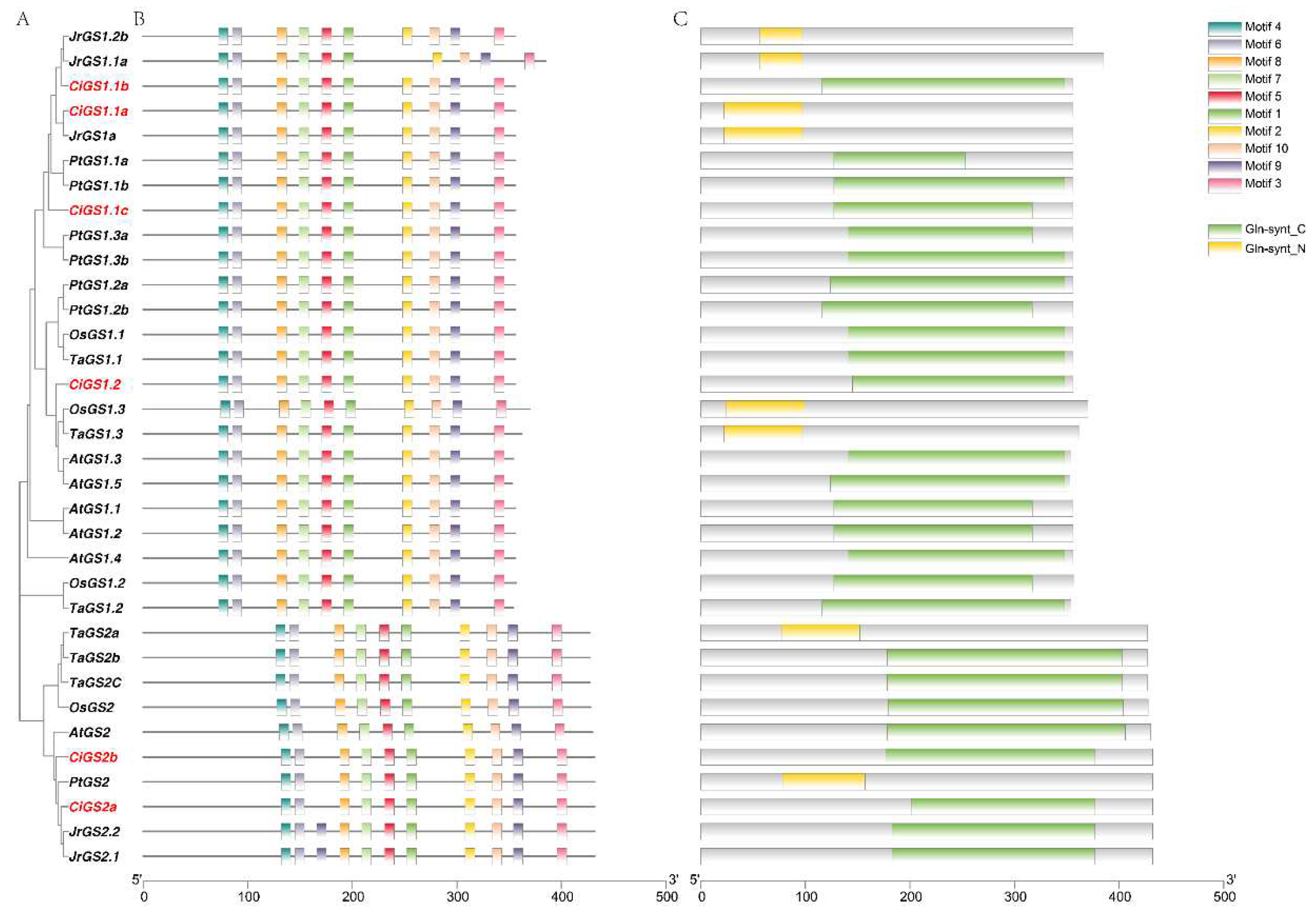
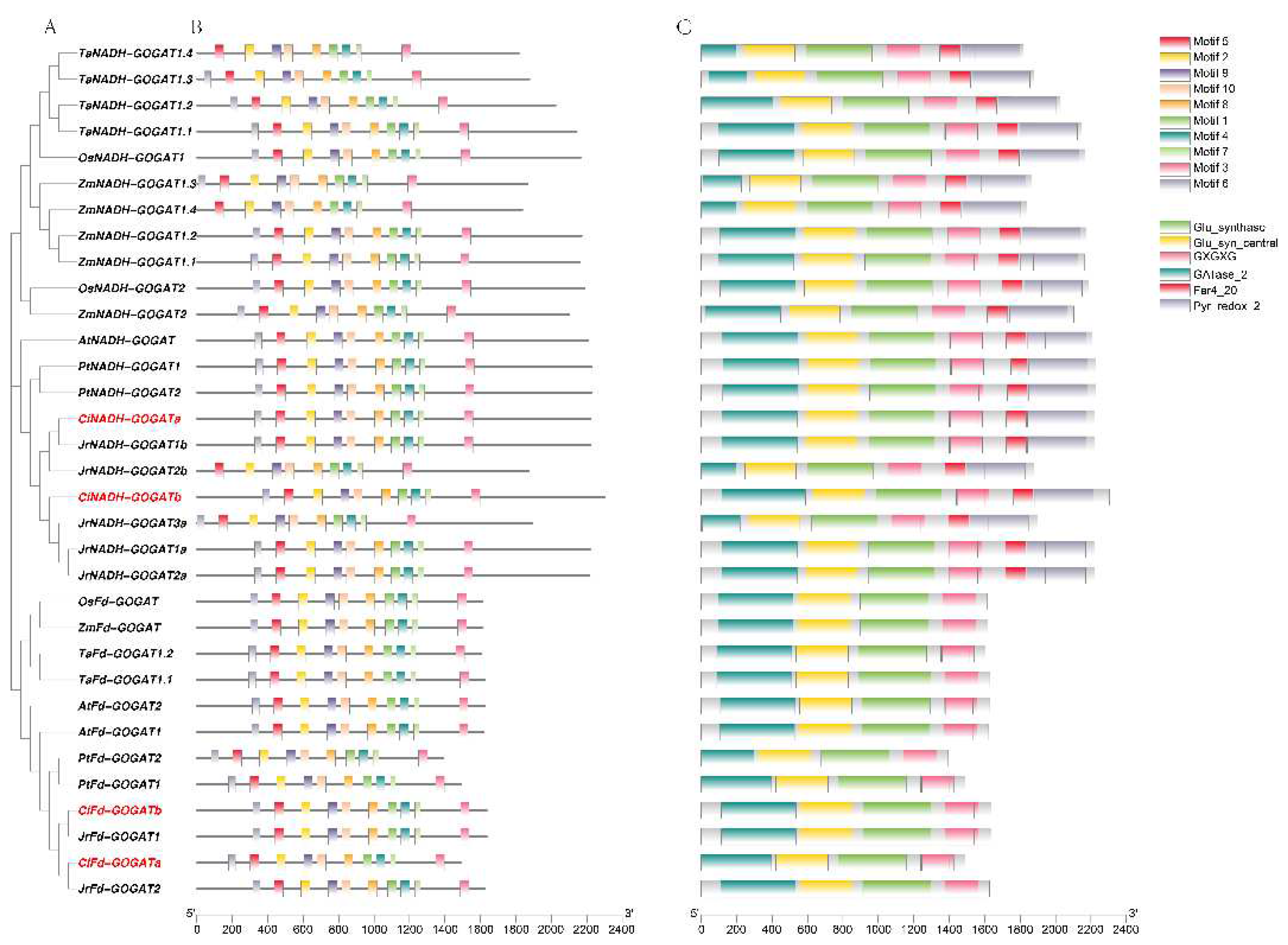
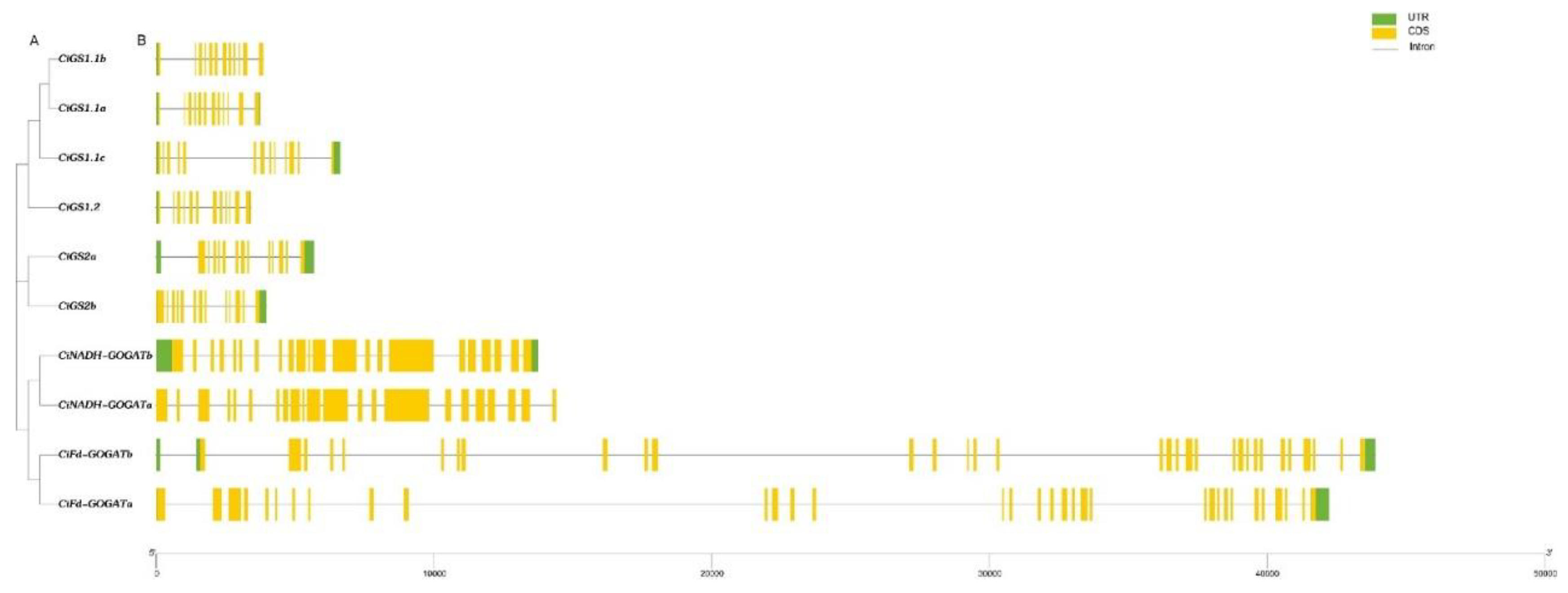
2.3. Conserved Motif, Conserved Domain and Gene Structural Analysis of GS and GOGAT
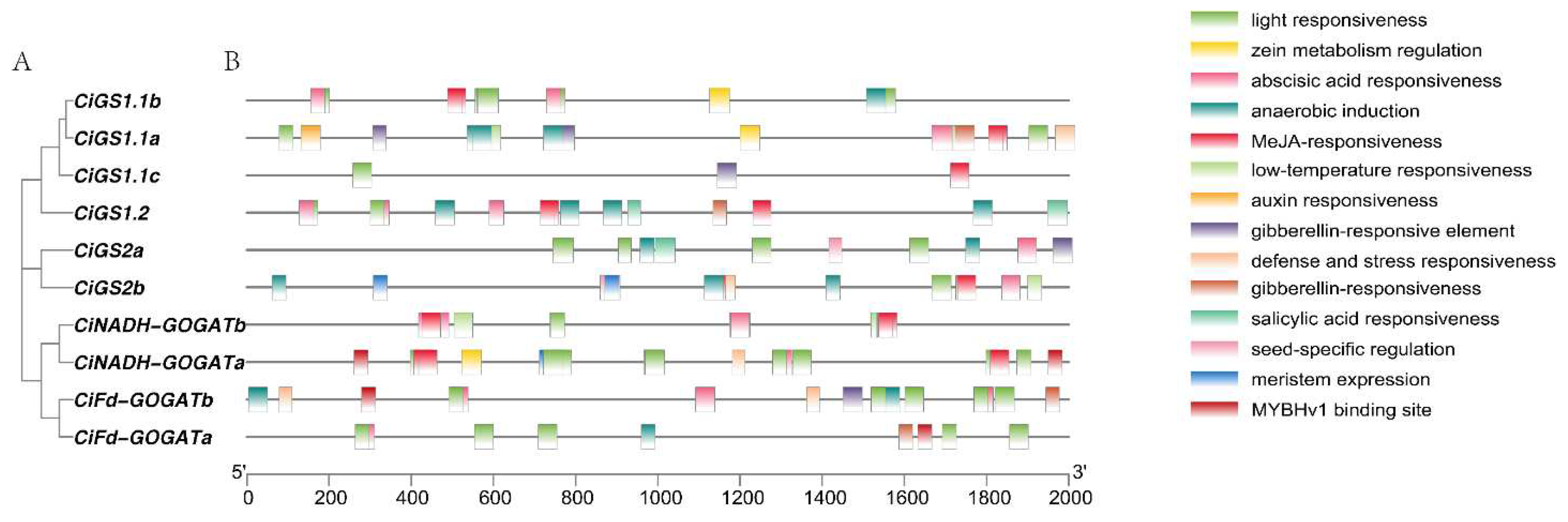
2.4. Analysis of Cis-Acting Elements in the Promoter Regions of GS and GOGAT Genes in Pecan
2.5. Duplication Events and Syntenic Analysis of CiGS and CiGOGAT Genes
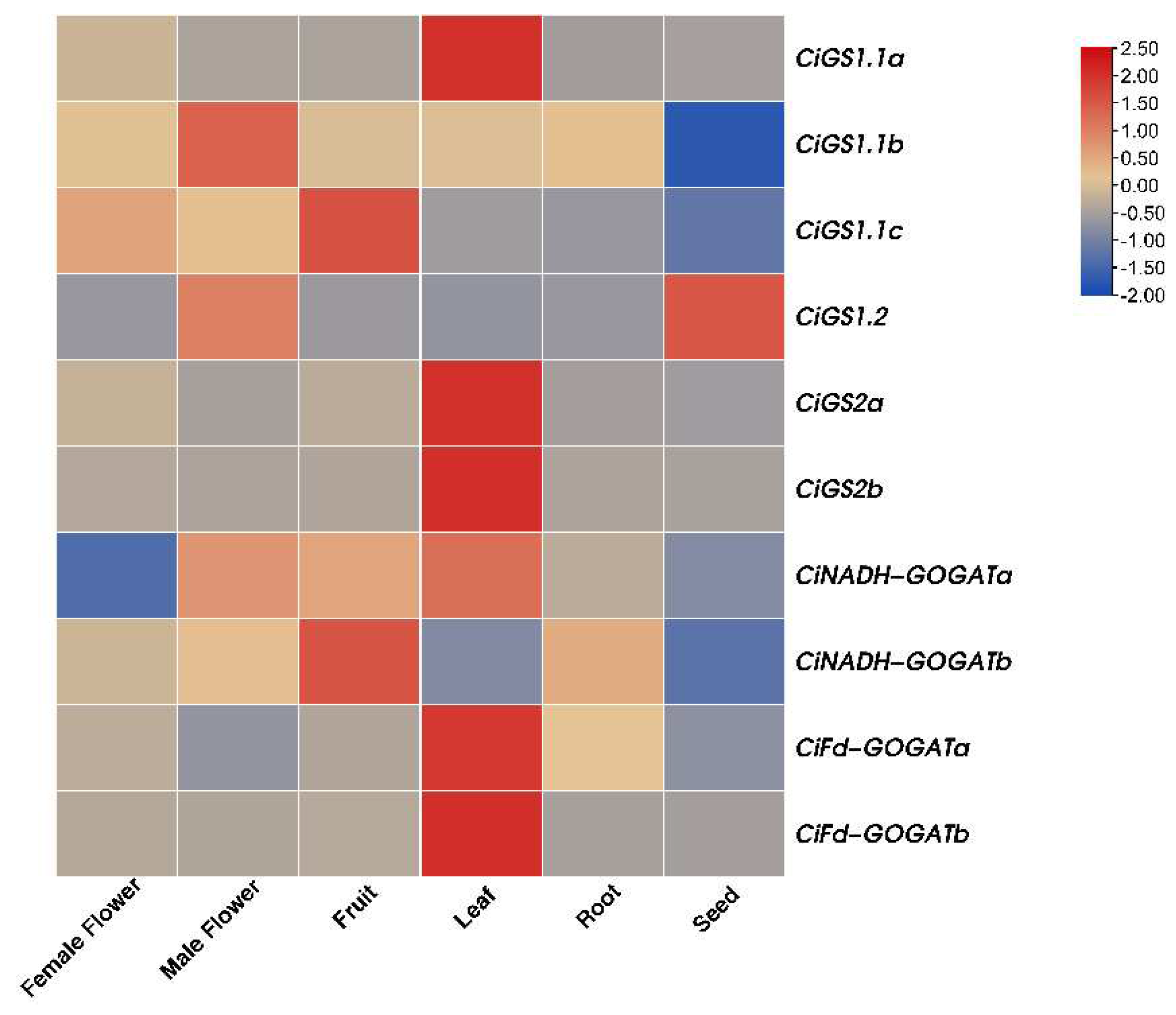
2.6. Tissue-Specific Expression Analysis of the CiGS and CiGOGAT Genes
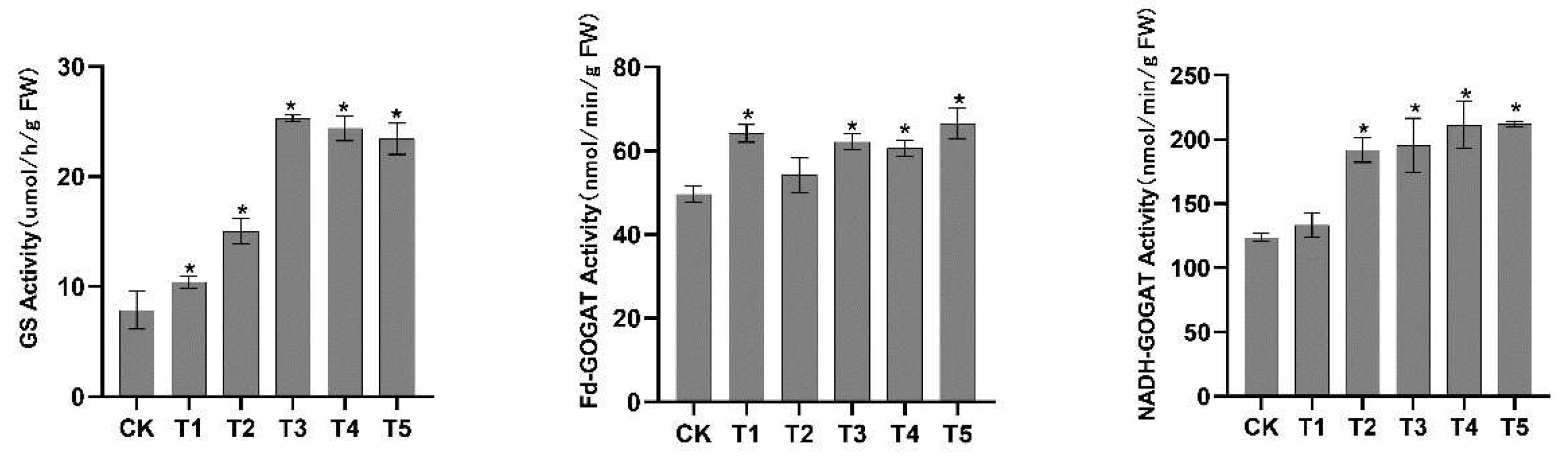
2.7. Effects of N Forms on GS and GOGAT Enzyme Activity of Pecan
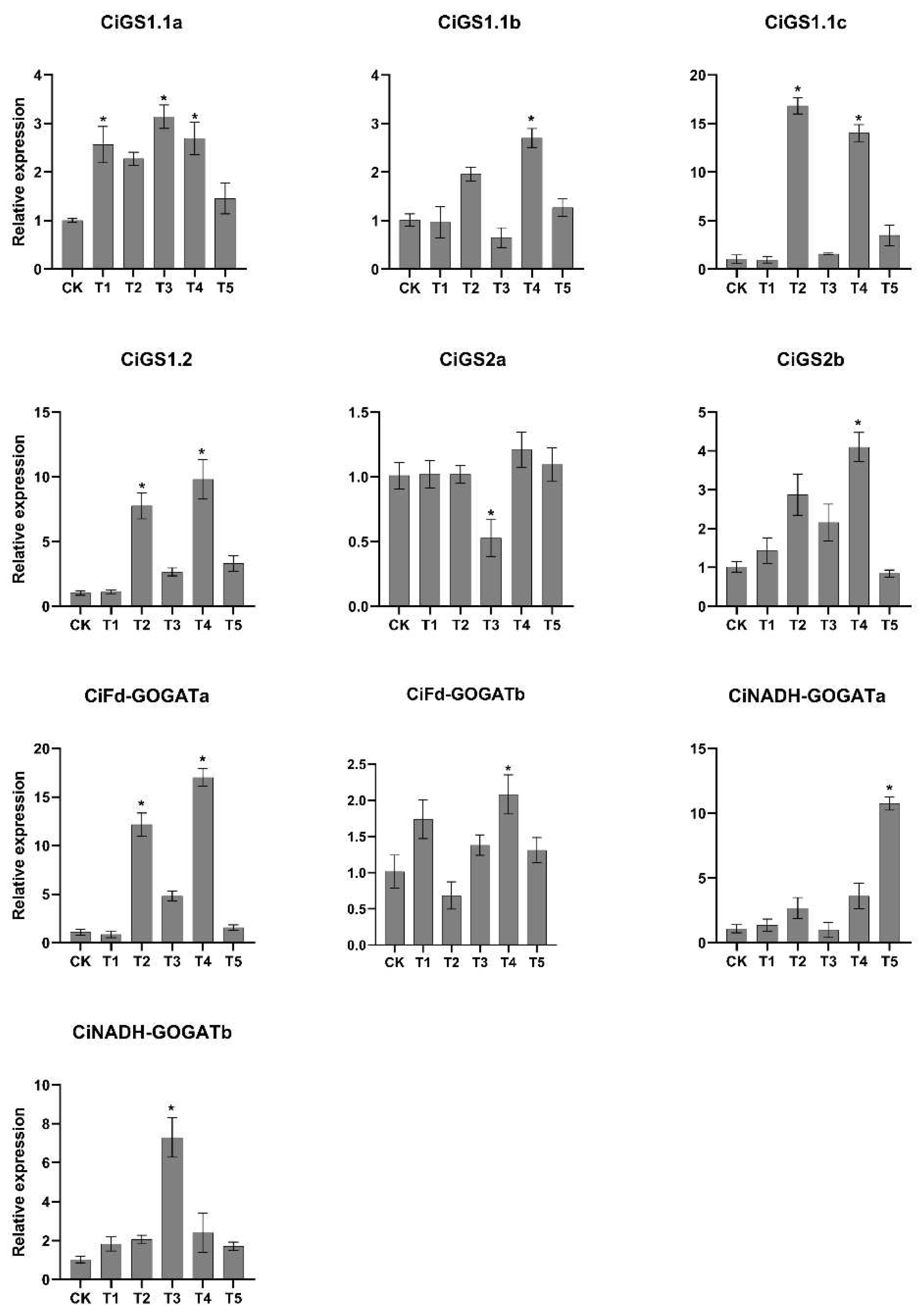
2.8. Effect of N Forms on GS and GOGAT Gene Expression Levels in Pecan
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Experimental Design
4.2. Identification of GS and GOGAT Gene Family Members of Pecan
4.3. Phylogenetic Analysis
4.4. Conserved Structural Domains, Conserved Motifs, and Gene Structural Analysis
4.5. Analysis of Promoter Cis-Acting Elements
4.6. Gene Duplication Analysis and Ka/Ks Value Calculation
4.7. Tissue-Specific Expression Analysis of the CiGS and CiGOGAT Genes
4.8. RNA Extraction and qRT-PCR Analysis
4.9. Determination of GS and GOGAT enzyme activity
4.10. Data analysis
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Guha, T.; Gopal, G.; Mukherjee, A.; Kundu, R. Fe3O4-Urea Nanocomposites as a Novel Nitrogen Fertilizer for Improving Nutrient Utilization Efficiency and Reducing Environmental Pollution. Environ Pollut 2022, 292 (Pt A), 118301. [Google Scholar] [CrossRef]
- Chen, M.; Zhu, K.; Tan, P.; Liu, J.; Xie, J.; Yao, X.; Chu, G.; Peng, F. Ammonia–Nitrate Mixture Dominated by NH4+–N Promoted Growth, Photosynthesis and Nutrient Accumulation in Pecan (Carya Illinoinensis). Forests 2021, 12, 1808. [Google Scholar] [CrossRef]
- Boczulak, S. A.; Hawkins, B. J.; Roy, R. Temperature Effects on Nitrogen Form Uptake by Seedling Roots of Three Contrasting Conifers. Tree Physiology 2014, 34, 513–523. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho Fernandes, G.; Turchetto-Zolet, A. C.; Pereira Passaglia, L. M. Glutamine Synthetase Evolutionary History Revisited: Tracing Back beyond the Last Universal Common Ancestor. Evolution 2022, 76, 605–622. [Google Scholar] [CrossRef]
- Bernard, S. M.; Habash, D. Z. The Importance of Cytosolic Glutamine Synthetase in Nitrogen Assimilation and Recycling. New Phytologist 2009, 182, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Clermont, K.; Graham, C. J.; Lloyd, S. W.; Grimm, C. C.; Randall, J. J.; Mattison, C. P. Proteomic Analysis of Pecan (Carya Illinoinensis) Nut Development. Foods 2023, 12, 866. [Google Scholar] [CrossRef]
- Wang, L.; Chen, F.; Lan, Y.; Liu, H.; Wu, M.; Yan, H.; Xiang, Y. Genome-Wide Identification of B3 Superfamily in Pecan (Carya Illinoensis): In Silico and Experimental Analyses. Scientia Horticulturae 2023, 307, 111533. [Google Scholar] [CrossRef]
- Liu, J.; Tang, Y.; Bao, J.; Wang, H.; Peng, F.; Chen, M.; Tan, P. Pecan Plantation Age Influences the Structures, Ecological Networks, and Functions of Soil Microbial Communities. Land Degradation & Development 2022, 33, 3294–3309. [Google Scholar] [CrossRef]
- Reyes-Vázquez, N. del C.; Rosa, L. A. de la; Morales-Landa, J. L.; García-Fajardo, J. A.; García-Cruz, M. Á. Phytochemical Content and Potential Health Applications of Pecan [Carya Illinoinensis (Wangenh) K. Koch] Nutshell. Current Topics in Medicinal Chemistry 22, 150–167.
- Juhaimi, F. A.; Özcan, M. M.; Uslu, N.; Doğu, S. Pecan Walnut (Carya Illinoinensis (Wangenh.) K. Koch) Oil Quality and Phenolic Compounds as Affected by Microwave and Conventional Roasting. J Food Sci Technol 2017, 54, 4436–4441. [Google Scholar] [CrossRef]
- Zhu, K.; Fan, P.; Liu, H.; Tan, P.; Ma, W.; Mo, Z.; Zhao, J.; Chu, G.; Peng, F. Insight into the CBL and CIPK Gene Families in Pecan (Carya Illinoinensis): Identification, Evolution and Expression Patterns in Drought Response. BMC Plant Biol 2022, 22, 221. [Google Scholar] [CrossRef]
- Lebedev, V. G.; Korobova, A. V.; Shendel, G. V.; Kudoyarova, G. R.; Shestibratov, K. A. Effect of Glutamine Synthetase Gene Overexpression in Birch (Betula Pubescens) Plants on Auxin Content and Rooting in Vitro. Dokl Biochem Biophys 2018, 480, 143–145. [Google Scholar] [CrossRef] [PubMed]
- Pascual, L.; Solé-Medina, A.; Faci, I.; Giraldo, P.; Ruiz, M.; Benavente, E. Development and Marker-Trait Relationships of Functional Markers for Glutamine Synthetase GS1 and GS2 Homoeogenes in Bread Wheat. Mol Breeding 2023, 43, 8. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Sun, Q.; Lu, X.; Zhang, L.; Yuan, Y.; Gong, C.; He, X.; Ma, W.; Mu, P. Identification of the Glutamine Synthetase (GS) Gene Family in Four Wheat Species and Functional Analysis of Ta4D. GSe in Arabidopsis Thaliana. Plant Mol Biol 2022, 110, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Moreira, E.; Coimbra, S.; Melo, P. Glutamine Synthetase: An Unlikely Case of Functional Redundancy in Arabidopsis Thaliana. Plant Biol (Stuttg) 2022, 24, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Gene expression, cellular localisation and function of glutamine synthetase isozymes in wheat (Triticum aestivum L.) - PubMed. https://pubmed.ncbi.nlm.nih.gov/18288574/.
- Fujita, T.; Beier, M. P.; Tabuchi-Kobayashi, M.; Hayatsu, Y.; Nakamura, H.; Umetsu-Ohashi, T.; Sasaki, K.; Ishiyama, K.; Murozuka, E.; Kojima, M.; Sakakibara, H.; Sawa, Y.; Miyao, A.; Hayakawa, T.; Yamaya, T.; Kojima, S. Cytosolic Glutamine Synthetase GS1;3 Is Involved in Rice Grain Ripening and Germination. Front. Plant Sci. 2022, 13, 835835. [Google Scholar] [CrossRef] [PubMed]
- Chloroplast and cytosolic glutamine synthetase are encoded by homologous nuclear genes which are differentially expressed in vivo - PubMed. https://pubmed.ncbi.nlm.nih.gov/2898472/.
- Swarbreck, S. M.; Defoin-Platel, M.; Hindle, M.; Saqi, M.; Habash, D. Z. New Perspectives on Glutamine Synthetase in Grasses. Journal of Experimental Botany 2011, 62, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Nigro, D.; Blanco, A.; Anderson, O. D.; Gadaleta, A. Characterization of Ferredoxin-Dependent Glutamine-Oxoglutarate Amidotransferase (Fd-GOGAT) Genes and Their Relationship with Grain Protein Content QTL in Wheat. PLoS One 2014, 9, e103869. [Google Scholar] [CrossRef] [PubMed]
- Peterman, T. K.; Goodman, H. M. The Glutamine Synthetase Gene Family of Arabidopsis Thaliana: Light-Regulation and Differential Expression in Leaves, Roots and Seeds. Mol Gen Genet 1991, 230, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Marchive, C.; Roudier, F.; Castaings, L.; Bréhaut, V.; Blondet, E.; Colot, V.; Meyer, C.; Krapp, A. Nuclear Retention of the Transcription Factor NLP7 Orchestrates the Early Response to Nitrate in Plants. Nat Commun 2013, 4, 1713. [Google Scholar] [CrossRef]
- Jor, K. W.; Blackwell, R. D.; Lea, P. J. Assimilation of Nitrogen in Mutants Lacking Enzymes of the Glutamate Synthase Cycle. Journal of Experimental Botany 1992, 43, 139–145. [Google Scholar] [CrossRef]
- García-Gutiérrez, Á.; Cánovas, F. M.; Ávila, C. Glutamate Synthases from Conifers: Gene Structure and Phylogenetic Studies. BMC Genomics 2018, 19, 65. [Google Scholar] [CrossRef] [PubMed]
- Teh, C.-Y.; Shaharuddin, N. A.; Ho, C.-L.; Mahmood, M. Exogenous Proline Significantly Affects the Plant Growth and Nitrogen Assimilation Enzymes Activities in Rice (Oryza Sativa) under Salt Stress. Acta Physiol Plant 2016, 38, 151. [Google Scholar] [CrossRef]
- Lu, Y.; Luo, F.; Yang, M.; Li, X.; Lian, X. Suppression of Glutamate Synthase Genes Significantly Affects Carbon and Nitrogen Metabolism in Rice (Oryza Sativa L.). Sci. China Life Sci. 2011, 54, 651–663. [Google Scholar] [CrossRef]
- Cao, L.; Xu, C.; Sun, Y.; Niu, C.; Leng, X.; Hao, B.; Ma, J.; Liu, Z.; Xu, Z.; Yang, C.; Liu, G. Genome-Wide Identification of Glutamate Synthase Gene Family and Expression Patterns Analysis in Response to Carbon and Nitrogen Treatment in Populus. Gene 2023, 851, 146996. [Google Scholar] [CrossRef]
- Banerjee, S.; Subramanian, A.; Chattopadhyay, J.; Rup Sarkar, R. Exploring the Role of GS–GOGAT Cycle in Microcystin Synthesis and Regulation – a Model Based Analysis. Molecular BioSystems 2017, 13, 2603–2614. [Google Scholar] [CrossRef]
- Moons, A. Transcriptional Profiling of the PDR Gene Family in Rice Roots in Response to Plant Growth Regulators, Redox Perturbations and Weak Organic Acid Stresses. Planta 2008, 229, 53–71. [Google Scholar] [CrossRef]
- Mondal, R.; Kumar, A.; Chattopadhyay, S. K. Structural Property, Molecular Regulation, and Functional Diversity of Glutamine Synthetase in Higher Plants: A Data-Mining Bioinformatics Approach. The Plant Journal 2021, 108, 1565–1584. [Google Scholar] [CrossRef]
- Role of the SPS Gene Families in the Regulation of Sucrose Accumulation in Sugarcane | SpringerLink. https://link.springer.com/article/10.1007/s12355-016-0454-x. 1007.
- Zhao, Y.; Cai, M.; Zhang, X.; Li, Y.; Zhang, J.; Zhao, H.; Kong, F.; Zheng, Y.; Qiu, F. Genome-Wide Identification, Evolution and Expression Analysis of MTERF Gene Family in Maize. PLoS One 2014, 9, e94126. [Google Scholar] [CrossRef] [PubMed]
- Parveen, R.; Vaish, S.; Gupta, D.; Basantani, M. K. Bioinformatics Characterization of Patatin-Related Phospholipase A (PPLA) Gene Family in Agriculturally Important Crops Viz Vigna Radiata, Vigna Angularis, and Glycine Max. Biologia 2022, 77, 1429–1446. [Google Scholar] [CrossRef]
- Wang, H.; He, T.; Huang, C.; Wang, K.; Shi, D.; Si, X.; Xu, Y.; Lyu, S.; Huang, J.; Li, Y. Genome-Wide Identification of KCS Gene Family in Carya Illinoinensis and Their Roles under Abiotic Stress Conditions. Scientia Horticulturae 2023, 321, 112343. [Google Scholar] [CrossRef]
- García-Gutiérrez, Á.; Cánovas, F. M.; Ávila, C. Glutamate Synthases from Conifers: Gene Structure and Phylogenetic Studies. BMC Genomics 2018, 19, 65. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, J.; Pereira, S.; Cánovas, F.; Salema, R. Glutamine Synthetase of Potato (Solanum Tuberosum L. Cv. Désirée) Plants: Cell- and Organ-Specific Expression and Differential Developmental Regulation Reveal Specific Roles in Nitrogen Assimilation and Mobilization. Journal of Experimental Botany 2005, 56, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A. Glutamate Synthase and Amino Acid Synthesis in Higher Plants. In Advances in Botanical Research; Elsevier, 2021; Vol. 100, pp 129–144. [CrossRef]
- Anderson, M. P.; Vance, C. P.; Heichel, G. H.; Miller, S. S. Purification and Characterization of NADH-Glutamate Synthase from Alfalfa Root Nodules. Plant Physiol. 1989, 90, 351–358. [Google Scholar] [CrossRef] [PubMed]
- del Amor, F. M.; Piñero, M. C.; Otálora-Alcón, G.; Pérez-Jimenez, M.; Marín-Miñano, M. Effect of Different Nitrogen Forms and CO2 Enrichment on the Nutrient Uptake and Water Relations of Pepper Plants (Capsicum Annuun L.). Procedia Environmental Sciences 2015, 29, 203–204. [Google Scholar] [CrossRef]
- Wan, X.; Wu, W.; Shah, F. Nitrogen Fertilizer Management for Mitigating Ammonia Emission and Increasing Nitrogen Use Efficiencies by 15N Stable Isotopes in Winter Wheat. Sci Total Environ 2021, 790, 147587. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Huang, X.; Shi, W.; Kronzucker, H. J.; Hou, L.; Yang, H.; Song, Q.; Liu, J.; Shi, J.; Yang, Q.; Zou, N. Coordination of Nitrogen Uptake and Assimilation Favours the Growth and Competitiveness of Moso Bamboo over Native Tree Species in High-NH4+ Environments. Journal of Plant Physiology 2021, 266, 153508. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Luo, Y.; Li, C.; Chang, Y.; Jin, M.; Li, Y.; Wang, Z. Effects of Nitrogen Forms on Nitrogen Utilization, Yield, and Quality of Two Wheat Varieties with Different Gluten Characteristics. European Journal of Agronomy 2023, 149, 126919. [Google Scholar] [CrossRef]
- Xu, G.; Jiang, M.; Lu, D.; Wang, H.; Chen, M. Nitrogen Forms Affect the Root Characteristic, Photosynthesis, Grain Yield, and Nitrogen Use Efficiency of Rice under Different Irrigation Regimes. Crop Sci. 2020, 60, 2594–2610. [Google Scholar] [CrossRef]
- Schulz, H.; Härtling, S.; Stange, C. F. Species-specific Differences in Nitrogen Uptake and Utilization by Six European Tree Species. Z. Pflanzenernähr. Bodenk. 2011, 174, 28–37. [Google Scholar] [CrossRef]
- Duan, Y.; Yang, H.; Yang, H.; Wu, Y.; Fan, S.; Wu, W.; Lyu, L.; Li, W. Integrative Physiological, Metabolomic and Transcriptomic Analysis Reveals Nitrogen Preference and Carbon and Nitrogen Metabolism in Blackberry Plants. Journal of Plant Physiology 2023, 280, 153888. [Google Scholar] [CrossRef]
- Iqbal, A.; Dong, Q.; Wang, X.; Gui, H.-P.; Zhang, H.; Pang, N.; Zhang, X.; Song, M. Nitrogen Preference and Genetic Variation of Cotton Genotypes for Nitrogen Use Efficiency. Journal of the Science of Food and Agriculture 2020, 100, 2761–2773. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Nakamura, T.; Tsuchiya, T. Advantages of NH4+ on Growth, Nitrogen Uptake and Root Respiration of Phragmites Australis. Plant Soil 2010, 331, 463–470. [Google Scholar] [CrossRef]
- Liu, Y.; Von Wirén, N. Ammonium as a Signal for Physiological and Morphological Responses in Plants. Journal of Experimental Botany 2017, 68, 2581–2592. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liu, L.; Ma, N.; Yang, J.; Dong, Z.; Zhang, J.; Zhang, J.; Cai, M. Effect of Ammonia Stress on Carbon Metabolism in Tolerant Aquatic Plant—Myriophyllum Aquaticum. Environmental Pollution 2020, 263, 114412. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S. R.; Luciani, A.; Potter, S. C.; Qureshi, M.; Richardson, L. J.; Salazar, G. A.; Smart, A.; Sonnhammer, E. L. L.; Hirsh, L.; Paladin, L.; Piovesan, D.; Tosatto, S. C. E.; Finn, R. D. The Pfam Protein Families Database in 2019. Nucleic Acids Res 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Finn, R. D.; Clements, J.; Eddy, S. R. HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res 2011, 39 (Web Server issue), W29–37. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, Z.; Zhai, Y.; Huang, H.; Vainstein, A.; Ma, H. Polygalacturonase Gene Family Analysis Identifies FcPG12 as a Key Player in Fig (Ficus Carica L.) Fruit Softening. BMC Plant Biology 2023, 23, 320. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H. R.; Frank, M. H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.-T.; Chen, L.-X.; Jin, J.; Du, Z.-K.; Li, J.-M. Genome-Wide Identification and Analysis of BZIP Gene Family Reveal Their Roles during Development and Drought Stress in Wheel Wingnut (Cyclocarya Paliurus). BMC Genomics 2022, 23, 743. [Google Scholar] [CrossRef]
- Ren, C.; Wang, H.; Zhou, Z.; Jia, J.; Zhang, Q.; Liang, C.; Li, W.; Zhang, Y.; Yu, G. Genome-Wide Identification of the B3 Gene Family in Soybean and the Response to Melatonin under Cold Stress. Front Plant Sci 2022, 13, 1091907. [Google Scholar] [CrossRef]
- Chen, M.; Zhu, K.; Xie, J.; Liu, J.; Tan, P.; Peng, F. Genome-Wide Identification and Expression Analysis of AMT and NRT Gene Family in Pecan (Carya Illinoinensis) Seedlings Revealed a Preference for NH4+-N. Int J Mol Sci 2022, 23, 13314. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed]
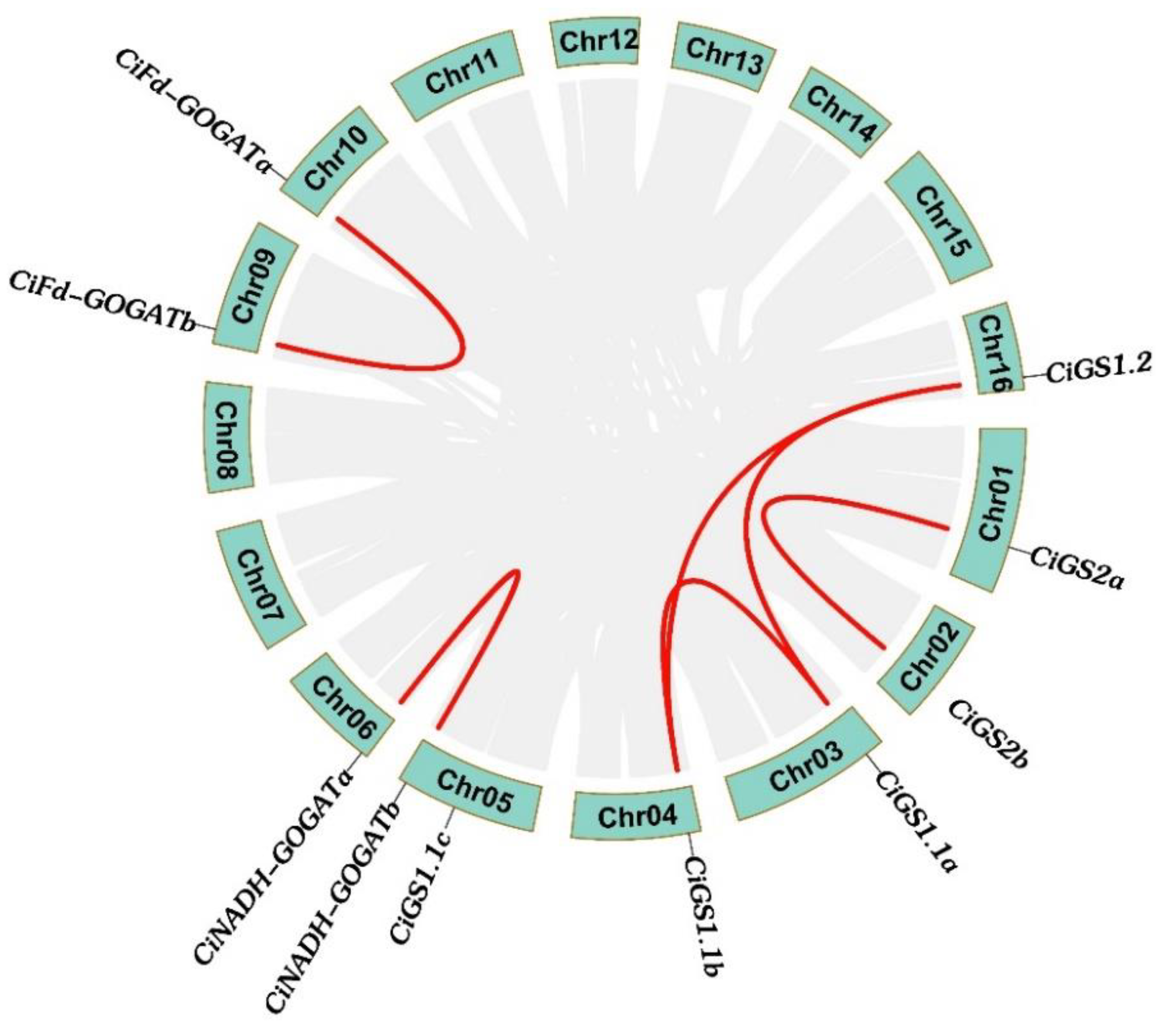
| Gene Pairs | Ka | Ks | Ka/Ks |
|---|---|---|---|
| CiGS1.1a/CiGS1.1b | 0.0346 | 0.3328 | 0.1039 |
| CiGS1.1a/CiGS1.1c | 0.0702 | 2.1672 | 0.0323 |
| CiGS1.1b/CiGS1.1c | 0.0628 | 1.6209 | 0.0387 |
| CiGS1.2/CiGS1.1a | 0.0678 | 1.0862 | 0.0624 |
| CiGS1.2/CiGS1.1b | 0.0689 | 1.0739 | 0.0641 |
| CiGS1.2/CiGS1.1c | 0.0905 | —— | —— |
| CiGS2a/CiGS2b | 0.0288 | 0.2568 | 0.1123 |
| CiNADH-GOGATb/CiNADH-GOGATa | 0.0462 | 0.3583 | 0.1291 |
| CiFd-GOGATa/CiFd-GOGATb | 0.0310 | 0.2917 | 0.1064 |
| Gene name | Forward primer (5’-3’) | Reverse primer (5’-3’) |
| CiGS1.1a | AATTGACAAGCTTGGCCGGA | CGATTGGCGACACCCCATAA |
| CiGS1.1b | CCCAAGCCAATTCAGGGTGAT | CCTCAGCCCAAGCTTTCCAA |
| CiGS1.1c | TTGCCGAGGAACCCTGGTAT | AATGCCTTGTCTGCCCCTAC |
| CiGS1.2 | CGCTAAAATCGCCTGTTGGG | ACCCGATCCACCGATCCATA |
| CiGS2a | CATCCGCCATTCCTGATCTGA | CCCCACATCTTTGCTGTCGT |
| CiGS2b | TATTGTAAGGGCTTCCCCCAC | CTGTGCCATTTTCACCTCGG |
| CiFd-GOGATa | GACGTGCAAGTACCGCCTT | CCAACTTTGCAACCTTCGGT |
| CiFd-GOGATb | GAGGAGCTTCCCGCATTTTC | CAAGTTTGCAACCCTCGGTC |
| CiNADH-GOGATa | TGAGCAGAAAGTTGAGGCAGA | GATTCACCCTCTTCTACCTTATTGG |
| CiNADH-GOGATb | GGGAATTCTAATCAGAAGGCAGA | CCTGTATTGAACACCCTCACGA |
| Actin | GCTGAACGGGAAATTGTC | AGAGATGGCTGGAAGAGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





