Submitted:
26 October 2023
Posted:
27 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
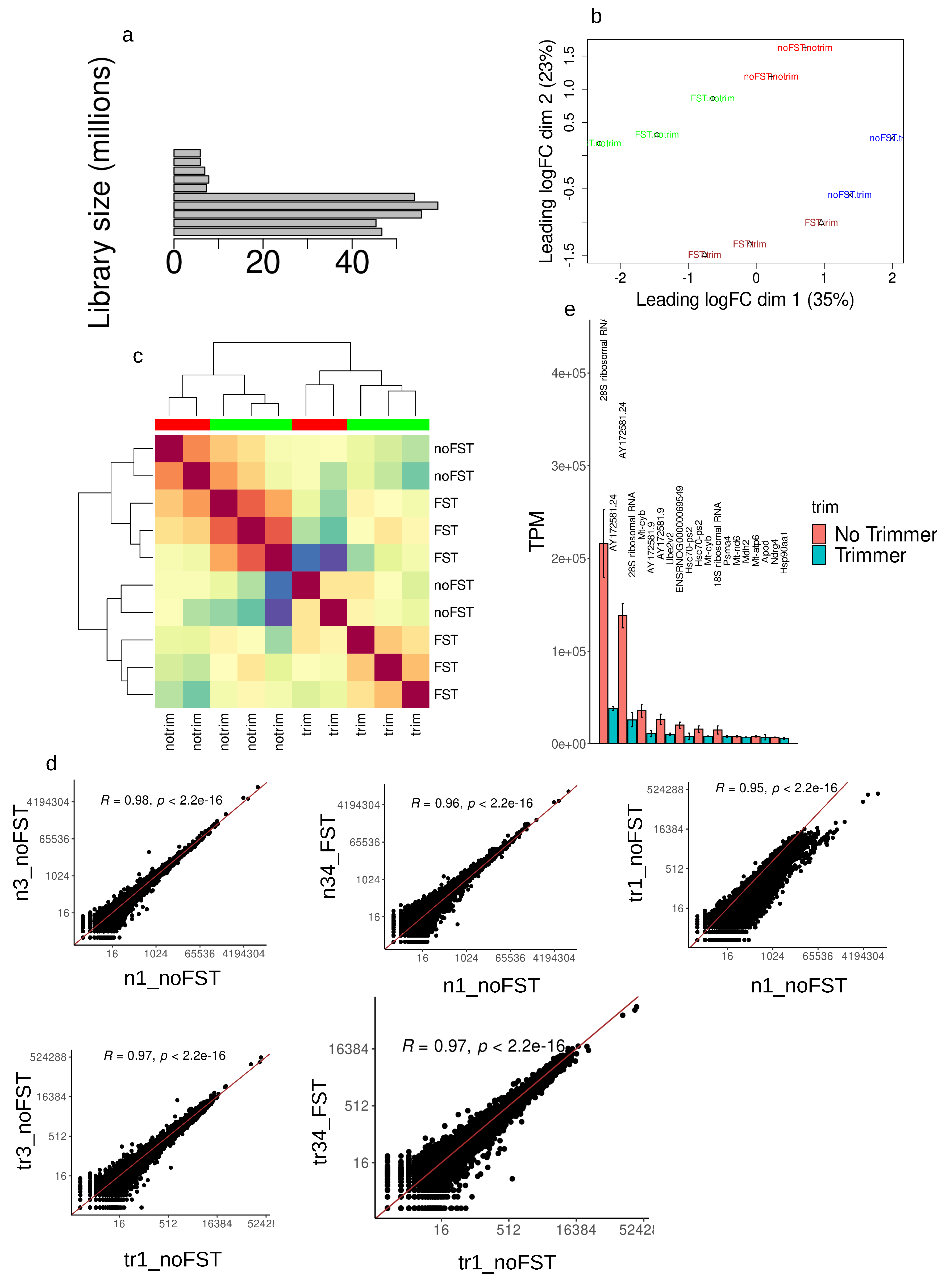
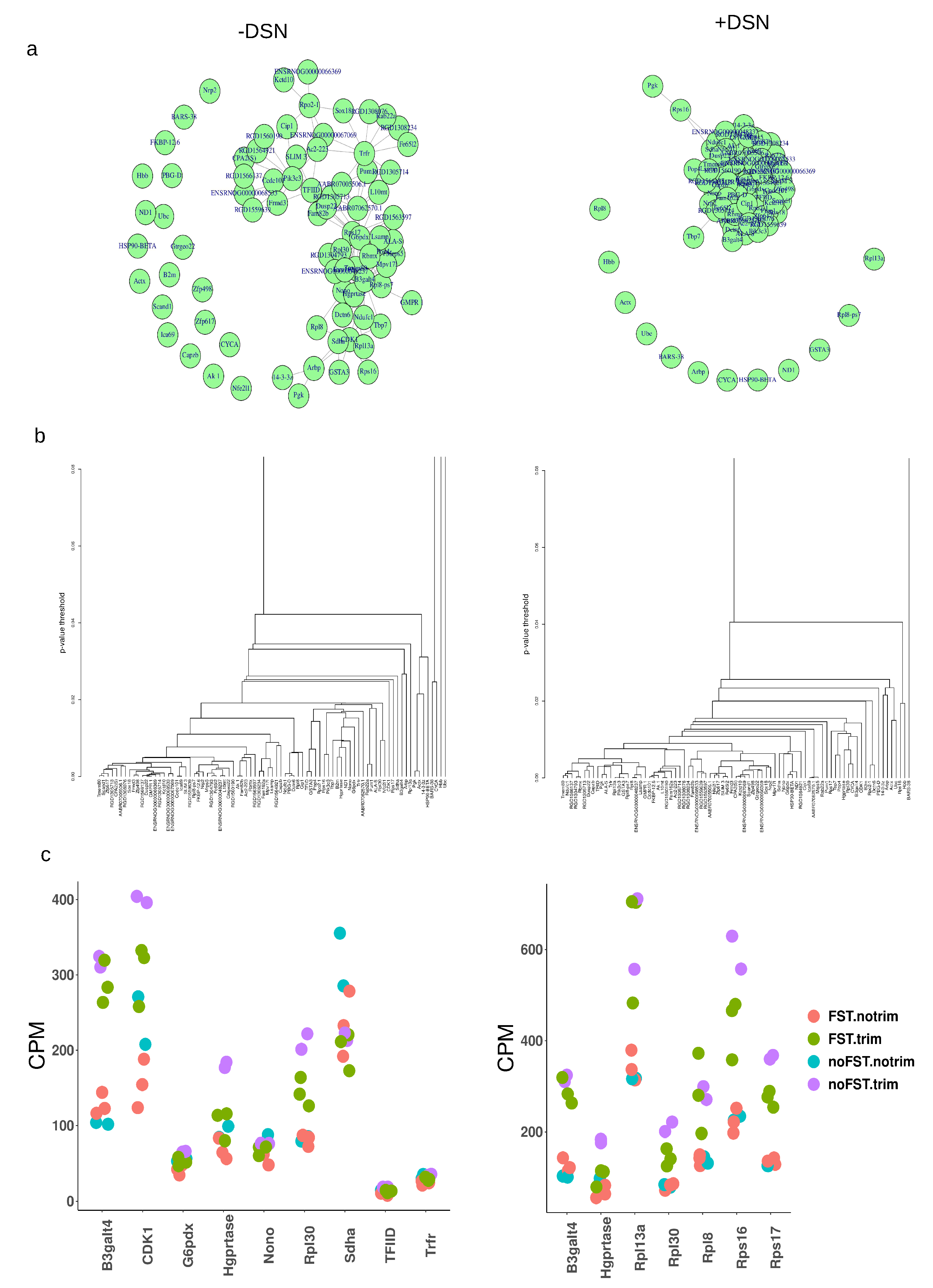
2.1. Library characterization and consistency validation
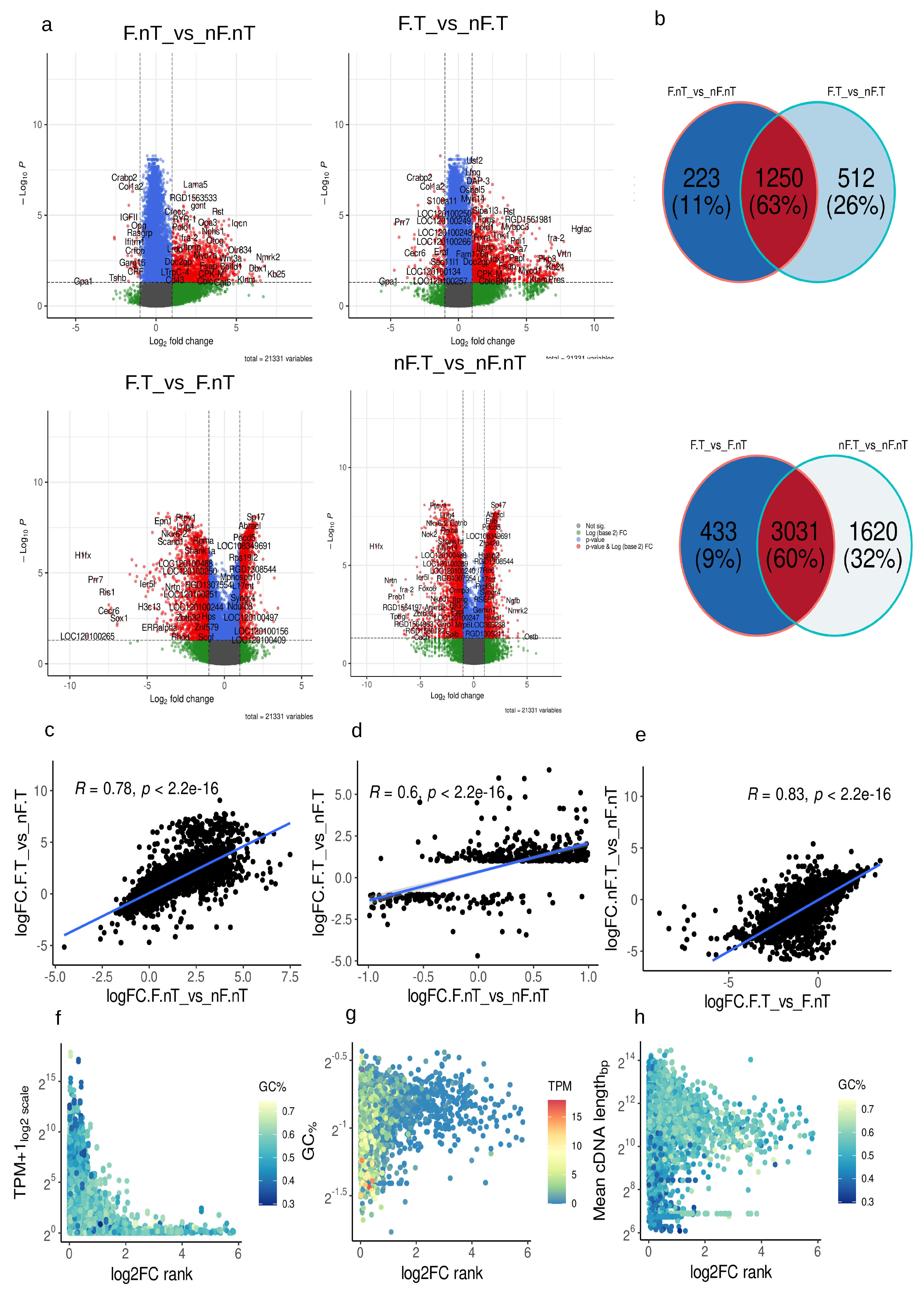
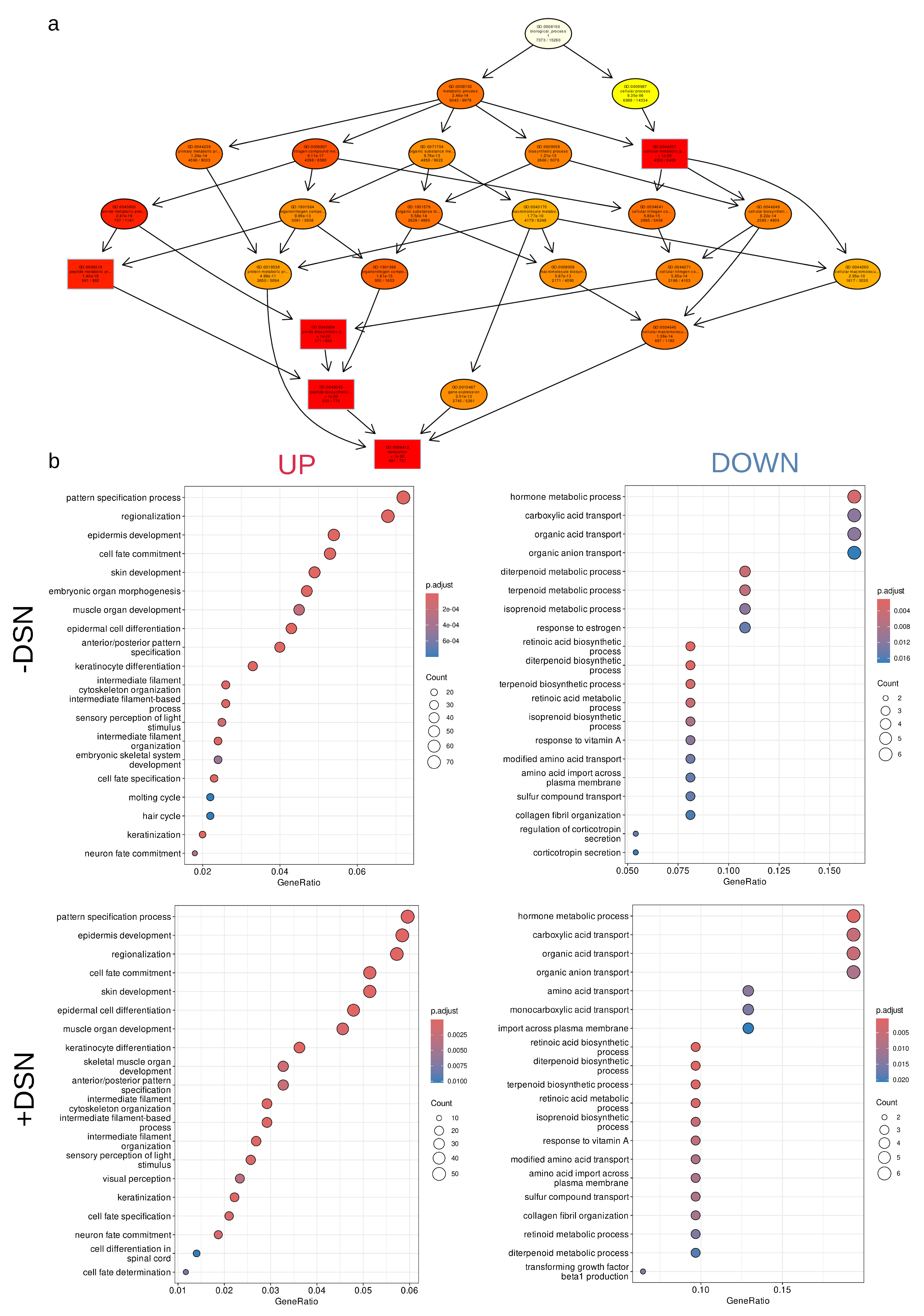
2.2. Transcriptomic alteration and functional annotation
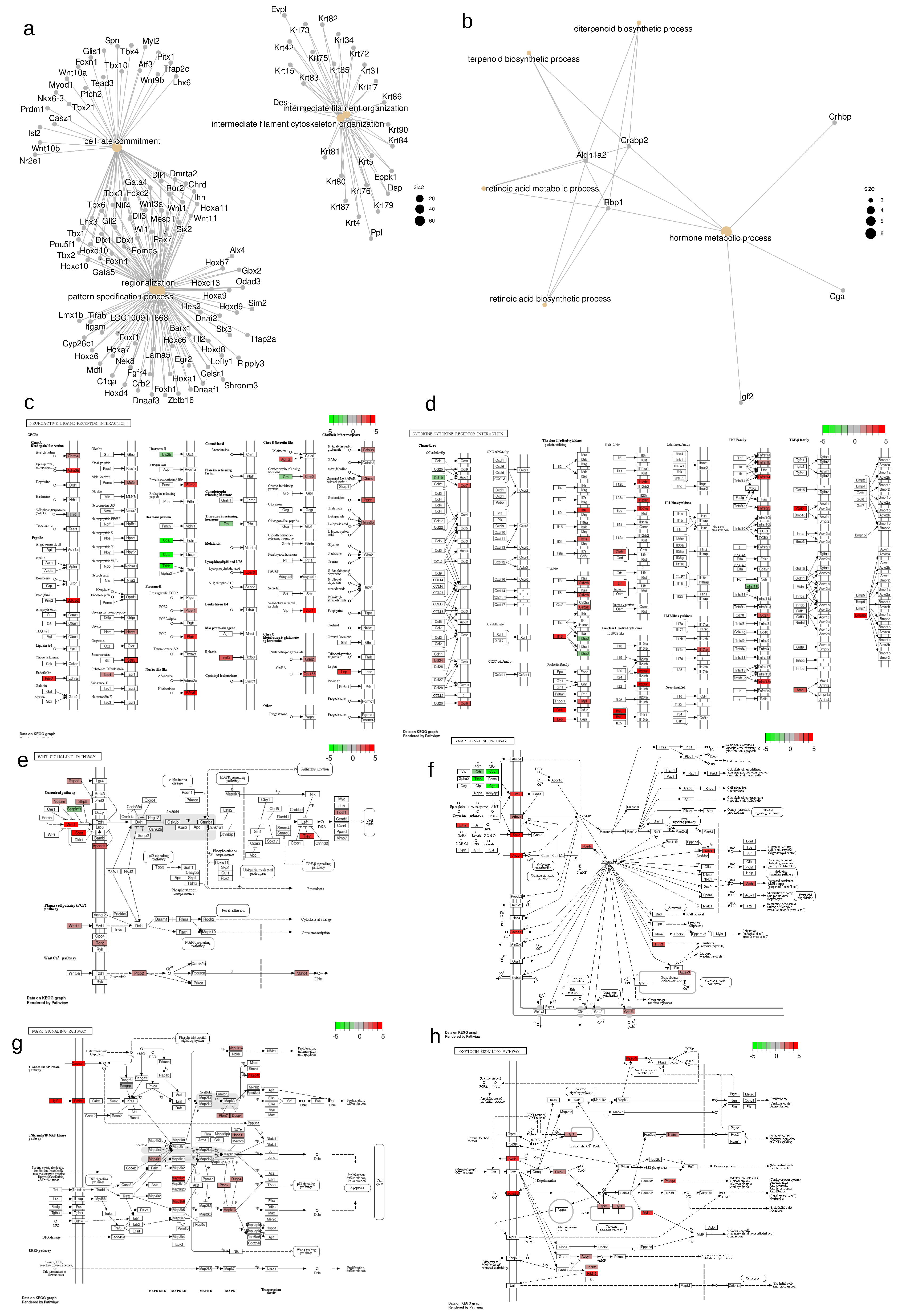
2.3. Changes in neuroactive ligand-receptor interaction
2.4. Changes in Cytokine-cytokine receptor interaction
2.5. Changes in Wnt signaling pathway
2.6. Changes in Calcium Signaling, cAMP response and MAPK pathways
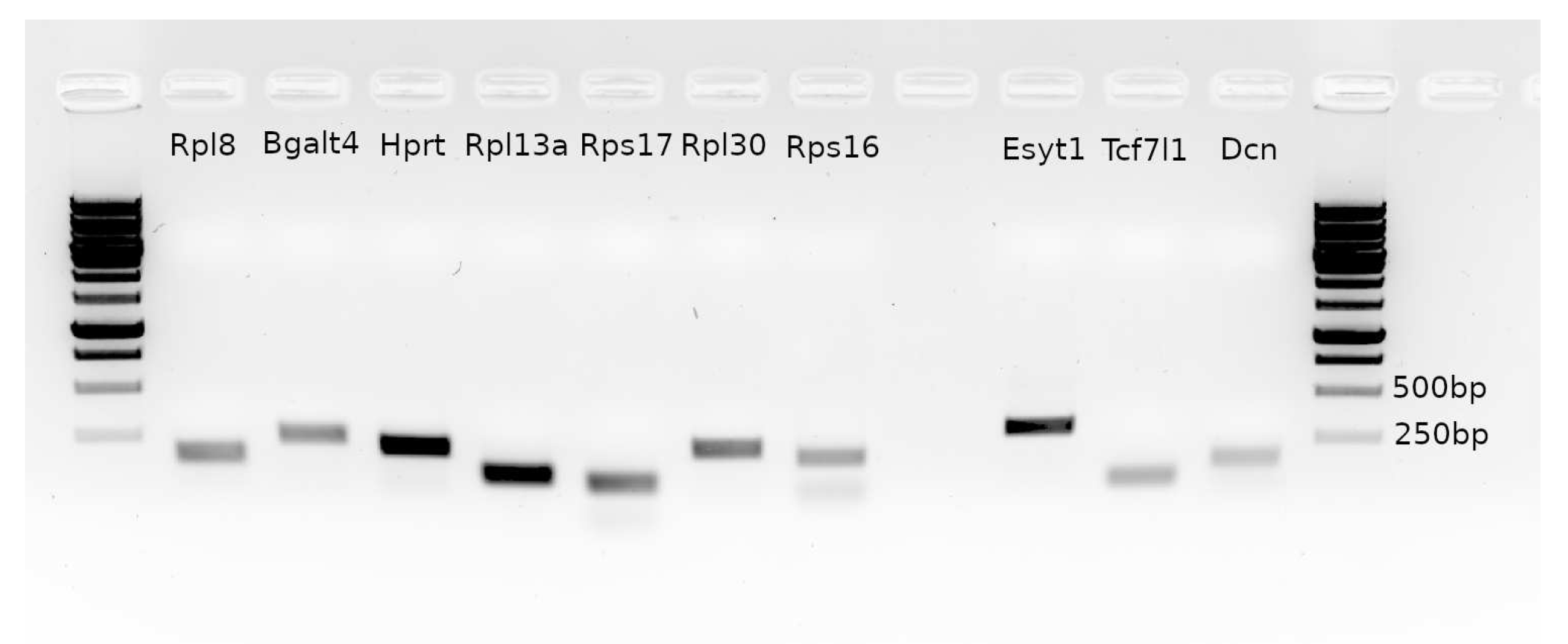
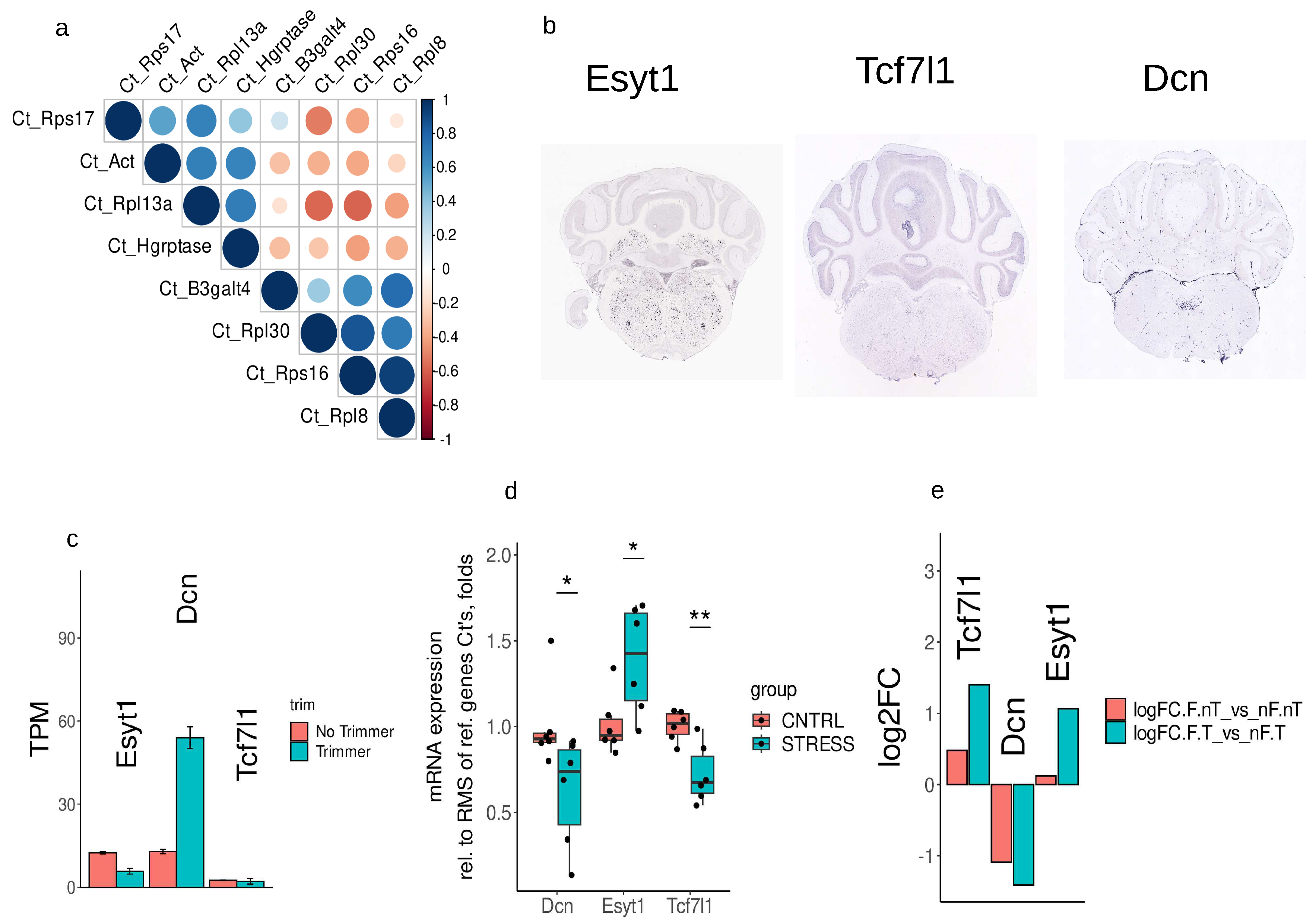
2.7. Choosing reference gene for RT-qPCR
2.8. Real-time RT-qPCR analysis
3. Discussion
4. Materials and Methods
4.1. Animals and Experimental Design
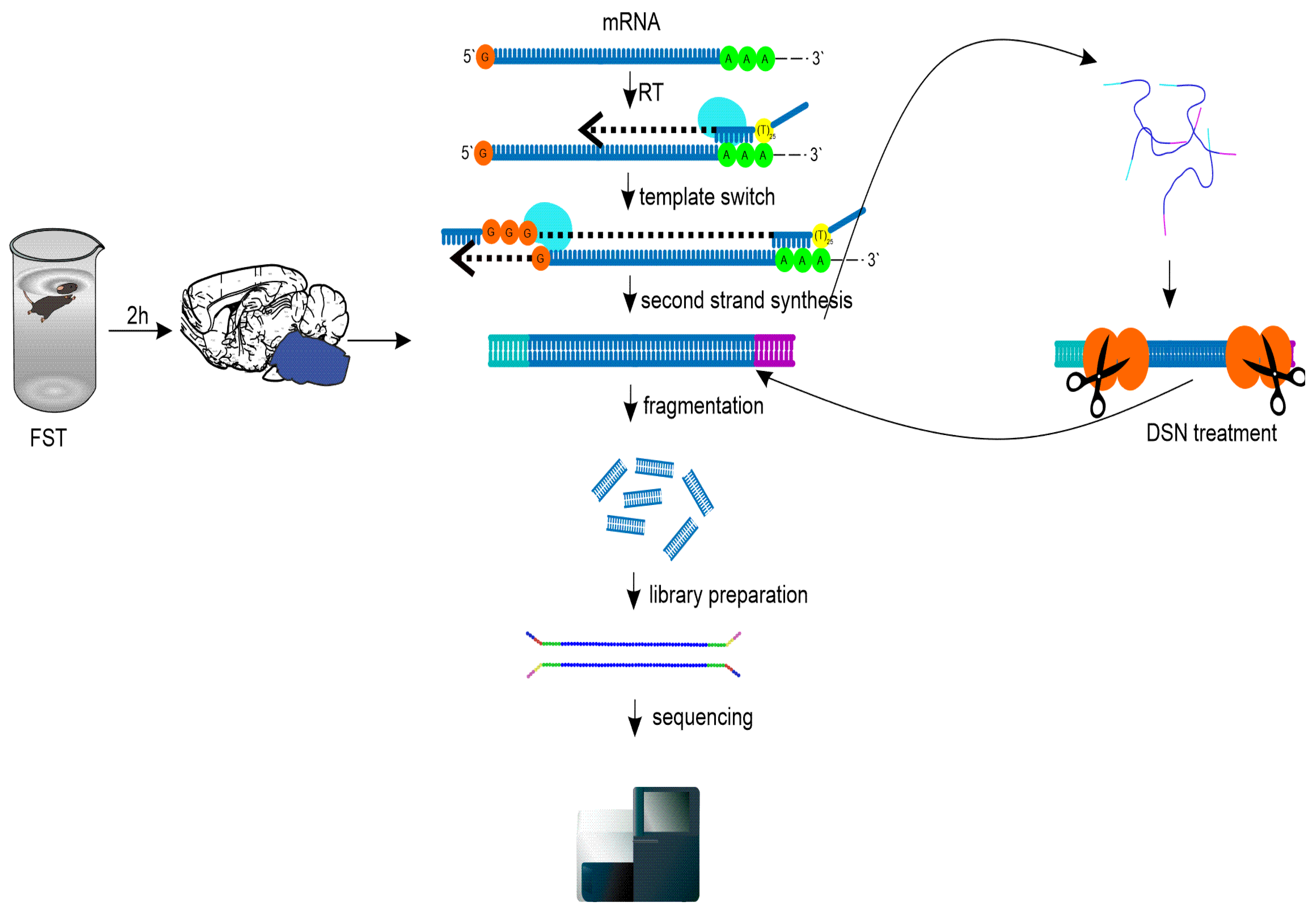
4.2. Force swim test and sample collections
4.3. Sequencing libraries preparation and DSN treatment
4.4. Bioinformatics analysis
4.5. Real-time RT-qPCR analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Supplementary Materials
| Name | Seqence 5′-3′ | product sze, bp | Tm, °C |
|---|---|---|---|
| Rps17F | AGACTGTGAAGAAGGCGGC | 81 | 64 |
| Rps17R | CGCGCTTGTTGGTGTGAAA | 81 | 64 |
| Rps16F | AGAGCTGTGCCTGCTTCTG | 180 | 64 |
| Rps16R | CTACATGCCTGGGGTTGGG | 180 | 64 |
| Rpl8F | CTGTTTGGGGGCTCTCTGG | 163 | 62 |
| Rpl8R | TAGGATGGTCACCTGCGGA | 163 | 62 |
| Rpl13aF | AGGTGGTGGTTGTACGCTG | 109 | 62 |
| Rpl13aR | CGAGACGGGTTGGTGTTCA | 109 | 62 |
| B3galt4F | TGTGGGAGTAAGCGCAAGG | 240 | 62 |
| B3galt4R | ATGAACCGGCACCTCAAGG | 240 | 62 |
| Rpl30F | TGCATGCATGGTCCCCATT | 214 | 64 |
| Rpl30R | GTTCCTGGGACCCAAGAGC | 214 | 64 |
| HgprtaseF | ATGTCGACCCTCAGTCCCA | 209 | 62 |
| HgprtaseR | CCCTTCAGCACACAGAGGG | 209 | 62 |
| Esyt1F | CTGACTTCTCGCCTTGCCTC | 299 | 61.7 |
| Esyt1R | GCTGTATTTGGCATGAGCTACG | 299 | 61.7 |
| Tcf7l1F | CCCCTTGTGCTGTGGTAGG | 75 | 61.7 |
| Tcf7l1R | GGAGGGCAGAGAGTCAGGA | 75 | 61.7 |
| DcnF | TGGAGCCTTGCAGGGAATG | 148 | 63.8 |
| DcnR | TTTCAGGCTGGCTGCATCA | 148 | 63.8 |
| -actin | Rn00667869_m1(Thermo) | - | 60 |
References
- Lanshakov, D.A.; Sukhareva, E.V.; Bulygina, V.V.; Bannova, A.V.; Shaburova, E.V.; Kalinina, T.S. Single neonatal dexamethasone administration has long-lasting outcome on depressive-like behaviour, Bdnf, Nt-3, p75ngfr and sorting receptors (SorCS1-3) stress reactive expression. Scientific Reports 2021, 11, 8092. [Google Scholar] [CrossRef] [PubMed]
- Chaves, T.; Fazekas, C.L.; Horváth, K.; Correia, P.; Szabó, A.; Török, B.; Bánrévi, K.; Zelena, D. Stress Adaptation and the Brainstem with Focus on Corticotropin-Releasing Hormone. International Journal of Molecular Sciences 2021, 22, 9090. [Google Scholar] [CrossRef] [PubMed]
- Sattin, D.; Leonardi, M.; Picozzi, M. The autonomic nervous system and the brainstem: A fundamental role or the background actors for consciousness generation? Hypothesis, evidence, and future directions for rehabilitation and theoretical approaches. Brain and Behavior 2020, 10, e01474. [Google Scholar] [CrossRef] [PubMed]
- Giustino, T.F.; Maren, S. Noradrenergic Modulation of Fear Conditioning and Extinction. Frontiers in Behavioral Neuroscience 2018, 12, 43. [Google Scholar] [CrossRef]
- Ross, J.A.; Van Bockstaele, E.J. The Locus Coeruleus- Norepinephrine System in Stress and Arousal: Unraveling Historical, Current, and Future Perspectives. Frontiers in Psychiatry 2021, 11, 601519. [Google Scholar] [CrossRef] [PubMed]
- Chaoui, N.; Anarghou, H.; Laaroussi, M.; Essaidi, O.; Najimi, M.; Chigr, F.; Biological Engineering Laboratory, Faculty of Sciences and Techniques, Sultan Moulay Slimane University, Beni Mellal, Morocco. Long lasting effect of acute restraint stress on behavior and brain anti-oxidative status. AIMS Neuroscience 2022, 9, 57–75. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Cho, Y.J.; Won, S.; Lee, J.E.; Jin Yu, H.; Kim, S.; Schroth, G.P.; Luo, S.; Chun, J. Duplex-specific nuclease efficiently removes rRNA for prokaryotic RNA-seq. Nucleic Acids Research 2011, 39, e140. [Google Scholar] [CrossRef]
- Zhulidov, P.A.; Bogdanova, E.A.; Shcheglov, A.S.; Vagner, L.L.; Khaspekov, G.L.; Kozhemyako, V.B.; Matz, M.V.; Meleshkevitch, E.; Moroz, L.L.; Lukyanov, S.A.; Shagin, D.A. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Research 2004, 32, e37. [Google Scholar] [CrossRef]
- Baserga, S.J.; Linnenbach, A.J.; Malcolm, S.; Ghosh, P.; Malcolm, A.D.; Takeshita, K.; Forget, B.G.; Benz, E.J. Polyadenylation of a human mitochondrial ribosomal RNA transcript detected by molecular cloning. Gene 1985, 35, 305–312. [Google Scholar] [CrossRef]
- Slomovic, S.; Laufer, D.; Geiger, D.; Schuster, G. Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Research 2006, 34, 2966–2975. [Google Scholar] [CrossRef]
- Margiotta, A.; Bucci, C. Role of Intermediate Filaments in Vesicular Traffic. Cells 2016, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Curis, E.; Courtin, C.; Geoffroy, P.A.; Laplanche, J.L.; Saubaméa, B.; Marie-Claire, C. Determination of sets of covariating gene expression using graph analysis on pairwise expression ratios. Bioinformatics 2019, 35, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Libersat, F.; Pflueger, H.J. Monoamines and the Orchestration of Behavior. BioScience 2004, 54, 17. [Google Scholar] [CrossRef]
- Von Ziegler, L.M.; Floriou-Servou, A.; Waag, R.; Das Gupta, R.R.; Sturman, O.; Gapp, K.; Maat, C.A.; Kockmann, T.; Lin, H.Y.; Duss, S.N.; Privitera, M.; Hinte, L.; Von Meyenn, F.; Zeilhofer, H.U.; Germain, P.L.; Bohacek, J. Multiomic profiling of the acute stress response in the mouse hippocampus. Nature Communications 2022, 13, 1824. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, C.; Andermann, M.L.; Scammell, T.E. Control of arousal by the orexin neurons. Current Opinion in Neurobiology 2013, 23, 752–759. [Google Scholar] [CrossRef] [PubMed]
- Inutsuka, A.; Yamanaka, A. The physiological role of orexin/hypocretin neurons in the regulation of sleep/wakefulness and neuroendocrine functions. Frontiers in Endocrinology 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, G.B.; Lakis, G.A.; Zhoba, H. Sleep-wake and arousal dysfunctions in post-traumatic stress disorder: Role of orexin systems. Brain Research Bulletin 2022, 186, 106–122. [Google Scholar] [CrossRef]
- Fu, C.Y.; Tang, X.L.; Yang, Q.; Chen, Q.; Wang, R. Effects of rat/mouse hemokinin-1, a mammalian tachykinin peptide, on the antinociceptive activity of pethidine administered at the peripheral and supraspinal level. Behavioural Brain Research 2007, 184, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Page, N.M.; Bell, N.J.; Gardiner, S.M.; Manyonda, I.T.; Brayley, K.J.; Strange, P.G.; Lowry, P.J. Characterization of the endokinins: Human tachykinins with cardiovascular activity. Proceedings of the National Academy of Sciences 2003, 100, 6245–6250. [Google Scholar] [CrossRef]
- Golias, C.; Charalabopoulos, A.; Stagikas, D.; Charalabopoulos, K.; Batistatou, A. The kinin system–bradykinin: biological effects and clinical implications. Multiple role of the kinin system–bradykinin. Hippokratia 2007, 11, 124–128. [Google Scholar]
- Zhang, Y.; Crofton, E.J.; Smith, T.E.; Koshy, S.; Li, D.; Green, T.A. Manipulation of retinoic acid signaling in the nucleus accumbens shell alters rat emotional behavior. Behavioural Brain Research 2019, 376, 112177. [Google Scholar] [CrossRef] [PubMed]
- Babenko, V.N.; Shishkina, G.T.; Lanshakov, D.A.; Sukhareva, E.V.; Dygalo, N.N. LPS Administration Impacts Glial Immune Programs by Alternative Splicing. Biomolecules 2022, 12, 277. [Google Scholar] [CrossRef] [PubMed]
- Shishkina, G.T.; Kalinina, T.S.; Lanshakov, D.A.; Bulygina, V.V.; Komysheva, N.P.; Bannova, A.V.; Drozd, U.S.; Dygalo, N.N. Genes Involved by Dexamethasone in Prevention of Long-Term Memory Impairment Caused by Lipopolysaccharide-Induced Neuroinflammation. Biomedicines 2023, 11, 2595. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Li, J.; Huang, Y.; Wei, P.; Miao, W.; Yang, Y.; Gao, Y. Interleukin 13 promotes long-term recovery after ischemic stroke by inhibiting the activation of STAT3. Journal of Neuroinflammation 2022, 19, 112. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, Y.; Wang, C.; Han, T.; Liu, H.; Sun, L.; Hong, J.; Hashimoto, M.; Wei, J. The reciprocal interactions between microglia and T cells in Parkinson’s disease: a double-edged sword. Journal of Neuroinflammation 2023, 20, 33. [Google Scholar] [CrossRef]
- Parkhurst, C.N.; Yang, G.; Ninan, I.; Savas, J.N.; Yates, J.R.; Lafaille, J.J.; Hempstead, B.L.; Littman, D.R.; Gan, W.B. Microglia Promote Learning-Dependent Synapse Formation through Brain-Derived Neurotrophic Factor. Cell 2013, 155, 1596–1609. [Google Scholar] [CrossRef] [PubMed]
- Paolicelli, R.C.; Bolasco, G.; Pagani, F.; Maggi, L.; Scianni, M.; Panzanelli, P.; Giustetto, M.; Ferreira, T.A.; Guiducci, E.; Dumas, L.; Ragozzino, D.; Gross, C.T. Synaptic Pruning by Microglia Is Necessary for Normal Brain Development. Science 2011, 333, 1456–1458. [Google Scholar] [CrossRef] [PubMed]
- Sugama, S.; Takenouchi, T.; Hashimoto, M.; Ohata, H.; Takenaka, Y.; Kakinuma, Y. Stress-induced microglial activation occurs through β-adrenergic receptor: noradrenaline as a key neurotransmitter in microglial activation. Journal of Neuroinflammation 2019, 16, 266. [Google Scholar] [CrossRef]
- Martins, L.; Seoane-Collazo, P.; Contreras, C.; González-García, I.; Martínez-Sánchez, N.; González, F.; Zalvide, J.; Gallego, R.; Diéguez, C.; Nogueiras, R.; Tena-Sempere, M.; López, M. A Functional Link between AMPK and Orexin Mediates the Effect of BMP8B on Energy Balance. Cell Reports 2016, 16, 2231–2242. [Google Scholar] [CrossRef]
- Teo, S.; Salinas, P.C. Wnt-Frizzled Signaling Regulates Activity-Mediated Synapse Formation. Frontiers in Molecular Neuroscience 2021, 14, 683035. [Google Scholar] [CrossRef]
- Tang, S.J. Synaptic Activity-Regulated Wnt Signaling in Synaptic Plasticity, Glial Function and Chronic Pain. CNS & Neurological Disorders - Drug Targets 2014, 13, 737–744. [Google Scholar] [CrossRef]
- Bem, J.; Brożko, N.; Chakraborty, C.; Lipiec, M.A.; Koziński, K.; Nagalski, A.; Szewczyk, M.; Wiśniewska, M.B. Wnt/β-catenin signaling in brain development and mental disorders: keeping TCF7L2 in mind. FEBS Letters 2019, 593, 1654–1674. [Google Scholar] [CrossRef] [PubMed]
- Balle, F.; Kaiserslautern, T.U. (Eds.) Tagungsband / Young Researcher Symposium (YRS) 2013; Fraunhofer Verlag: Stuttgart, 2013. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: a fast spliced aligner with low memory requirements. Nature Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Research 2019, 47, e47–e47. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Research 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; Fu, X.; Liu, S.; Bo, X.; Yu, G. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





