Submitted:
26 October 2023
Posted:
27 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Arginine in Protein Structure and Intrinsic Disorder
3. Cation-π Interactions Often Involve Arginine
4. Importance of Arginine in Post-Translational Modifications of Proteins
5. Crosslinks of Arginines and Their Biological Importance
6. Arginine in the Active Sites of Enzymes
7. Metabolic Importance of Arginine
8. Arginine in Protein Interactions
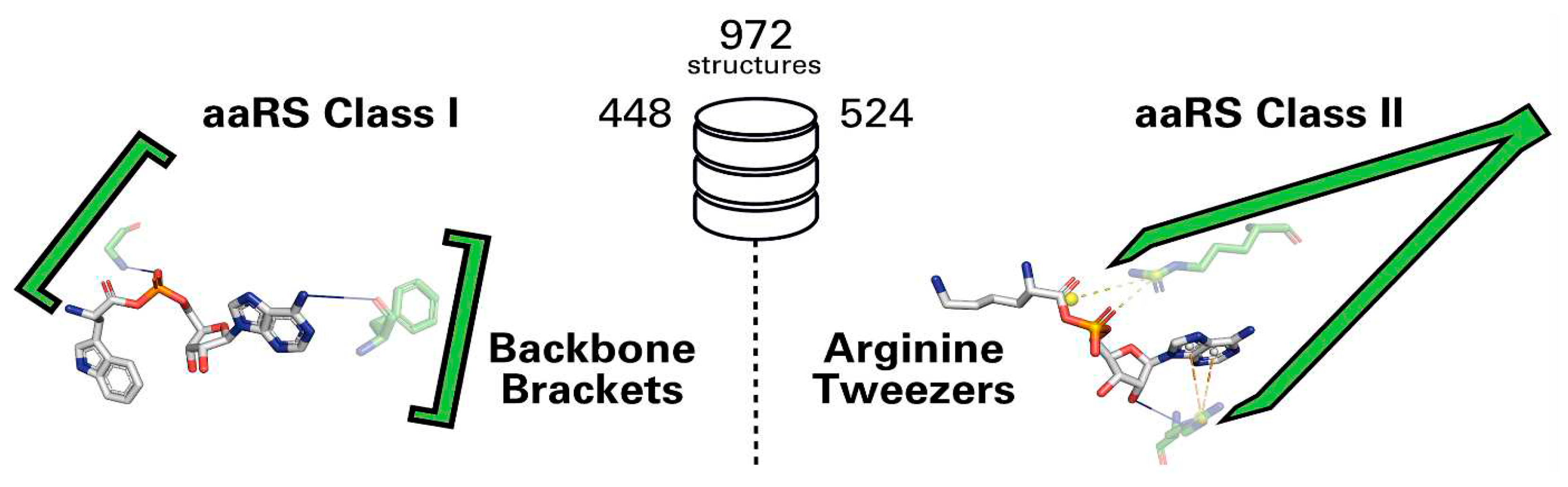
8.1. Arginine Tweezers
8.2. Sprocket Arginine Residues in Histones
8.3. Rings, Strings, and Stacks Made of the Arginine Residues at Protein-Protein Interfaces
9. Arginine Applications in Biotechnology
9.1. Arginine as an Excipient in Drug Formulations
9.2. Arg as a Suppressor of Protein Aggregation during Protein Refolding
9.3. Arginine Containing Cationic Cell-Penetrating Peptides in Drug Design
9.4. Arginine in Ion-Exchange, Hydrophobic Interaction, and Affinity Chromatography
10. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fitch, C.A.; Platzer, G.; Okon, M.; Garcia-Moreno, B.E.; McIntosh, L.P. Arginine: Its pKa value revisited. Protein Sci. 2015, 24, 752–761. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, T.; Tsumoto, K.; Kita, Y.; Chang, B.; Ejima, D. Biotechnology applications of amino acids in protein purification and formulations. Amino Acids 2007, 33, 587–605. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.X.; Cao, Y.; Tang, Y.L.; Wang, J.H.; Ding, Y.H.; Tan, H.; Chen, Z.L.; Fang, R.Q.; Yin, J.; Chen, R.C.; et al. Improving mass spectrometry analysis of protein structures with arginine-selective chemical cross-linkers. Nat. Commun. 2019, 10, 3911. [Google Scholar] [CrossRef] [PubMed]
- Vacic, V.; Uversky, V.N.; Dunker, A.K.; Lonardi, S. Composition Profiler: A tool for discovery and visualization of amino acid composition differences. BMC Bioinform. 2007, 8, 211. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A.; Apweiler, R.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. The Universal Protein Resource (UniProt). Nucleic Acids Res. 2005, 33, D154–D159. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Vihinen, M. Spectrum of disease-causing mutations in protein secondary structures. BMC Struct. Biol. 2007, 7, 56. [Google Scholar] [CrossRef]
- Petsko, G.A.; Ringe, D. Protein Structure and Function; New Science Press: 2004.
- Stryer, L. Biochemistry, 3rd ed.; W H Freeman: New York, 1988. [Google Scholar]
- Harms, M.J.; Schlessman, J.L.; Sue, G.R.; Garcia-Moreno, B. Arginine residues at internal positions in a protein are always charged. Proc. Natl. Acad. Sci. USA 2011, 108, 18954–18959. [Google Scholar] [CrossRef] [PubMed]
- Brinda, K.V.; Vishveshwara, S. A network representation of protein structures: Implications for protein stability. Biophys. J. 2005, 89, 4159–4170. [Google Scholar] [CrossRef]
- Chen, H.; Gu, F.; Huang, Z. Improved Chou-Fasman method for protein secondary structure prediction. BMC Bioinform. 2006, 7 (Suppl. 4), S14. [Google Scholar] [CrossRef]
- Kumar, S.; Bansal, M. Geometrical and sequence characteristics of alpha-helices in globular proteins. Biophys. J. 1998, 75, 1935–1944. [Google Scholar] [CrossRef]
- Pace, C.N.; Scholtz, J.M. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998, 75, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Meuzelaar, H.; Vreede, J.; Woutersen, S. Influence of Glu/Arg, Asp/Arg, and Glu/Lys Salt Bridges on alpha-Helical Stability and Folding Kinetics. Biophys. J. 2016, 110, 2328–2341. [Google Scholar] [CrossRef] [PubMed]
- Huyghues-Despointes, B.M.; Scholtz, J.M.; Baldwin, R.L. Helical peptides with three pairs of Asp-Arg and Glu-Arg residues in different orientations and spacings. Protein Sci. 1993, 2, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Munoz, V.; Serrano, L. Elucidating the folding problem of helical peptides using empirical parameters. Nat. Struct. Biol. 1994, 1, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Meuzelaar, H.; Tros, M.; Huerta-Viga, A.; van Dijk, C.N.; Vreede, J.; Woutersen, S. Solvent-Exposed Salt Bridges Influence the Kinetics of alpha-Helix Folding and Unfolding. J. Phys. Chem. Lett. 2014, 5, 900–904. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M.; Obradovic, Z.; Mathura, V.; Braun, W.; Garner, E.C.; Young, J.; Takayama, S.; Brown, C.J.; Dunker, A.K. The protein non-folding problem: Amino acid determinants of intrinsic order and disorder. Pac. Symp. Biocomput. 2001, 89–100. [Google Scholar]
- Radivojac, P.; Iakoucheva, L.M.; Oldfield, C.J.; Obradovic, Z.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder and functional proteomics. Biophys. J. 2007, 92, 1439–1456. [Google Scholar] [CrossRef]
- Bhowmick, J.; Chandra, S.; Varadarajan, R. Deep mutational scanning to probe specificity determinants in proteins. In Structure and Intrinsic Disorder in Enzymology; Elsevier: 2023; pp. 31–71.
- Smith, C.A.; Chen, L.; Frankel, A.D. Using peptides as models of RNA-protein interactions. Methods Enzym. 2000, 318, 423–438. [Google Scholar] [CrossRef]
- Oliva, R. Protamines and male infertility. Hum. Reprod. Update 2006, 12, 417–435. [Google Scholar] [CrossRef]
- Miescher, F. Das Protamin, eine neue organische Base aus den Samenfäden des Rheinlachses. Berichte Der Dtsch. Chem. Ges. 1874, 7, 376–379. [Google Scholar] [CrossRef]
- Balhorn, R. The protamine family of sperm nuclear proteins. Genome Biol. 2007, 8, 227. [Google Scholar] [CrossRef] [PubMed]
- Hud, N.V.; Milanovich, F.P.; Balhorn, R. Evidence of novel secondary structure in DNA-bound protamine is revealed by Raman spectroscopy. Biochemistry 1994, 33, 7528–7535. [Google Scholar] [CrossRef]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Brown, C.J.; Uversky, V.N.; Dunker, A.K. Comparing and combining predictors of mostly disordered proteins. Biochemistry 2005, 44, 1989–2000. [Google Scholar] [CrossRef]
- Mohan, A.; Sullivan, W.J., Jr.; Radivojac, P.; Dunker, A.K.; Uversky, V.N. Intrinsic disorder in pathogenic and non-pathogenic microbes: Discovering and analyzing the unfoldomes of early-branching eukaryotes. Mol. Biosyst. 2008, 4, 328–340. [Google Scholar] [CrossRef]
- Huang, F.; Oldfield, C.; Meng, J.; Hsu, W.L.; Xue, B.; Uversky, V.N.; Romero, P.; Dunker, A.K. Subclassifying disordered proteins by the CH-CDF plot method. Pac. Symp. Biocomput. 2012, 128–139. [Google Scholar]
- Dayhoff, G.W., 2nd; Uversky, V.N. Rapid prediction and analysis of protein intrinsic disorder. Protein Sci. 2022, 31, e4496. [Google Scholar] [CrossRef]
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J. Cell. Biochem. 2011, 112, 3256–3267. [Google Scholar] [CrossRef]
- Uversky, V.N. Analyzing IDPs in interactomes. Intrinsically Disordered Proteins: Methods and Protocols.
- Oliva, R.; Dixon, G.H. Vertebrate protamine genes and the histone-to-protamine replacement reaction. Prog. Nucleic Acid Res. Mol. Biol. 1991, 40, 25–94. [Google Scholar] [CrossRef]
- Balhorn, R.; Cosman, M.; Thornton, K.; Krishnan, V.; Corzett, M.; Bench, G.; Kramer, C.; Hud, N.; Allen, M.; Prieto, M. Protamine Mediated Condensation of DNA in Mammalian Sperm; Cache River Press: 1999.
- Bench, G.S.; Friz, A.M.; Corzett, M.H.; Morse, D.H.; Balhorn, R. DNA and total protamine masses in individual sperm from fertile mammalian subjects. Cytometry 1996, 23, 263–271. [Google Scholar] [CrossRef]
- Marshall, M.S.; Steele, R.P.; Thanthiriwatte, K.S.; Sherrill, C.D. Potential energy curves for cation-pi interactions: Off-axis configurations are also attractive. J. Phys. Chem. A 2009, 113, 13628–13632. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Lee, S.E.; Jeong, S.; Lee, J.; Park, D.; Chang, S. Multivalent electrostatic pi-cation interaction between synaptophysin and synapsin is responsible for the coacervation. Mol. Brain 2021, 14, 137. [Google Scholar] [CrossRef] [PubMed]
- Harel, M.; Schalk, I.; Ehret-Sabatier, L.; Bouet, F.; Goeldner, M.; Hirth, C.; Axelsen, P.H.; Silman, I.; Sussman, J.L. Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proc. Natl. Acad. Sci. USA 1993, 90, 9031–9035. [Google Scholar] [CrossRef]
- Michael, L.A.; Chenault, J.A.; Miller, B.R., 3rd; Knolhoff, A.M.; Nagan, M.C. Water, shape recognition, salt bridges, and cation-pi interactions differentiate peptide recognition of the HIV rev-responsive element. J. Mol. Biol. 2009, 392, 774–786. [Google Scholar] [CrossRef]
- Hohlweg, W.; Wagner, G.E.; Hofbauer, H.F.; Sarkleti, F.; Setz, M.; Gubensak, N.; Lichtenegger, S.; Falsone, S.F.; Wolinski, H.; Kosol, S.; et al. A cation-pi interaction in a transmembrane helix of vacuolar ATPase retains the proton-transporting arginine in a hydrophobic environment. J. Biol. Chem. 2018, 293, 18977–18988. [Google Scholar] [CrossRef] [PubMed]
- Gallivan, J.P.; Dougherty, D.A. Cation-pi interactions in structural biology. Proc. Natl. Acad. Sci. USA 1999, 96, 9459–9464. [Google Scholar] [CrossRef]
- Kumar, K.; Woo, S.M.; Siu, T.; Cortopassi, W.A.; Duarte, F.; Paton, R.S. Cation-pi interactions in protein-ligand binding: Theory and data-mining reveal different roles for lysine and arginine. Chem. Sci. 2018, 9, 2655–2665. [Google Scholar] [CrossRef]
- Bergsma, S.; Poulios, E.; Charalampogiannis, N.; Andraws, O.; Achinas, S. Cation-pi Interaction as a Key Player in Healthcare: A Mini-Review. Digit. Med. Healthc. Technol. 2022. [Google Scholar] [CrossRef]
- Kruger, D.M.; Neubacher, S.; Grossmann, T.N. Protein-RNA interactions: Structural characteristics and hotspot amino acids. RNA 2018, 24, 1457–1465. [Google Scholar] [CrossRef]
- Barik, A.C.N.; Pilla, S.P.; Bahadur, R.P. Molecular architecture of protein-RNA recognition sites. J. Biomol. Struct. Dyn. 2015, 33, 2738–2751. [Google Scholar] [CrossRef]
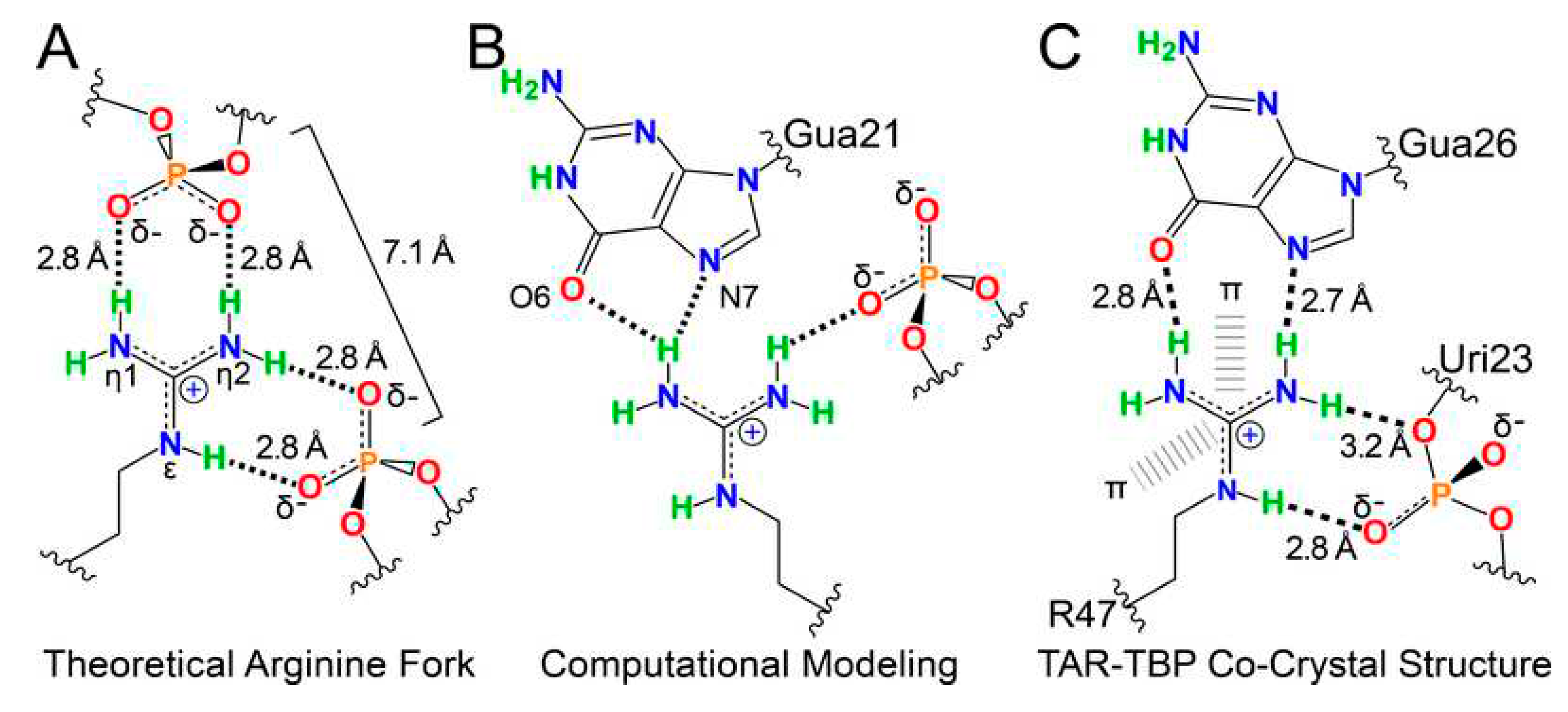
- Calnan, B.J.; Tidor, B.; Biancalana, S.; Hudson, D.; Frankel, A.D. Arginine-mediated RNA recognition: The arginine fork. Science 1991, 252, 1167–1171. [Google Scholar] [CrossRef]
- Chavali, S.S.; Cavender, C.E.; Mathews, D.H.; Wedekind, J.E. Arginine Forks Are a Widespread Motif to Recognize Phosphate Backbones and Guanine Nucleobases in the RNA Major Groove. J. Am. Chem. Soc. 2020, 142, 19835–19839. [Google Scholar] [CrossRef]
- Leclerc, F.; Karplus, M. MCSS-based predictions of RNA binding sites. Theor. Chem. Acc. 1999, 101, 131–137. [Google Scholar] [CrossRef]
- Belashov, I.A.; Crawford, D.W.; Cavender, C.E.; Dai, P.; Beardslee, P.C.; Mathews, D.H.; Pentelute, B.L.; McNaughton, B.R.; Wedekind, J.E. Structure of HIV TAR in complex with a Lab-Evolved RRM provides insight into duplex RNA recognition and synthesis of a constrained peptide that impairs transcription. Nucleic Acids Res. 2018, 46, 6401–6415. [Google Scholar] [CrossRef]
- Nott, T.J.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.D.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 2015, 57, 936–947. [Google Scholar] [CrossRef]
- Lin, Y.; Currie, S.L.; Rosen, M.K. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 2017, 292, 19110–19120. [Google Scholar] [CrossRef]
- Wang, J.; Choi, J.M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174, 688–699. [Google Scholar] [CrossRef]
- Sudhof, T.C.; Lottspeich, F.; Greengard, P.; Mehl, E.; Jahn, R. A synaptic vesicle protein with a novel cytoplasmic domain and four transmembrane regions. Science 1987, 238, 1142–1144. [Google Scholar] [CrossRef]
- Park, D.; Wu, Y.; Lee, S.E.; Kim, G.; Jeong, S.; Milovanovic, D.; De Camilli, P.; Chang, S. Cooperative function of synaptophysin and synapsin in the generation of synaptic vesicle-like clusters in non-neuronal cells. Nat. Commun. 2021, 12, 263. [Google Scholar] [CrossRef]
- Ueda, T.; Greengard, P. Adenosine 3':5'-monophosphate-regulated phosphoprotein system of neuronal membranes. I. Solubilization, purification, and some properties of an endogenous phosphoprotein. J. Biol. Chem. 1977, 252, 5155–5163. [Google Scholar] [CrossRef]
- Ferrari, L.; Stucchi, R.; Konstantoulea, K.; van de Kamp, G.; Kos, R.; Geerts, W.J.C.; van Bezouwen, L.S.; Forster, F.G.; Altelaar, M.; Hoogenraad, C.C.; et al. Arginine pi-stacking drives binding to fibrils of the Alzheimer protein Tau. Nat. Commun. 2020, 11, 571. [Google Scholar] [CrossRef]
- Hong, Y.; Najafi, S.; Casey, T.; Shea, J.E.; Han, S.I.; Hwang, D.S. Hydrophobicity of arginine leads to reentrant liquid-liquid phase separation behaviors of arginine-rich proteins. Nat. Commun. 2022, 13, 7326. [Google Scholar] [CrossRef]
- Das, S.; Lin, Y.H.; Vernon, R.M.; Forman-Kay, J.D.; Chan, H.S. Comparative roles of charge, pi, and hydrophobic interactions in sequence-dependent phase separation of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA 2020, 117, 28795–28805. [Google Scholar] [CrossRef]
- Schuster, B.S.; Dignon, G.L.; Tang, W.S.; Kelley, F.M.; Ranganath, A.K.; Jahnke, C.N.; Simpkins, A.G.; Regy, R.M.; Hammer, D.A.; Good, M.C.; et al. Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Proc. Natl. Acad. Sci. USA 2020, 117, 11421–11431. [Google Scholar] [CrossRef]
- Greig, J.A.; Nguyen, T.A.; Lee, M.; Holehouse, A.S.; Posey, A.E.; Pappu, R.V.; Jedd, G. Arginine-Enriched Mixed-Charge Domains Provide Cohesion for Nuclear Speckle Condensation. Mol. Cell 2020, 77, 1237–1250. [Google Scholar] [CrossRef]
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; Van Den Bosch, L.; et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018, 28, 420–435. [Google Scholar] [CrossRef]
- Vernon, R.M.; Chong, P.A.; Tsang, B.; Kim, T.H.; Bah, A.; Farber, P.; Lin, H.; Forman-Kay, J.D. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife 2018, 7. [Google Scholar] [CrossRef]
- Krainer, G.; Welsh, T.J.; Joseph, J.A.; Espinosa, J.R.; Wittmann, S.; de Csillery, E.; Sridhar, A.; Toprakcioglu, Z.; Gudiskyte, G.; Czekalska, M.A.; et al. Reentrant liquid condensate phase of proteins is stabilized by hydrophobic and non-ionic interactions. Nat. Commun. 2021, 12, 1085. [Google Scholar] [CrossRef]
- Prather, L.J.; Weerasekare, G.M.; Sima, M.; Quinn, C.; Stewart, R.J. Aqueous Liquid-Liquid Phase Separation of Natural and Synthetic Polyguanidiniums. Polymers 2019, 11. [Google Scholar] [CrossRef]
- Vazdar, M.; Heyda, J.; Mason, P.E.; Tesei, G.; Allolio, C.; Lund, M.; Jungwirth, P. Arginine “Magic”: Guanidinium Like-Charge Ion Pairing from Aqueous Salts to Cell Penetrating Peptides. Acc Chem. Res. 2018, 51, 1455–1464. [Google Scholar] [CrossRef]
- Gao, B.; Wyttenbach, T.; Bowers, M.T. Protonated arginine and protonated lysine: Hydration and its effect on the stability of salt-bridge structures. J. Phys. Chem. B 2009, 113, 9995–10000. [Google Scholar] [CrossRef]
- Vondrasek, J.; Mason, P.E.; Heyda, J.; Collins, K.D.; Jungwirth, P. The molecular origin of like-charge arginine-arginine pairing in water. J. Phys. Chem. B 2009, 113, 9041–9045. [Google Scholar] [CrossRef]
- Daoud, H.; Suhail, H.; Sabbagh, M.; Belzil, V.; Szuto, A.; Dionne-Laporte, A.; Khoris, J.; Camu, W.; Salachas, F.; Meininger, V.; et al. C9orf72 hexanucleotide repeat expansions as the causative mutation for chromosome 9p21-linked amyotrophic lateral sclerosis and frontotemporal dementia. Arch. Neurol. 2012, 69, 1159–1163. [Google Scholar] [CrossRef]
- Renton, A.E.; Majounie, E.; Waite, A.; Simon-Sanchez, J.; Rollinson, S.; Gibbs, J.R.; Schymick, J.C.; Laaksovirta, H.; van Swieten, J.C.; Myllykangas, L.; et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011, 72, 257–268. [Google Scholar] [CrossRef]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef]
- Gijselinck, I.; Cruts, M.; Van Broeckhoven, C. The Genetics of C9orf72 Expansions. Cold Spring Harb. Perspect. Med. 2018, 8. [Google Scholar] [CrossRef]
- Ash, P.E.; Bieniek, K.F.; Gendron, T.F.; Caulfield, T.; Lin, W.L.; Dejesus-Hernandez, M.; van Blitterswijk, M.M.; Jansen-West, K.; Paul, J.W., 3rd; Rademakers, R.; et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 2013, 77, 639–646. [Google Scholar] [CrossRef]
- Mori, K.; Weng, S.M.; Arzberger, T.; May, S.; Rentzsch, K.; Kremmer, E.; Schmid, B.; Kretzschmar, H.A.; Cruts, M.; Van Broeckhoven, C.; et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 2013, 339, 1335–1338. [Google Scholar] [CrossRef]
- Gendron, T.F.; Bieniek, K.F.; Zhang, Y.J.; Jansen-West, K.; Ash, P.E.; Caulfield, T.; Daughrity, L.; Dunmore, J.H.; Castanedes-Casey, M.; Chew, J.; et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013, 126, 829–844. [Google Scholar] [CrossRef]
- Shi, K.Y.; Mori, E.; Nizami, Z.F.; Lin, Y.; Kato, M.; Xiang, S.; Wu, L.C.; Ding, M.; Yu, Y.; Gall, J.G.; et al. Toxic PR(n) poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc. Natl. Acad. Sci. USA 2017, 114, E1111–E1117. [Google Scholar] [CrossRef]
- Tran, H.; Almeida, S.; Moore, J.; Gendron, T.F.; Chalasani, U.; Lu, Y.; Du, X.; Nickerson, J.A.; Petrucelli, L.; Weng, Z.; et al. Differential Toxicity of Nuclear RNA Foci versus Dipeptide Repeat Proteins in a Drosophila Model of C9ORF72 FTD/ALS. Neuron 2015, 87, 1207–1214. [Google Scholar] [CrossRef]
- Jovicic, A.; Mertens, J.; Boeynaems, S.; Bogaert, E.; Chai, N.; Yamada, S.B.; Paul, J.W., 3rd; Sun, S.; Herdy, J.R.; Bieri, G.; et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 2015, 18, 1226–1229. [Google Scholar] [CrossRef]
- Wen, X.; Tan, W.; Westergard, T.; Krishnamurthy, K.; Markandaiah, S.S.; Shi, Y.; Lin, S.; Shneider, N.A.; Monaghan, J.; Pandey, U.B.; et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 2014, 84, 1213–1225. [Google Scholar] [CrossRef]
- Mizielinska, S.; Gronke, S.; Niccoli, T.; Ridler, C.E.; Clayton, E.L.; Devoy, A.; Moens, T.; Norona, F.E.; Woollacott, I.O.C.; Pietrzyk, J.; et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 2014, 345, 1192–1194. [Google Scholar] [CrossRef]
- Yamakawa, M.; Ito, D.; Honda, T.; Kubo, K.; Noda, M.; Nakajima, K.; Suzuki, N. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum. Mol. Genet. 2015, 24, 1630–1645. [Google Scholar] [CrossRef]
- Mizielinska, S.; Lashley, T.; Norona, F.E.; Clayton, E.L.; Ridler, C.E.; Fratta, P.; Isaacs, A.M. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 2013, 126, 845–857. [Google Scholar] [CrossRef]
- Darling, A.L.; Breydo, L.; Rivas, E.G.; Gebru, N.T.; Zheng, D.; Baker, J.D.; Blair, L.J.; Dickey, C.A.; Koren, J., 3rd; Uversky, V.N. Repeated repeat problems: Combinatorial effect of C9orf72-derived dipeptide repeat proteins. Int. J. Biol. Macromol. 2019, 127, 136–145. [Google Scholar] [CrossRef]
- Tao, Z.; Wang, H.; Xia, Q.; Li, K.; Li, K.; Jiang, X.; Xu, G.; Wang, G.; Ying, Z. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum. Mol. Genet. 2015, 24, 2426–2441. [Google Scholar] [CrossRef]
- Kwon, I.; Xiang, S.; Kato, M.; Wu, L.; Theodoropoulos, P.; Wang, T.; Kim, J.; Yun, J.; Xie, Y.; McKnight, S.L. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 2014, 345, 1139–1145. [Google Scholar] [CrossRef]
- Moens, T.G.; Niccoli, T.; Wilson, K.M.; Atilano, M.L.; Birsa, N.; Gittings, L.M.; Holbling, B.V.; Dyson, M.C.; Thoeng, A.; Neeves, J.; et al. C9orf72 arginine-rich dipeptide proteins interact with ribosomal proteins in vivo to induce a toxic translational arrest that is rescued by eIF1A. Acta Neuropathol. 2019, 137, 487–500. [Google Scholar] [CrossRef]
- Hartmann, H.; Hornburg, D.; Czuppa, M.; Bader, J.; Michaelsen, M.; Farny, D.; Arzberger, T.; Mann, M.; Meissner, F.; Edbauer, D. Proteomics and C9orf72 neuropathology identify ribosomes as poly-GR/PR interactors driving toxicity. Life Sci. Alliance 2018, 1, e201800070. [Google Scholar] [CrossRef]
- Lee, K.H.; Zhang, P.; Kim, H.J.; Mitrea, D.M.; Sarkar, M.; Freibaum, B.D.; Cika, J.; Coughlin, M.; Messing, J.; Molliex, A.; et al. C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell 2016, 167, 774–788. [Google Scholar] [CrossRef]
- Odeh, H.M.; Shorter, J. Arginine-rich dipeptide-repeat proteins as phase disruptors in C9-ALS/FTD. Emerg. Top. Life Sci. 2020, 4, 293–305. [Google Scholar] [CrossRef]
- Park, J.; Wu, Y.; Shao, W.; Gendron, T.F.; van der Spek, S.J.F.; Sultanakhmetov, G.; Basu, A.; Castellanos Otero, P.; Jones, C.J.; Jansen-West, K.; et al. Poly(GR) interacts with key stress granule factors promoting its assembly into cytoplasmic inclusions. Cell Rep. 2023, 42, 112822. [Google Scholar] [CrossRef]
- Cook, C.N.; Wu, Y.; Odeh, H.M.; Gendron, T.F.; Jansen-West, K.; Del Rosso, G.; Yue, M.; Jiang, P.; Gomes, E.; Tong, J.; et al. C9orf72 poly(GR) aggregation induces TDP-43 proteinopathy. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- White, M.R.; Mitrea, D.M.; Zhang, P.; Stanley, C.B.; Cassidy, D.E.; Nourse, A.; Phillips, A.H.; Tolbert, M.; Taylor, J.P.; Kriwacki, R.W. C9orf72 Poly(PR) Dipeptide Repeats Disturb Biomolecular Phase Separation and Disrupt Nucleolar Function. Mol. Cell 2019, 74, 713–728. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Guo, L.; Gonzales, P.K.; Gendron, T.F.; Wu, Y.; Jansen-West, K.; O’Raw, A.D.; Pickles, S.R.; Prudencio, M.; Carlomagno, Y.; et al. Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science 2019, 363. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Gendron, T.F.; Ebbert, M.T.W.; O’Raw, A.D.; Yue, M.; Jansen-West, K.; Zhang, X.; Prudencio, M.; Chew, J.; Cook, C.N.; et al. Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat. Med. 2018, 24, 1136–1142. [Google Scholar] [CrossRef]
- Boeynaems, S.; Bogaert, E.; Kovacs, D.; Konijnenberg, A.; Timmerman, E.; Volkov, A.; Guharoy, M.; De Decker, M.; Jaspers, T.; Ryan, V.H.; et al. Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics. Mol. Cell 2017, 65, 1044–1055. [Google Scholar] [CrossRef]
- Harrison, P.M. Intrinsic disorder and posttranslational modification: An evolutionary perspective. In Structure and Intrinsic Disorder in Enzymology; Elsevier: 2023; pp. 377–396.
- Uversky, V.N. Posttranslational Modifications. In Brenner's Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: London, UK; Walham, MA, USA; San Diego, CA, USA, 2013; Volume 5, pp. 425–430. [Google Scholar]
- Darling, A.L.; Uversky, V.N. Intrinsic Disorder and Posttranslational Modifications: The Darker Side of the Biological Dark Matter. Front. Genet. 2018, 9, 158. [Google Scholar] [CrossRef]
- Gupta, M.N.; Uversky, V.N. Moonlighting enzymes: When cellular context defines specificity. Cell. Mol. Life Sci. 2023, 80, 130. [Google Scholar] [CrossRef]
- Xie, H.; Vucetic, S.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J. Proteome Res. 2007, 6, 1917–1932. [Google Scholar] [CrossRef]
- Pejaver, V.; Hsu, W.L.; Xin, F.; Dunker, A.K.; Uversky, V.N.; Radivojac, P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014, 23, 1077–1093. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsic disorder-based protein interactions and their modulators. Curr. Pharm. Des. 2013, 19, 4191–4213. [Google Scholar] [CrossRef]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [Google Scholar] [CrossRef]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Showing your ID: Intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 2005, 18, 343–384. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef]
- Tsikas, D. Post-translational modifications (PTM): Analytical approaches, signaling, physiology and pathophysiology-part I. Amino Acids 2021, 53, 485–487. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Kornhauser, J.M.; Tkachev, S.; Zhang, B.; Skrzypek, E.; Murray, B.; Latham, V.; Sullivan, M. PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012, 40, D261–D270. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.A.; Cole, P.A. The Chemical Biology of Reversible Lysine Post-translational Modifications. Cell Chem. Biol. 2020, 27, 953–969. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Conrad, R.J.; Verdin, E.; Ott, M. Lysine Acetylation Goes Global: From Epigenetics to Metabolism and Therapeutics. Chem. Rev. 2018, 118, 1216–1252. [Google Scholar] [CrossRef] [PubMed]
- Catrina, A.; Krishnamurthy, A.; Rethi, B. Current view on the pathogenic role of anti-citrullinated protein antibodies in rheumatoid arthritis. RMD Open 2021, 7. [Google Scholar] [CrossRef] [PubMed]
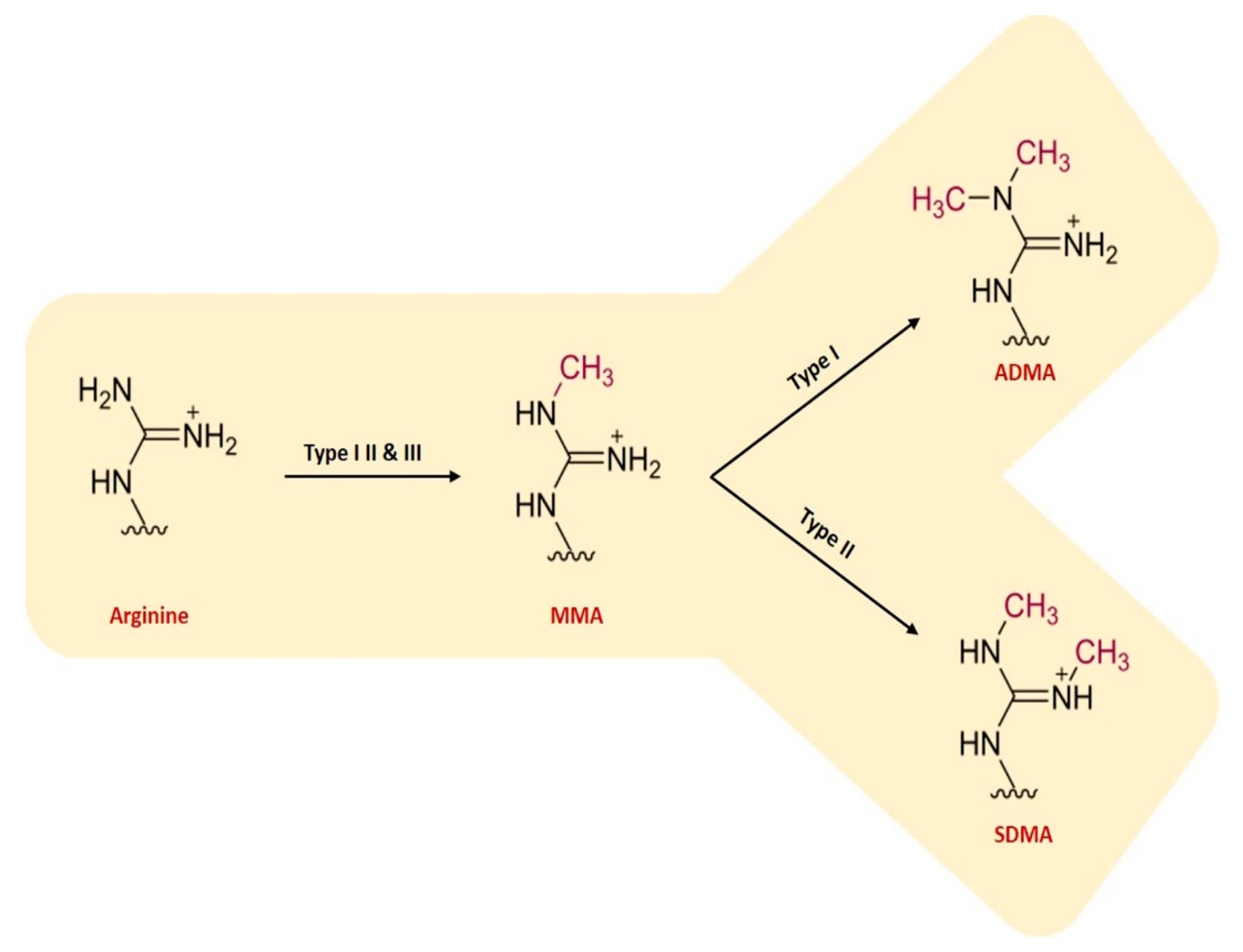
- Samuel, S.F.; Barry, A.; Greenman, J.; Beltran-Alvarez, P. Arginine methylation: The promise of a ‘silver bullet’ for brain tumours? Amino Acids 2021, 53, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Katz, J.E.; Dlakic, M.; Clarke, S. Automated identification of putative methyltransferases from genomic open reading frames. Mol. Cell. Proteom. 2003, 2, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.; Gu, H.; Zhou, J.; Mulhern, D.; Wang, Y.; Lee, K.A.; Yang, V.; Aguiar, M.; Kornhauser, J.; Jia, X.; et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteom. 2014, 13, 372–387. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
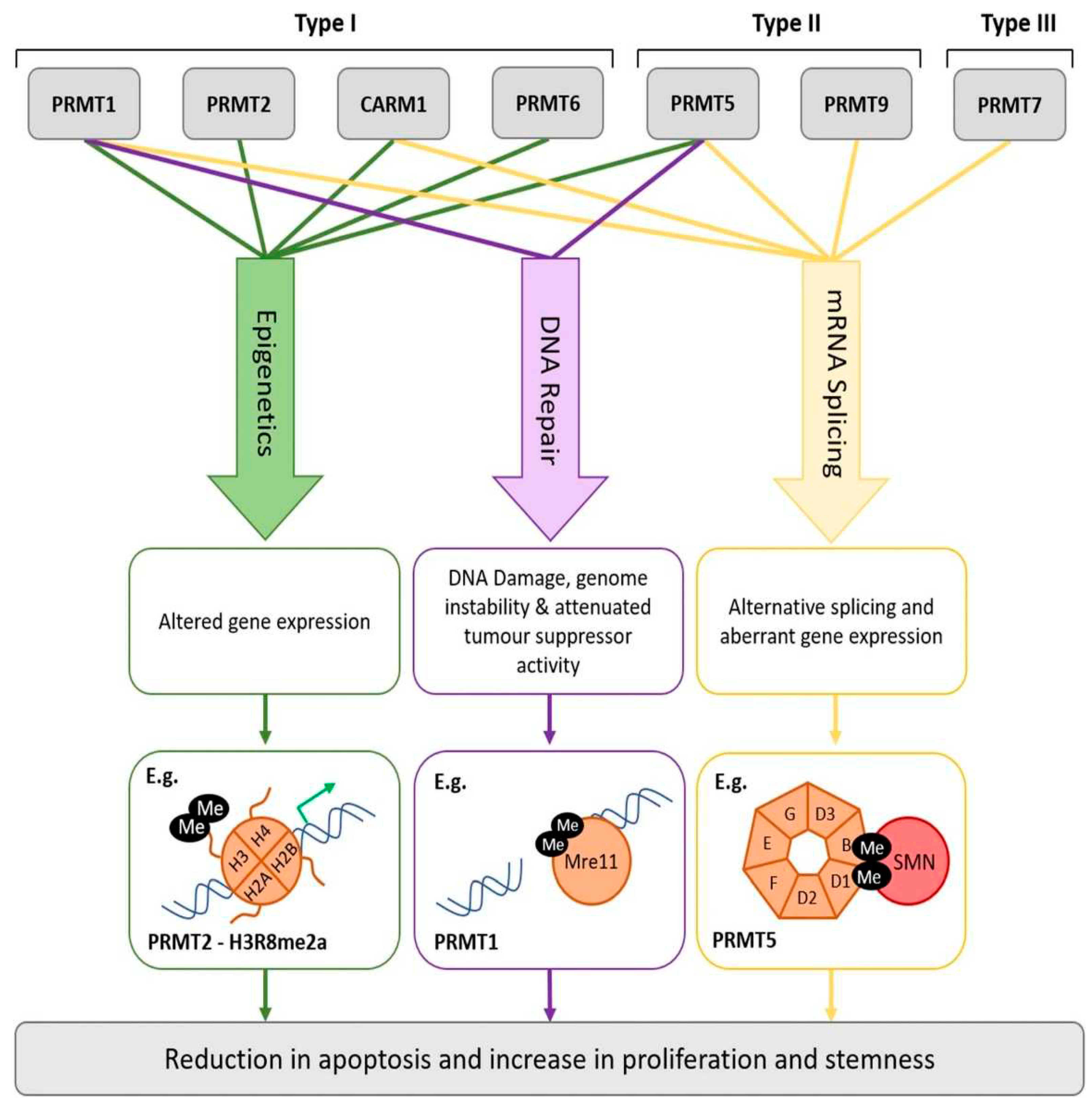
- Yang, Y.; Bedford, M.T. Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 2013, 13, 37–50. [Google Scholar] [CrossRef]
- Post, A.; Bollenbach, A.; Bakker, S.J.L.; Tsikas, D. Whole-body arginine dimethylation is associated with all-cause mortality in adult renal transplant recipients. Amino Acids 2021, 53, 541–554. [Google Scholar] [CrossRef]
- Said, M.Y.; Bollenbach, A.; Minovic, I.; van Londen, M.; Frenay, A.R.; de Borst, M.H.; van den Berg, E.; Kayacelebi, A.A.; Tsikas, D.; van Goor, H.; et al. Plasma ADMA, urinary ADMA excretion, and late mortality in renal transplant recipients. Amino Acids 2019, 51, 913–927. [Google Scholar] [CrossRef] [PubMed]
- Said, M.Y.; Douwes, R.M.; van Londen, M.; Minovic, I.; Frenay, A.R.; de Borst, M.H.; van den Berg, E.; Heiner-Fokkema, M.R.; Kayacelebi, A.A.; Bollenbach, A.; et al. Effect of renal function on homeostasis of asymmetric dimethylarginine (ADMA): Studies in donors and recipients of renal transplants. Amino Acids 2019, 51, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Frenay, A.R.; van den Berg, E.; de Borst, M.H.; Beckmann, B.; Tsikas, D.; Feelisch, M.; Navis, G.; Bakker, S.J.; van Goor, H. Plasma ADMA associates with all-cause mortality in renal transplant recipients. Amino Acids 2015, 47, 1941–1949. [Google Scholar] [CrossRef] [PubMed]
- Fliser, D.; Kronenberg, F.; Kielstein, J.T.; Morath, C.; Bode-Boger, S.M.; Haller, H.; Ritz, E. Asymmetric dimethylarginine and progression of chronic kidney disease: The mild to moderate kidney disease study. J. Am. Soc. Nephrol. 2005, 16, 2456–2461. [Google Scholar] [CrossRef] [PubMed]
- Achan, V.; Broadhead, M.; Malaki, M.; Whitley, G.; Leiper, J.; MacAllister, R.; Vallance, P. Asymmetric dimethylarginine causes hypertension and cardiac dysfunction in humans and is actively metabolized by dimethylarginine dimethylaminohydrolase. Arter. Thromb. Vasc. Biol. 2003, 23, 1455–1459. [Google Scholar] [CrossRef]
- Tain, Y.L.; Hsu, C.N. Toxic Dimethylarginines: Asymmetric Dimethylarginine (ADMA) and Symmetric Dimethylarginine (SDMA). Toxins 2017, 9. [Google Scholar] [CrossRef]
- Busch, M.; Fleck, C.; Wolf, G.; Stein, G. Asymmetrical (ADMA) and symmetrical dimethylarginine (SDMA) as potential risk factors for cardiovascular and renal outcome in chronic kidney disease—Possible candidates for paradoxical epidemiology? Amino Acids 2006, 30, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Marsden, A.J.; Riley, D.R.J.; Birkett, S.; Rodriguez-Barucg, Q.; Guinn, B.A.; Carroll, S.; Ingle, L.; Sathyapalan, T.; Beltran-Alvarez, P. Love is in the hair: Arginine methylation of human hair proteins as novel cardiovascular biomarkers. Amino Acids 2022, 54, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.N.; Perwez, M.; Sardar, M. Protein crosslinking: Uses in chemistry, biology and biotechnology. Biocatal. Biotransform. 2020, 38, 178–201. [Google Scholar] [CrossRef]
- Mukherjee, J.; Majumder, A.B.; Gupta, M.N. Adding an appropriate amino acid during crosslinking results in more stable crosslinked enzyme aggregates. Anal. Biochem. 2016, 507, 27–32. [Google Scholar] [CrossRef]
- Collier, T.A.; Nash, A.; Birch, H.L.; de Leeuw, N.H. Intra-molecular lysine-arginine derived advanced glycation end-product cross-linking in Type I collagen: A molecular dynamics simulation study. Biophys. Chem. 2016, 218, 42–46. [Google Scholar] [CrossRef]
- Thorpe, C.T.; Streeter, I.; Pinchbeck, G.L.; Goodship, A.E.; Clegg, P.D.; Birch, H.L. Aspartic acid racemization and collagen degradation markers reveal an accumulation of damage in tendon collagen that is enhanced with aging. J. Biol. Chem. 2010, 285, 15674–15681. [Google Scholar] [CrossRef] [PubMed]
- Biemel, K.M.; Friedl, D.A.; Lederer, M.O. Identification and quantification of major maillard cross-links in human serum albumin and lens protein. Evidence for glucosepane as the dominant compound. J. Biol. Chem. 2002, 277, 24907–24915. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Martin, E.; Gallego-Munoz, P.; Ibares-Frias, L.; Marcos, S.; Perez-Merino, P.; Fernandez, I.; Kochevar, I.E.; Martinez-Garcia, M.C. Rose Bengal and Green Light Versus Riboflavin-UVA Cross-Linking: Corneal Wound Repair Response. Investig. Ophthalmol. Vis. Sci. 2018, 59, 4821–4830. [Google Scholar] [CrossRef] [PubMed]
- Seiler, T.G.; Engler, M.; Beck, E.; Birngruber, R.; Kochevar, I.E. Interface Bonding With Corneal Crosslinking (CXL) After LASIK Ex Vivo. Investig. Ophthalmol. Vis. Sci. 2017, 58, 6292–6298. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Munoz, P.; Ibares-Frias, L.; Lorenzo, E.; Marcos, S.; Perez-Merino, P.; Bekesi, N.; Kochevar, I.E.; Martinez-Garcia, M.C. Corneal Wound Repair After Rose Bengal and Green Light Crosslinking: Clinical and Histologic Study. Investig. Ophthalmol. Vis. Sci. 2017, 58, 3471–3480. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Alt, C.; Webb, R.H.; Melki, S.; Kochevar, I.E. Corneal Crosslinking With Rose Bengal and Green Light: Efficacy and Safety Evaluation. Cornea 2016, 35, 1234–1241. [Google Scholar] [CrossRef]
- Bekesi, N.; Kochevar, I.E.; Marcos, S. Corneal Biomechanical Response Following Collagen Cross-Linking with Rose Bengal-Green Light and Riboflavin-UVA. Investig. Ophthalmol. Vis. Sci. 2016, 57, 992–1001. [Google Scholar] [CrossRef]
- Wertheimer, C.M.; Mendes, B.; Pei, Q.; Brandt, K.; Kochevar, I.E. Arginine as an Enhancer in Rose Bengal Photosensitized Corneal Crosslinking. Transl. Vis. Sci. Technol. 2020, 9, 24. [Google Scholar] [CrossRef]
- Wertheimer, C.M.; Elhardt, C.; Kaminsky, S.M.; Pham, L.; Pei, Q.; Mendes, B.; Afshar, S.; Kochevar, I.E. Enhancing Rose Bengal-Photosensitized Protein Crosslinking in the Cornea. Investig. Ophthalmol. Vis. Sci. 2019, 60, 1845–1852. [Google Scholar] [CrossRef]
- Cherfan, D.; Verter, E.E.; Melki, S.; Gisel, T.E.; Doyle, F.J., Jr.; Scarcelli, G.; Yun, S.H.; Redmond, R.W.; Kochevar, I.E. Collagen cross-linking using rose bengal and green light to increase corneal stiffness. Investig. Ophthalmol. Vis. Sci. 2013, 54, 3426–3433. [Google Scholar] [CrossRef] [PubMed]
- Udhayakumar, S.; Shankar, K.G.; Sowndarya, S.; Venkatesh, S.; Muralidharan, C.; Rose, C. L-arginine intercedes bio-crosslinking of a collagen–chitosan 3D-hybrid scaffold for tissue engineering and regeneration: In silico, in vitro, and in vivo studies. RSC Adv. 2017, 7, 25070–25088. [Google Scholar] [CrossRef]
- Caruso, A.; Martinie, R.J.; Bushin, L.B.; Seyedsayamdost, M.R. Macrocyclization via an arginine-tyrosine crosslink broadens the reaction scope of radical S-adenosylmethionine enzymes. J. Am. Chem. Soc. 2019, 141, 16610–16614. [Google Scholar] [CrossRef] [PubMed]
- Riordan, J.F. Arginyl residues and anion binding sites in proteins. Mol. Cell. Biochem. 1979, 26, 71–92. [Google Scholar] [CrossRef]
- Leone, D.L.; Hubalek, M.; Pohl, R.; Sykorova, V.; Hocek, M. 1,3-Diketone-Modified Nucleotides and DNA for Cross-Linking with Arginine-Containing Peptides and Proteins. Angew. Chem. Int. Ed. Engl. 2021, 60, 17383–17387. [Google Scholar] [CrossRef]
- Means, G.; Feeney, R. Chemical Modification of Proteins; Hoiden-Day Inc.: San Francisco, CA, USA, 1971. [Google Scholar]
- Tyagi, R.; Gupta, M.N. Chemical modification and chemical crosslinking for enhancing thermostability of enzymes. In Thermostability of Enzymes; Gupta, M.N., Ed.; Springer: Berlin, 1993; pp. 146–160. [Google Scholar]
- Tyagi, R.; Gupta, M.N. Chemical modification and chemical crosslinking of proteins/enzyme stabilization. Biochemistry 1998, 63, 395–407. [Google Scholar]
- Zhou, Y.; Liu, G.; Huang, H.; Wu, J. Advances and impact of arginine-based materials in wound healing. J. Mater. Chem. B 2021, 9, 6738–6750. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Kuroyanagi, Y. Development of a wound dressing composed of hyaluronic acid sponge containing arginine and epidermal growth factor. J. Biomater. Sci. Polym. Ed. 2010, 21, 715–726. [Google Scholar] [CrossRef]
- Reesi, F.; Minaiyan, M.; Taheri, A. A novel lignin-based nanofibrous dressing containing arginine for wound-healing applications. Drug Deliv. Transl. Res. 2018, 8, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Durmus, A.S.; Tuzcu, M.; Ozdemir, O.; Orhan, C.; Sahin, N.; Ozercan, I.H.; Komorowski, J.R.; Ali, S.; Sahin, K. Arginine Silicate Inositol Complex Accelerates Cutaneous Wound Healing. Biol. Trace. Elem. Res. 2017, 177, 122–131. [Google Scholar] [CrossRef]
- Baron, R.I.; Culica, M.E.; Biliuta, G.; Bercea, M.; Gherman, S.; Zavastin, D.; Ochiuz, L.; Avadanei, M.; Coseri, S. Physical Hydrogels of Oxidized Polysaccharides and Poly(Vinyl Alcohol) for Wound Dressing Applications. Materials 2019, 12. [Google Scholar] [CrossRef] [PubMed]
- Antunes, B.P.; Moreira, A.F.; Gaspar, V.M.; Correia, I.J. Chitosan/arginine-chitosan polymer blends for assembly of nanofibrous membranes for wound regeneration. Carbohydr. Polym. 2015, 130, 104–112. [Google Scholar] [CrossRef]
- Wu, J.; Mutschler, M.A.; Chu, C.C. Synthesis and characterization of ionic charged water soluble arginine-based poly(ester amide). J. Mater. Sci. Mater. Med. 2011, 22, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Cotton, F.A.; la Cour, T.; Hazen, E.E., Jr.; Legg, M.J. The role of arginine residues at enzyme active sites. The interaction between guanidinium ions and p-nitro-phenyl phosphate and its effect on the rate of hydrolysis of the ester. Biochim. Biophys. Acta 1977, 481, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Adak, S.; Bandyopadhyay, D.; Bandyopadhyay, U.; Banerjee, R.K. An essential role of active site arginine residue in iodide binding and histidine residue in electron transfer for iodide oxidation by horseradish peroxidase. Mol. Cell. Biochem. 2001, 218, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.C.; Nowak, T. Arginine residues at the active site of avian liver phosphoenolpyruvate carboxykinase. J. Biol. Chem. 1989, 264, 3317–3324. [Google Scholar] [CrossRef] [PubMed]
- Walker, V. Ammonia metabolism and hyperammonemic disorders. Adv. Clin. Chem. 2014, 67, 73–150. [Google Scholar] [PubMed]
- Kumar, S.; Lennane, J.; Ratner, S. Argininosuccinate synthetase: Essential role of cysteine and arginine residues in relation to structure and mechanism of ATP activation. Proc. Natl. Acad. Sci. USA 1985, 82, 6745–6749. [Google Scholar] [CrossRef] [PubMed]
- Keenholtz, R.A.; Mouw, K.W.; Boocock, M.R.; Li, N.S.; Piccirilli, J.A.; Rice, P.A. Arginine as a general acid catalyst in serine recombinase-mediated DNA cleavage. J. Biol. Chem. 2013, 288, 29206–29214. [Google Scholar] [CrossRef]
- Pauff, J.M.; Hemann, C.F.; Junemann, N.; Leimkuhler, S.; Hille, R. The role of arginine 310 in catalysis and substrate specificity in xanthine dehydrogenase from Rhodobacter capsulatus. J. Biol. Chem. 2007, 282, 12785–12790. [Google Scholar] [CrossRef]
- Albert, S.; Will, E.; Gallwitz, D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 1999, 18, 5216–5225. [Google Scholar] [CrossRef]
- Chen, G.; Chen, X. Arginine residues in the active site of human phenol sulfotransferase (SULT1A1). J. Biol. Chem. 2003, 278, 36358–36364. [Google Scholar] [CrossRef]
- Sulfurtransferases: Essential Enzymes for Life; Nagahara, N. , Ed.; Elsevier: San Diego, CA, USA, 2023. [Google Scholar]
- Communi, D.; Lecocq, R.; Erneux, C. Arginine 343 and 350 are two active residues involved in substrate binding by human Type I D-myo-inositol 1,4,5,-trisphosphate 5-phosphatase. J. Biol. Chem. 1996, 271, 11676–11683. [Google Scholar] [CrossRef]
- Shi, J.; Mukhopadhyay, R.; Rosen, B.P. Identification of a triad of arginine residues in the active site of the ArsC arsenate reductase of plasmid R773. FEMS Microbiol. Lett. 2003, 227, 295–301. [Google Scholar] [CrossRef]
- Hsiao, J.C.; McGrath, A.P.; Kielmann, L.; Kalimuthu, P.; Darain, F.; Bernhardt, P.V.; Harmer, J.; Lee, M.; Meyers, K.; Maher, M.J.; et al. The central active site arginine in sulfite oxidizing enzymes alters kinetic properties by controlling electron transfer and redox interactions. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 19–27. [Google Scholar] [CrossRef]
- Murtas, G.; Sacchi, S.; Pollegioni, L. Substitution of Arginine 120 in Human D-Amino Acid Oxidase Favors FAD-Binding and Nuclear Mistargeting. Front. Mol. Biosci. 2019, 6, 125. [Google Scholar] [CrossRef]
- Sarkar, A.; Bhakta, S.; Chattopadhyay, S.; Dey, A. Role of distal arginine residue in the mechanism of heme nitrite reductases. Chem. Sci. 2023, 14, 7875–7886. [Google Scholar] [CrossRef]
- Morris, S.M., Jr. Arginine metabolism: Boundaries of our knowledge. J. Nutr. 2007, 137, 1602S–1609S. [Google Scholar] [CrossRef]
- Tong, B.C.; Barbul, A. Cellular and physiological effects of arginine. Mini Rev. Med. Chem. 2004, 4, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.R.; Hamilton-Reeves, J.; Martindale, R.G.; Sarav, M.; Ochoa Gautier, J.B. Acquired Amino Acid Deficiencies: A Focus on Arginine and Glutamine. Nutr. Clin. Pr. 2017, 32, 30S–47S. [Google Scholar] [CrossRef] [PubMed]
- Hristina, K.; Langerholc, T.; Trapecar, M. Novel metabolic roles of L-arginine in body energy metabolism and possible clinical applications. J. Nutr. Health Aging 2014, 18, 213–218. [Google Scholar] [CrossRef]
- Xiong, L.; Teng, J.L.; Botelho, M.G.; Lo, R.C.; Lau, S.K.; Woo, P.C. Arginine Metabolism in Bacterial Pathogenesis and Cancer Therapy. Int. J. Mol. Sci. 2016, 17, 363. [Google Scholar] [CrossRef]
- Erens, C.; Van Broeckhoven, J.; Bronckaers, A.; Lemmens, S.; Hendrix, S. The Dark Side of an Essential Amino Acid: L-Arginine in Spinal Cord Injury. J. Neurotrauma 2023, 40, 820–832. [Google Scholar] [CrossRef]
- Ma, C.; Liao, K.; Wang, J.; Li, T.; Liu, L. L-Arginine, as an essential amino acid, is a potential substitute for treating COPD via regulation of ROS/NLRP3/NF-kappaB signaling pathway. Cell Biosci. 2023, 13, 152. [Google Scholar] [CrossRef]
- Leblond, J.; Petitjean, A. Molecular tweezers: Concepts and applications. ChemPhysChem 2011, 12, 1043–1051. [Google Scholar] [CrossRef]
- Kaiser, F.; Bittrich, S.; Salentin, S.; Leberecht, C.; Haupt, V.J.; Krautwurst, S.; Schroeder, M.; Labudde, D. Backbone Brackets and Arginine Tweezers delineate Class I and Class II aminoacyl tRNA synthetases. PLoS Comput. Biol. 2018, 14, e1006101. [Google Scholar] [CrossRef]
- Schrader, T.; Bitan, G.; Klarner, F.G. Molecular tweezers for lysine and arginine—Powerful inhibitors of pathologic protein aggregation. Chem. Commun. 2016, 52, 11318–11334. [Google Scholar] [CrossRef]
- Mbarek, A.; Moussa, G.; Chain, J.L. Pharmaceutical Applications of Molecular Tweezers, Clefts and Clips. Molecules 2019, 24. [Google Scholar] [CrossRef]
- Hadrovic, I.; Rebmann, P.; Klarner, F.G.; Bitan, G.; Schrader, T. Molecular Lysine Tweezers Counteract Aberrant Protein Aggregation. Front. Chem. 2019, 7, 657. [Google Scholar] [CrossRef]
- Hodges, A.J.; Gallegos, I.J.; Laughery, M.F.; Meas, R.; Tran, L.; Wyrick, J.J. Histone Sprocket Arginine Residues Are Important for Gene Expression, DNA Repair, and Cell Viability in Saccharomyces cerevisiae. Genetics 2015, 200, 795–806. [Google Scholar] [CrossRef]
- Muthurajan, U.M.; Park, Y.J.; Edayathumangalam, R.S.; Suto, R.K.; Chakravarthy, S.; Dyer, P.N.; Luger, K. Structure and dynamics of nucleosomal DNA. Biopolymers 2003, 68, 547–556. [Google Scholar] [CrossRef]
- Davey, C.A.; Sargent, D.F.; Luger, K.; Maeder, A.W.; Richmond, T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002, 319, 1097–1113. [Google Scholar] [CrossRef]
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Magalhaes, A.; Maigret, B.; Hoflack, J.; Gomes, J.N.; Scheraga, H.A. Contribution of unusual arginine-arginine short-range interactions to stabilization and recognition in proteins. J. Protein Chem. 1994, 13, 195–215. [Google Scholar] [CrossRef]
- Neves, M.A.; Yeager, M.; Abagyan, R. Unusual arginine formations in protein function and assembly: Rings, strings, and stacks. J. Phys. Chem. B 2012, 116, 7006–7013. [Google Scholar] [CrossRef]
- Wang, W. Oral protein drug delivery. J. Drug Target 1996, 4, 195–232. [Google Scholar] [CrossRef]
- Wang, W. Lyophilization and development of solid protein pharmaceuticals. Int. J. Pharm. 2000, 203, 1–60. [Google Scholar] [CrossRef]
- Wang, W. Instability, stabilization, and formulation of liquid protein pharmaceuticals. Int. J. Pharm. 1999, 185, 129–188. [Google Scholar] [CrossRef]
- Garcia Garcia, C.; Patkar, S.S.; Wang, B.; Abouomar, R.; Kiick, K.L. Recombinant protein-based injectable materials for biomedical applications. Adv. Drug Deliv. Rev. 2023, 193, 114673. [Google Scholar] [CrossRef]
- Ryan, S.; Shortall, K.; Dully, M.; Djehedar, A.; Murray, D.; Butler, J.; Neilan, J.; Soulimane, T.; Hudson, S.P. Long acting injectables for therapeutic proteins. Colloids Surf. B Biointerfaces 2022, 217, 112644. [Google Scholar] [CrossRef]
- Ibeanu, N.; Egbu, R.; Onyekuru, L.; Javaheri, H.; Khaw, P.T.; Williams, G.R.; Brocchini, S.; Awwad, S. Injectables and Depots to Prolong Drug Action of Proteins and Peptides. Pharmaceutics 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Julicher, F.; Hyman, A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef] [PubMed]
- Brangwynne, C.P. Phase transitions and size scaling of membrane-less organelles. J. Cell Biol. 2013, 203, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Brangwynne, C.P.; Tompa, P.; Pappu, R.V. Polymer physics of intracellular phase transitions. Nat. Phys. 2015, 11, 899–904. [Google Scholar] [CrossRef]
- Uversky, V.N.; Kuznetsova, I.M.; Turoverov, K.K.; Zaslavsky, B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett. 2015, 589, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Dundr, M.; Misteli, T. Biogenesis of nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000711. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Brangwynne, C.P. Nuclear bodies: The emerging biophysics of nucleoplasmic phases. Curr. Opin. Cell Biol. 2015, 34, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Protein intrinsic disorder-based liquid-liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles. Adv. Colloid Interface Sci. 2017, 239, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef]
- Mitrea, D.M.; Kriwacki, R.W. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 2016, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wang, S.; Huang, Y.; He, X.; Cui, H.; Zhu, X.; Zheng, Y. Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell 2015, 163, 108–122. [Google Scholar] [CrossRef] [PubMed]
- Antifeeva, I.A.; Fonin, A.V.; Fefilova, A.S.; Stepanenko, O.V.; Povarova, O.I.; Silonov, S.A.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. Liquid-liquid phase separation as an organizing principle of intracellular space: Overview of the evolution of the cell compartmentalization concept. Cell. Mol. Life Sci. 2022, 79, 251. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.L.; Liu, Y.; Oldfield, C.J.; Uversky, V.N. Intrinsically Disordered Proteome of Human Membrane-Less Organelles. Proteomics 2018, 18, e1700193. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.X.; Nguemaha, V.; Mazarakos, K.; Qin, S. Why Do Disordered and Structured Proteins Behave Differently in Phase Separation? Trends Biochem. Sci. 2018, 43, 499–516. [Google Scholar] [CrossRef] [PubMed]
- Turoverov, K.K.; Kuznetsova, I.M.; Fonin, A.V.; Darling, A.L.; Zaslavsky, B.Y.; Uversky, V.N. Stochasticity of Biological Soft Matter: Emerging Concepts in Intrinsically Disordered Proteins and Biological Phase Separation. Trends Biochem. Sci. 2019, 44, 716–728. [Google Scholar] [CrossRef]
- Fonin, A.V.; Antifeeva, I.A.; Kuznetsova, I.M.; Turoverov, K.K.; Zaslavsky, B.Y.; Kulkarni, P.; Uversky, V.N. Biological soft matter: Intrinsically disordered proteins in liquid-liquid phase separation and biomolecular condensates. Essays Biochem. 2022, 66, 831–847. [Google Scholar] [CrossRef] [PubMed]
- Raut, A.S.; Kalonia, D.S. Opalescence in monoclonal antibody solutions and its correlation with intermolecular interactions in dilute and concentrated solutions. J. Pharm. Sci. 2015, 104, 1263–1274. [Google Scholar] [CrossRef]
- Oki, S.; Nishinami, S.; Shiraki, K. Arginine suppresses opalescence and liquid-liquid phase separation in IgG solutions. Int. J. Biol. Macromol. 2018, 118, 1708–1712. [Google Scholar] [CrossRef]
- Banks, D.D.; Cordia, J.F. Suppression of Electrostatic Mediated Antibody Liquid-Liquid Phase Separation by Charged and Noncharged Preferentially Excluded Excipients. Mol. Pharm. 2021, 18, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Pyne, P.; Mitra, R.K. Excipients Do Regulate Phase Separation in Lysozyme and Thus Also Its Hydration. J. Phys. Chem. Lett. 2022, 13, 931–938. [Google Scholar] [CrossRef]
- Tsumoto, K.; Umetsu, M.; Kumagai, I.; Ejima, D.; Philo, J.S.; Arakawa, T. Role of arginine in protein refolding, solubilization, and purification. Biotechnol. Prog. 2004, 20, 1301–1308. [Google Scholar] [CrossRef]
- Arakawa, T.; Tsumoto, K. The effects of arginine on refolding of aggregated proteins: Not facilitate refolding, but suppress aggregation. Biochem. Biophys. Res. Commun. 2003, 304, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Baynes, B.M.; Wang, D.I.; Trout, B.L. Role of arginine in the stabilization of proteins against aggregation. Biochemistry 2005, 44, 4919–4925. [Google Scholar] [CrossRef]
- Gupta, M.N.; Uversky, V.N. Macromolecular crowding: How it affects protein structure, disorder, and catalysis. In Structure and Intrinsic Disorder in Enzymology; Elsevier: 2023; pp. 353–376.
- Liu, Y.D.; Li, J.J.; Wang, F.W.; Chen, J.; Li, P.; Su, Z.G. A newly proposed mechanism for arginine-assisted protein refolding--not inhibiting soluble oligomers although promoting a correct structure. Protein Expr. Purif. 2007, 51, 235–242. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Y.; Wang, Y.; Ding, H.; Su, Z. Different effects of l-arginine on protein refolding: Suppressing aggregates of hydrophobic interaction, not covalent binding. Biotechnol. Prog. 2008, 24, 1365–1372. [Google Scholar] [CrossRef]
- Fass, D.; Thorpe, C. Chemistry and Enzymology of Disulfide Cross-Linking in Proteins. Chem. Rev. 2018, 118, 1169–1198. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, T.; Ejima, D.; Tsumoto, K.; Obeyama, N.; Tanaka, Y.; Kita, Y.; Timasheff, S.N. Suppression of protein interactions by arginine: A proposed mechanism of the arginine effects. Biophys. Chem. 2007, 127, 1–8. [Google Scholar] [CrossRef]
- Das, U.; Hariprasad, G.; Ethayathulla, A.S.; Manral, P.; Das, T.K.; Pasha, S.; Mann, A.; Ganguli, M.; Verma, A.K.; Bhat, R.; et al. Inhibition of protein aggregation: Supramolecular assemblies of arginine hold the key. PLoS ONE 2007, 2, e1176. [Google Scholar] [CrossRef]
- Kim, N.A.; Hada, S.; Thapa, R.; Jeong, S.H. Arginine as a protein stabilizer and destabilizer in liquid formulations. Int. J. Pharm. 2016, 513, 26–37. [Google Scholar] [CrossRef]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef]
- Wang, F.; Wang, Y.; Zhang, X.; Zhang, W.; Guo, S.; Jin, F. Recent progress of cell-penetrating peptides as new carriers for intracellular cargo delivery. J. Control Release 2014, 174, 126–136. [Google Scholar] [CrossRef]
- Peitz, M.; Pfannkuche, K.; Rajewsky, K.; Edenhofer, F. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: A tool for efficient genetic engineering of mammalian genomes. Proc. Natl. Acad. Sci. USA 2002, 99, 4489–4494. [Google Scholar] [CrossRef]
- Allen, J.; Pellois, J.P. Hydrophobicity is a key determinant in the activity of arginine-rich cell penetrating peptides. Sci. Rep. 2022, 12, 15981. [Google Scholar] [CrossRef]
- Arakawa, T.; Tsumoto, K.; Nagase, K.; Ejima, D. The effects of arginine on protein binding and elution in hydrophobic interaction and ion-exchange chromatography. Protein Expr. Purif. 2007, 54, 110–116. [Google Scholar] [CrossRef]
- Hall, T.; Kelly, G.M.; Emery, W.R. Use of mobile phase additives for the elution of bispecific and monoclonal antibodies from phenyl based hydrophobic interaction chromatography resins. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2018, 1096, 20–30. [Google Scholar] [CrossRef]
- Valente, J.F.A.; Sousa, A.; Azevedo, G.A.; Queiroz, J.A.; Sousa, F. Purification of supercoiled p53-encoding plasmid using an arginine-modified macroporous support. J. Chromatogr. A 2020, 1618, 460890. [Google Scholar] [CrossRef]
- Soares, A.; Queiroz, J.A.; Sousa, F.; Sousa, A. Purification of human papillomavirus 16 E6/E7 plasmid deoxyribonucleic acid-based vaccine using an arginine modified monolithic support. J. Chromatogr. A 2013, 1320, 72–79. [Google Scholar] [CrossRef]
- Valente, J.F.A.; Carreira, T.S.; Dias, J.R.; Sousa, F.; Alves, N. Arginine-Modified 3D-Printed Chromatographic Supports. Pharmaceutics 2022, 14. [Google Scholar] [CrossRef]
- Gupta, M.N. Methods for Affinity-Based Separations of Enzymes and Proteins; Springer Science & Business Media: 2002.
- Roy, I.; Gupta, M.N. Current trends in affinity-based separations of proteins/enzymes. Curr. Sci. 2000, 587–591. [Google Scholar]
- Mondal, K.; Gupta, M.N. The affinity concept in bioseparation: Evolving paradigms and expanding range of applications. Biomol. Eng. 2006, 23, 59–76. [Google Scholar] [CrossRef]
- Bartnicki, F.; Bonarek, P.; Kowalska, E.; Strzalka, W. The Argi system: One-step purification of proteins tagged with arginine-rich cell-penetrating peptides. Sci. Rep. 2017, 7, 2619. [Google Scholar] [CrossRef]
- Burg, M.B.; Ferraris, J.D. Intracellular organic osmolytes: Function and regulation. J. Biol. Chem. 2008, 283, 7309–7313. [Google Scholar] [CrossRef]
- Xu, M.; Liu, P.; Chen, J.; Peng, A.; Yang, T.; Zhang, X.; Xu, Z.; Rao, Z. Development of a Novel Biosensor-Driven Mutation and Selection System via in situ Growth of Corynebacterium crenatum for the Production of L-Arginine. Front. Bioeng. Biotechnol. 2020, 8, 175. [Google Scholar] [CrossRef]
- Stasyuk, N.; Gayda, G.; Demkiv, O.; Darmohray, L.; Gonchar, M.; Nisnevitch, M. Amperometric biosensors for L-arginine determination based on L-arginine oxidase and peroxidase-like nanozymes. Appl. Sci. 2021, 11, 7024. [Google Scholar] [CrossRef]
- Berketa, K.; Saiapina, O.; Fayura, L.; Sibirny, A.; Dzyadevych, S.; Soldatkin, O. Novel highly sensitive conductometric biosensor based on arginine deiminase from Mycoplasma hominis for determination of arginine. Sens. Actuators B Chem. 2022, 367, 132023. [Google Scholar] [CrossRef]
- Rahman, M.; Niu, J.; Cui, X.; Zhou, C.; Tang, N.; Jin, H.; Cui, D. Electrochemical Biosensor Based on l-Arginine and rGO-AuNSs Deposited on the Electrode Combined with DNA Probes for Ultrasensitive Detection of the Gastric Cancer-Related PIK3CA Gene of ctDNA. ACS Appl. Bio Mater. 2022, 5, 5094–5103. [Google Scholar] [CrossRef]
- Roy, I.; Gupta, M.N. White & grey biotechnologies for shaping a sustainable future. RSC Sustain. 2023, 1, 1722–1736. [Google Scholar]
- Li, S.Y.; Ng, I.S.; Chen, P.T.; Chiang, C.J.; Chao, Y.P. Biorefining of protein waste for production of sustainable fuels and chemicals. Biotechnol. Biofuels 2018, 11, 256. [Google Scholar] [CrossRef]
- De Schouwer, F.; Claes, L.; Vandekerkhove, A.; Verduyckt, J.; De Vos, D.E. Protein-rich biomass waste as a resource for future biorefineries: State of the art, challenges, and opportunities. ChemSusChem 2019, 12, 1272–1303. [Google Scholar] [CrossRef] [PubMed]
- Peydayesh, M.; Bagnani, M.; Soon, W.L.; Mezzenga, R. Turning Food Protein Waste into Sustainable Technologies. Chem. Rev. 2023, 123, 2112–2154. [Google Scholar] [CrossRef] [PubMed]
- Piercy, E.; Verstraete, W.; Ellis, P.R.; Banks, M.; Rockström, J.; Smith, P.; Witard, O.C.; Hallett, J.; Hogstrand, C.; Knott, G. A sustainable waste-to-protein system to maximise waste resource utilisation for developing food-and feed-grade protein solutions. Green Chem. 2023, 25, 808–832. [Google Scholar] [CrossRef]
- Maeta, S.; Nakakido, M.; Matsuura, H.; Sakai, N.; Hirata, K.; Kuroda, D.; Fukunaga, A.; Tsumoto, K. Arginine cluster introduction on framework region in anti-lysozyme antibody improved association rate constant by changing conformational diversity of CDR loops. Protein Sci. 2023, 32, e4745. [Google Scholar] [CrossRef]
- Gupta, M.N.; Uversky, V.N. Structure and Intrinsic Disorder in Enzymology; Elsevier: 2022.

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




