Submitted:
25 September 2025
Posted:
26 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Data
2.2. Description of the Model Used
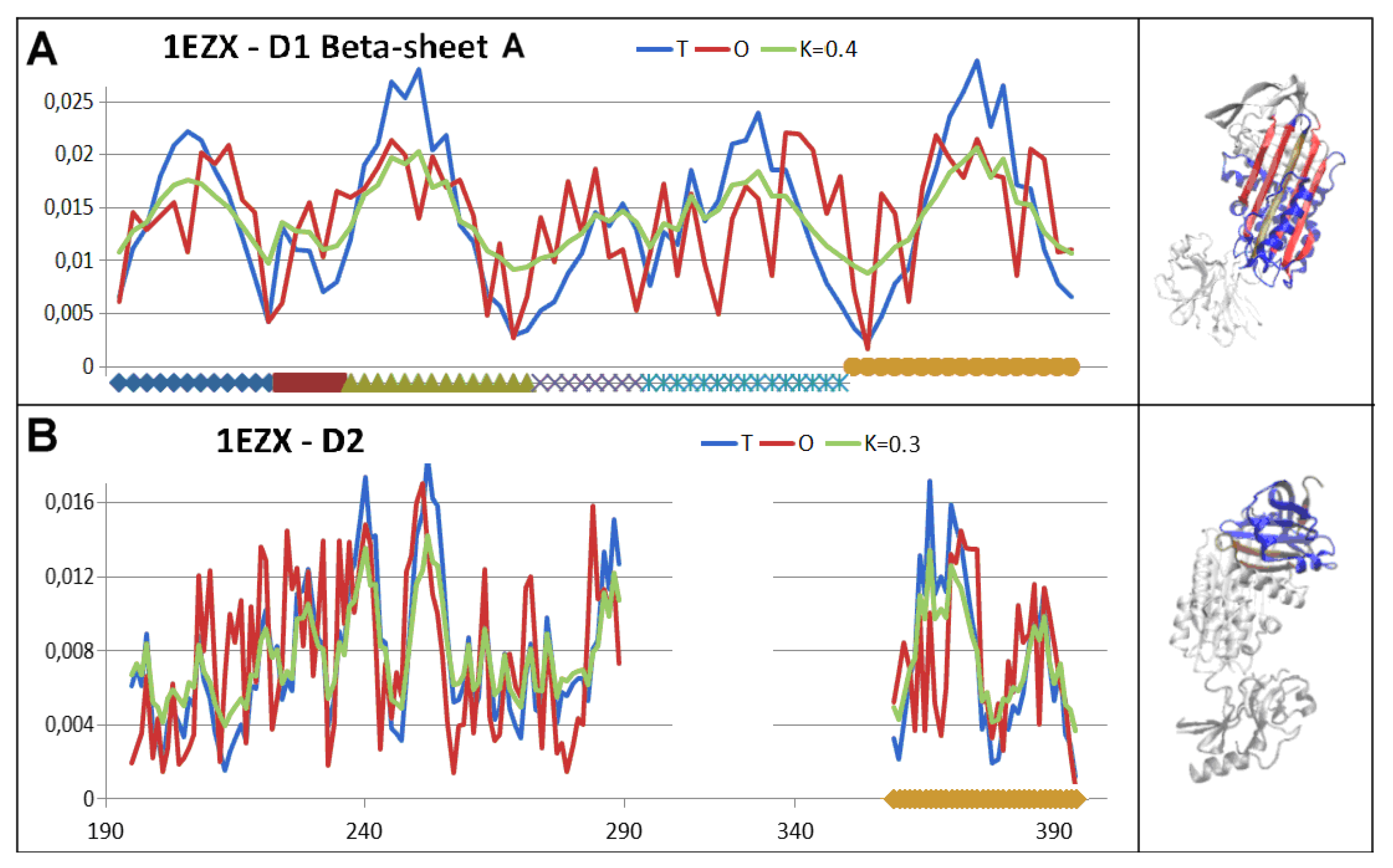
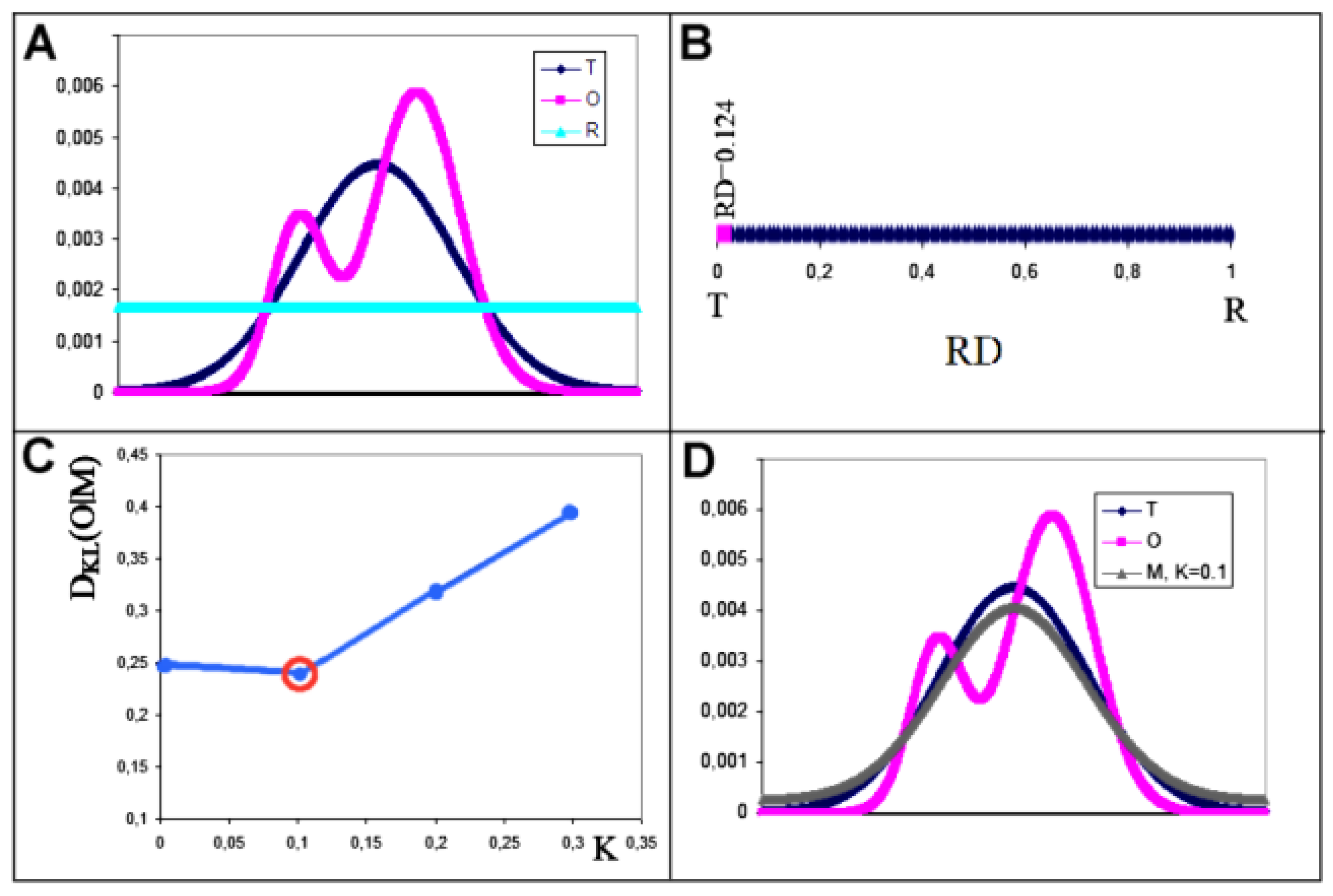
- RD indicates the degree of restoration of the micelle-like arrangement in the hydrophobicity distribution. The hydrophobicity distribution in the micelle structure (RD<0.5) was treated as idealised and consistent with a centric hydrophobic nucleus and a polar surface. The higher the RD value in the range [0,1], the more the hydrophobicity distribution in the protein body approaches an aligned distribution with a uniform layout of comparable hydrophobicity levels. This means deprivation of the hydrophobic nucleus.
- K indicates the degree to which the environment is different from the polar aquatic environment. The higher the K parameter value, the higher the contribution of non-aqueous factors to the formation of the protein structure from the perspective of hydrophobicity distribution.
3. Results
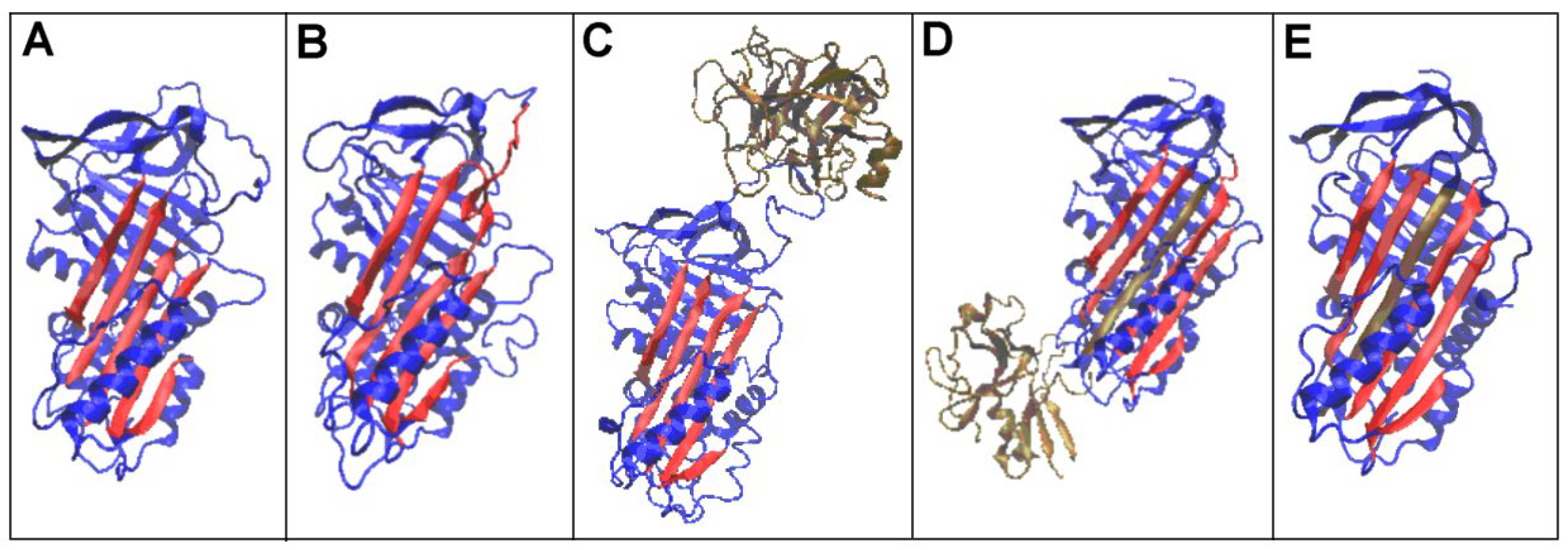
3.1. Status of Complexes and Chains Present in Them
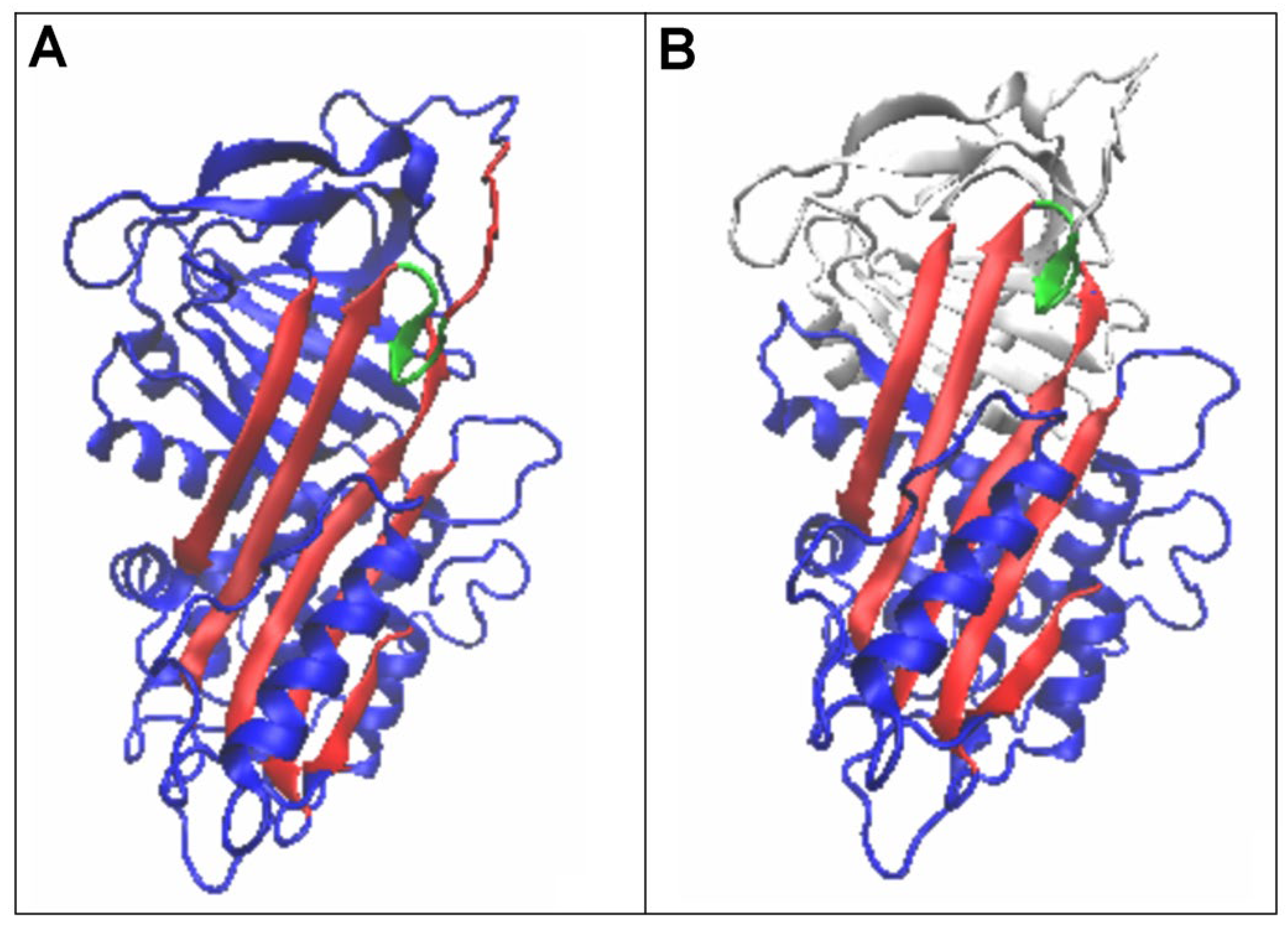
3.2. Structure of the Serpin Chain
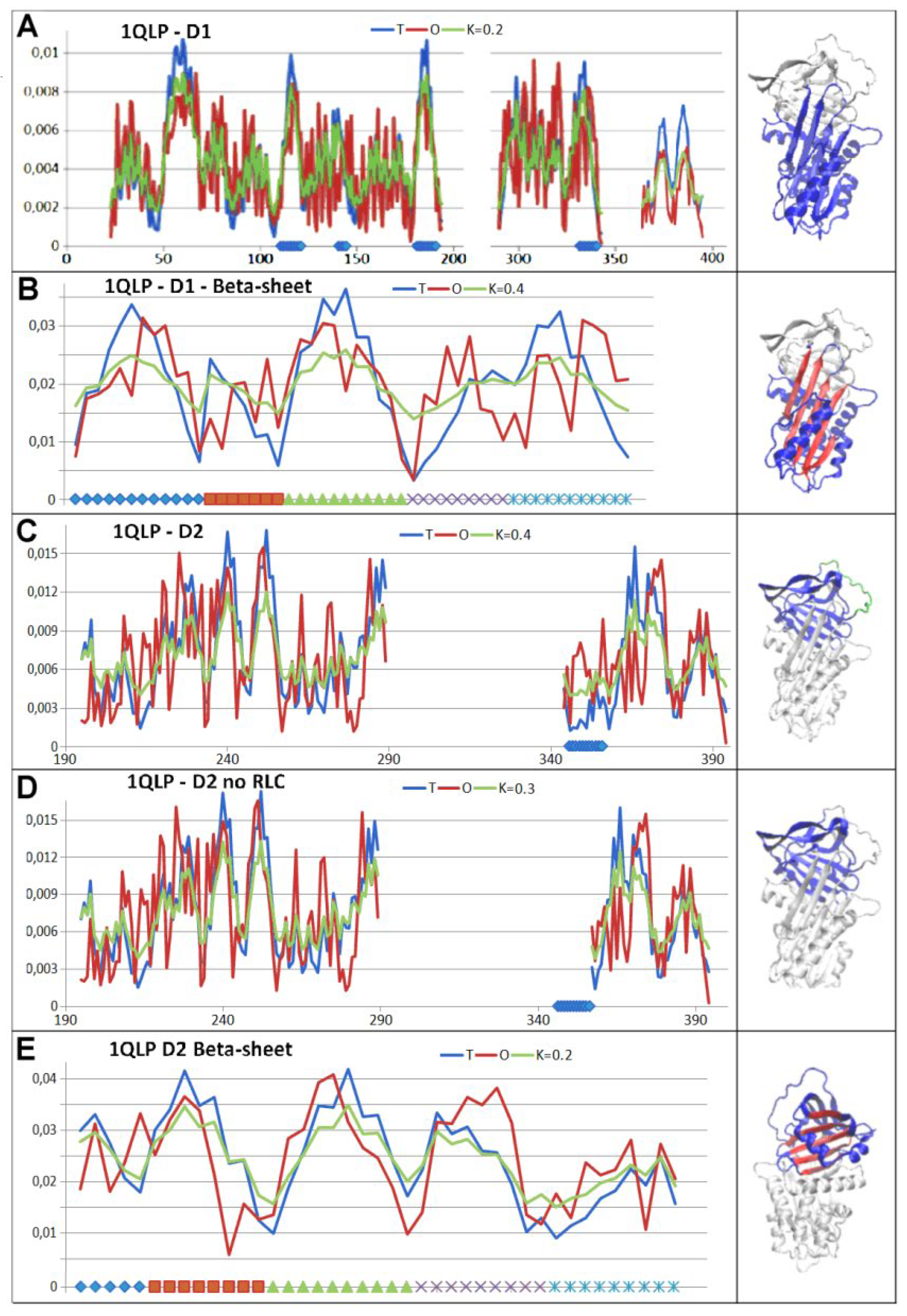
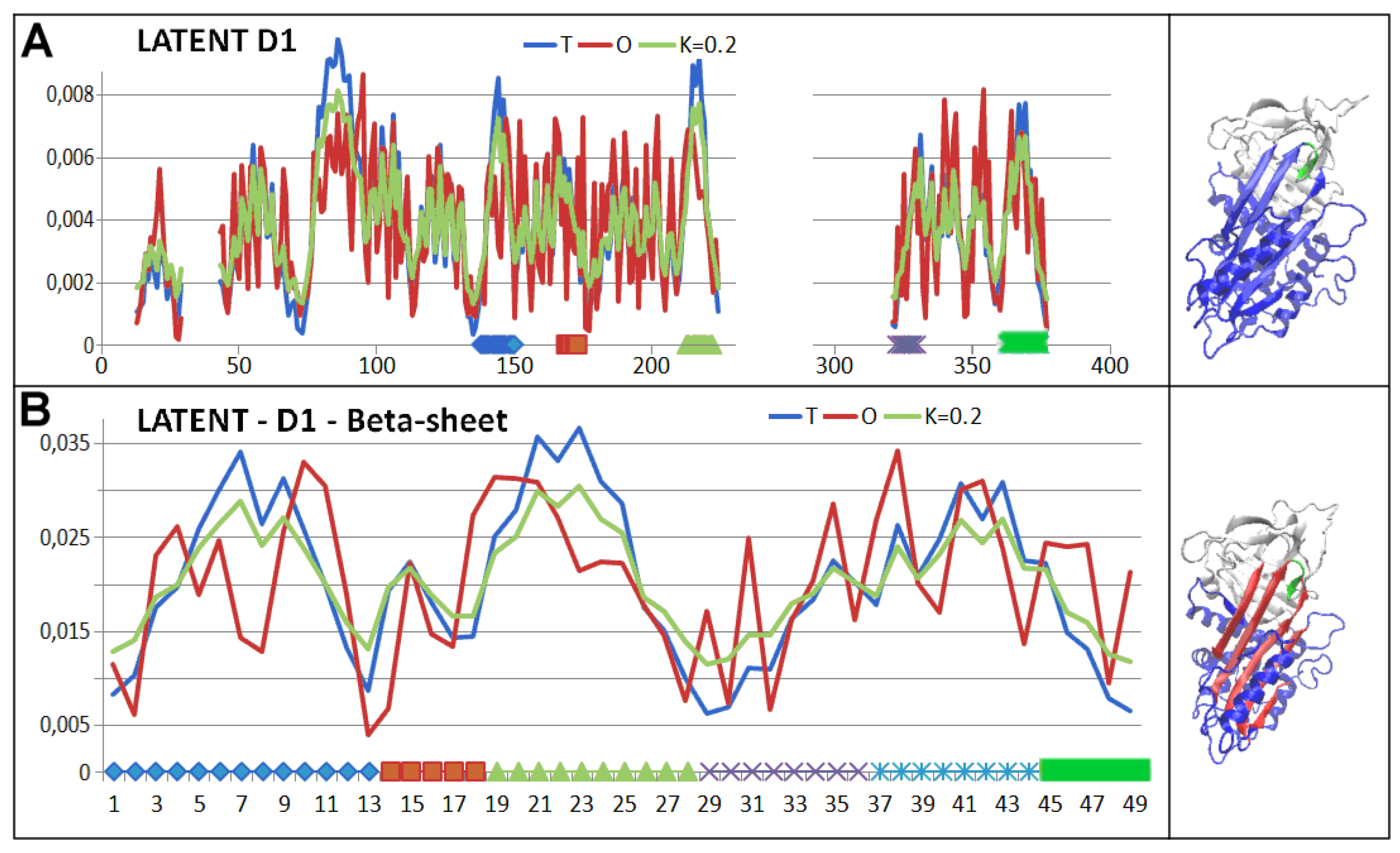
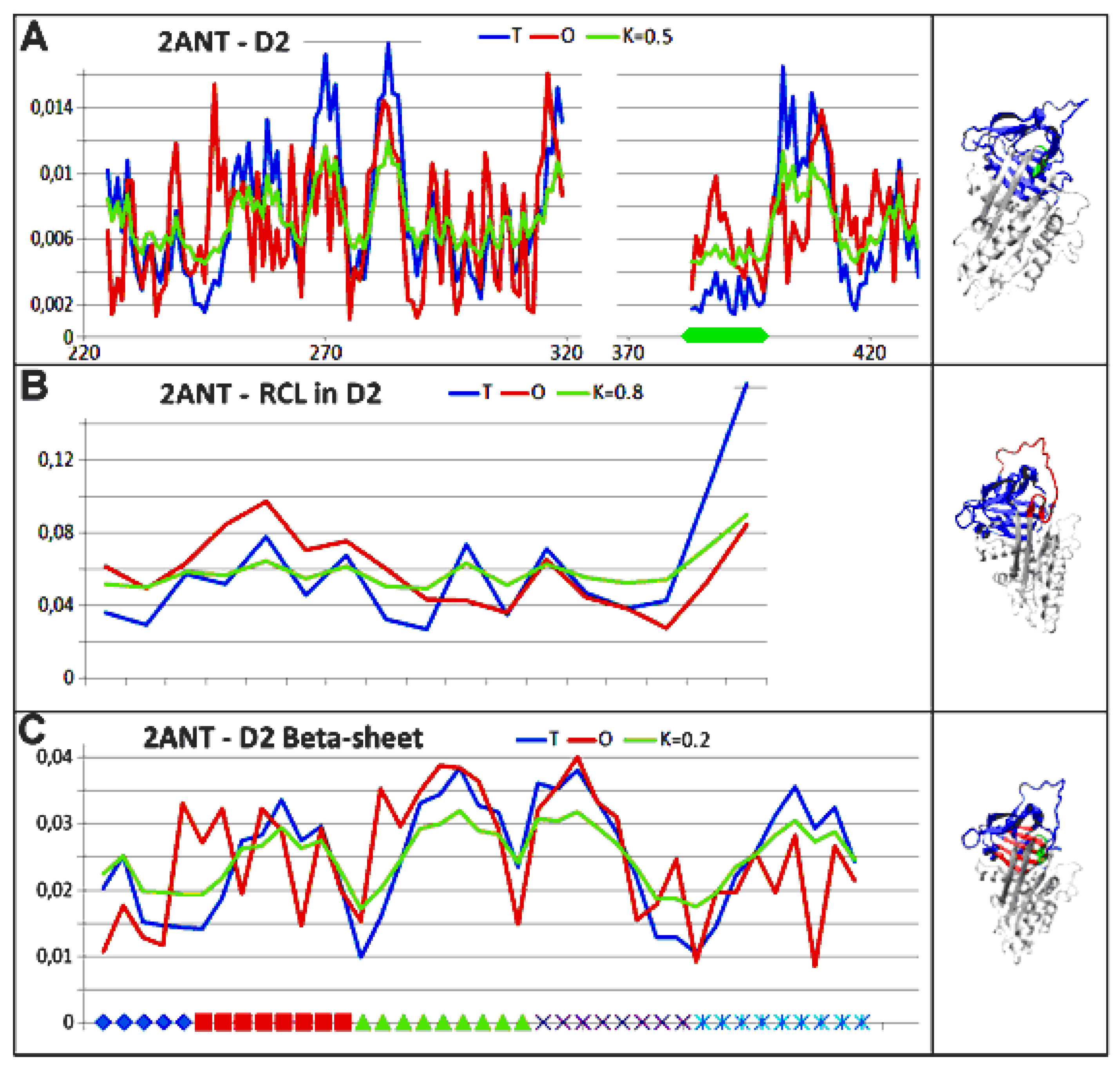
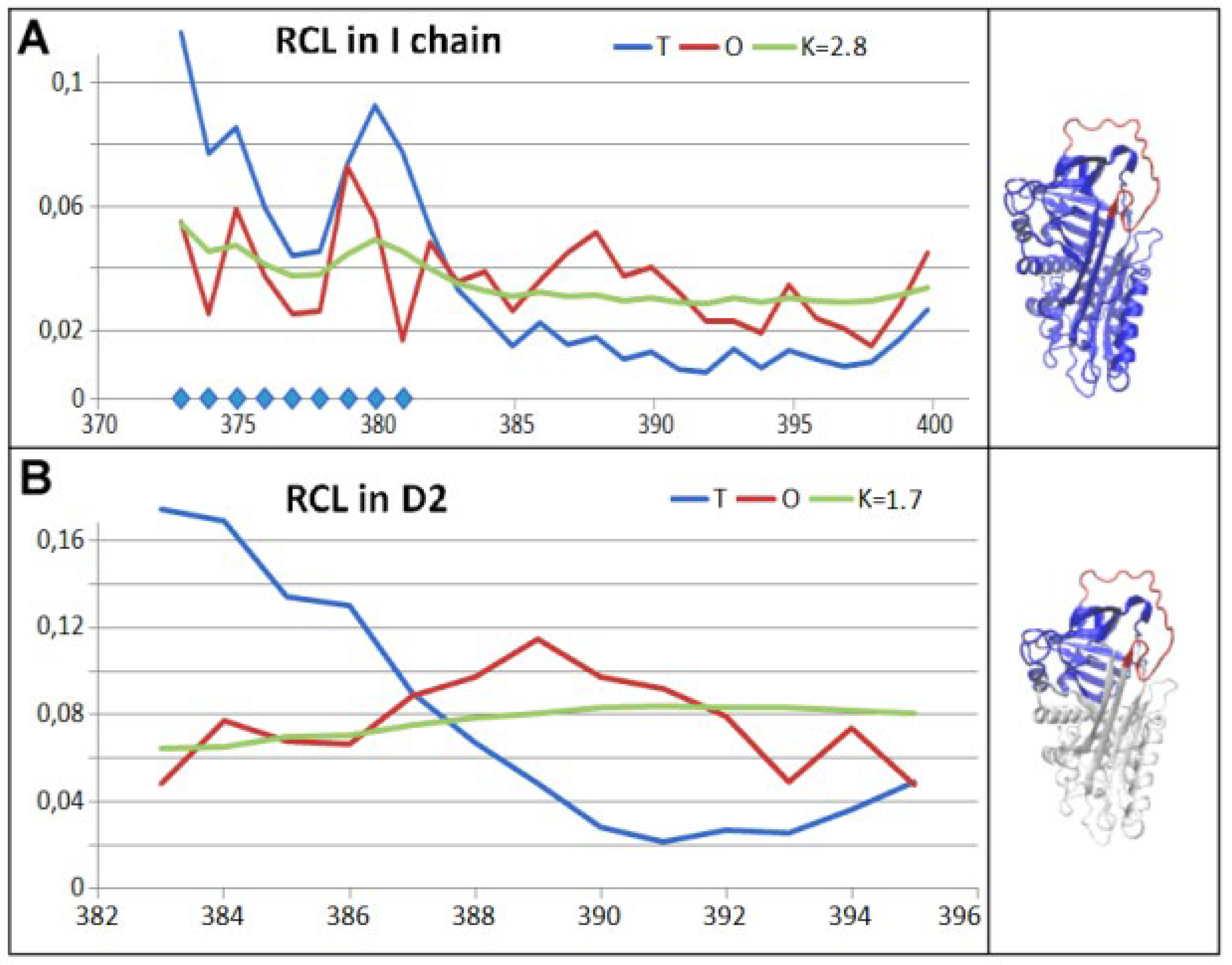
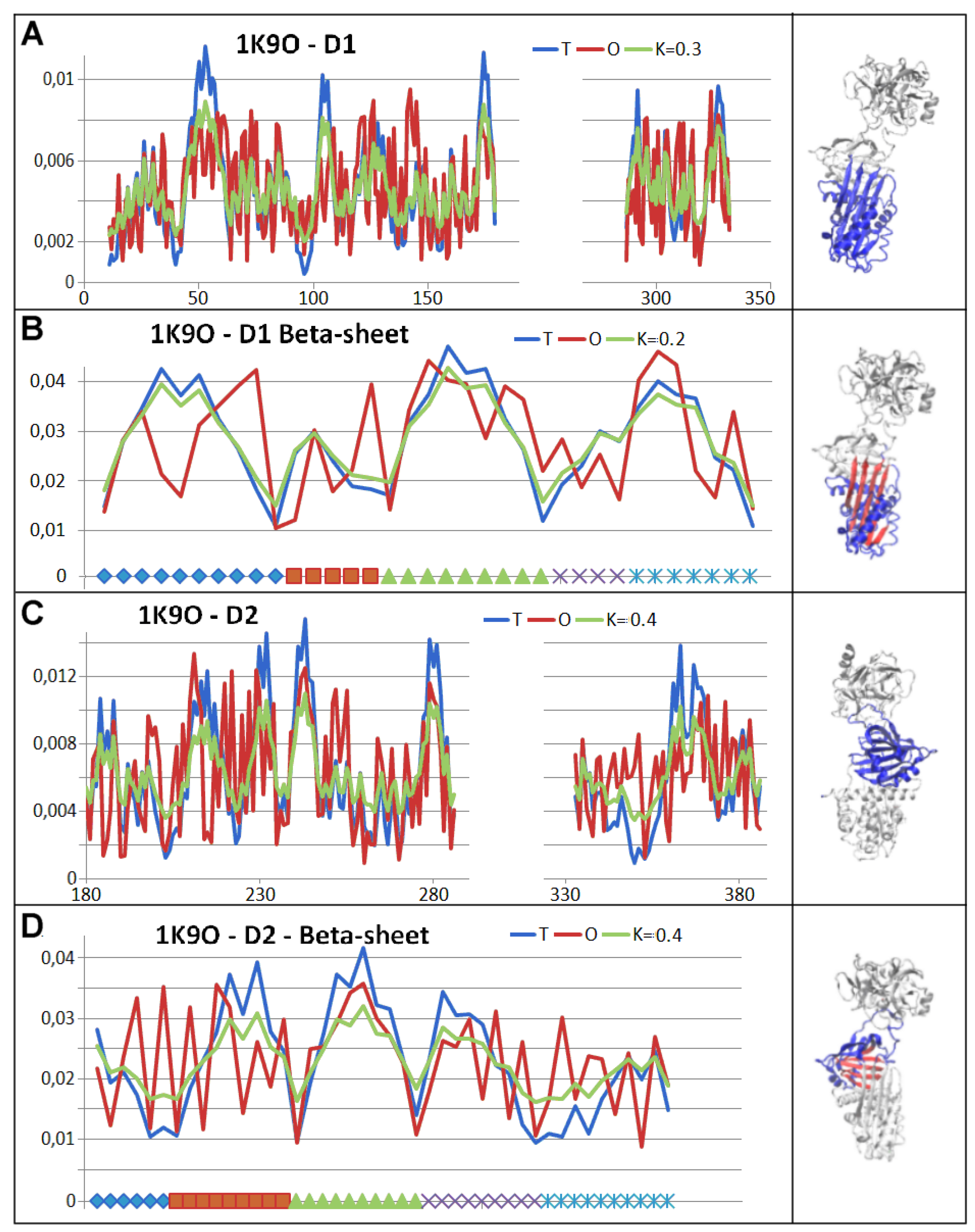
3.3. Status of the Domains
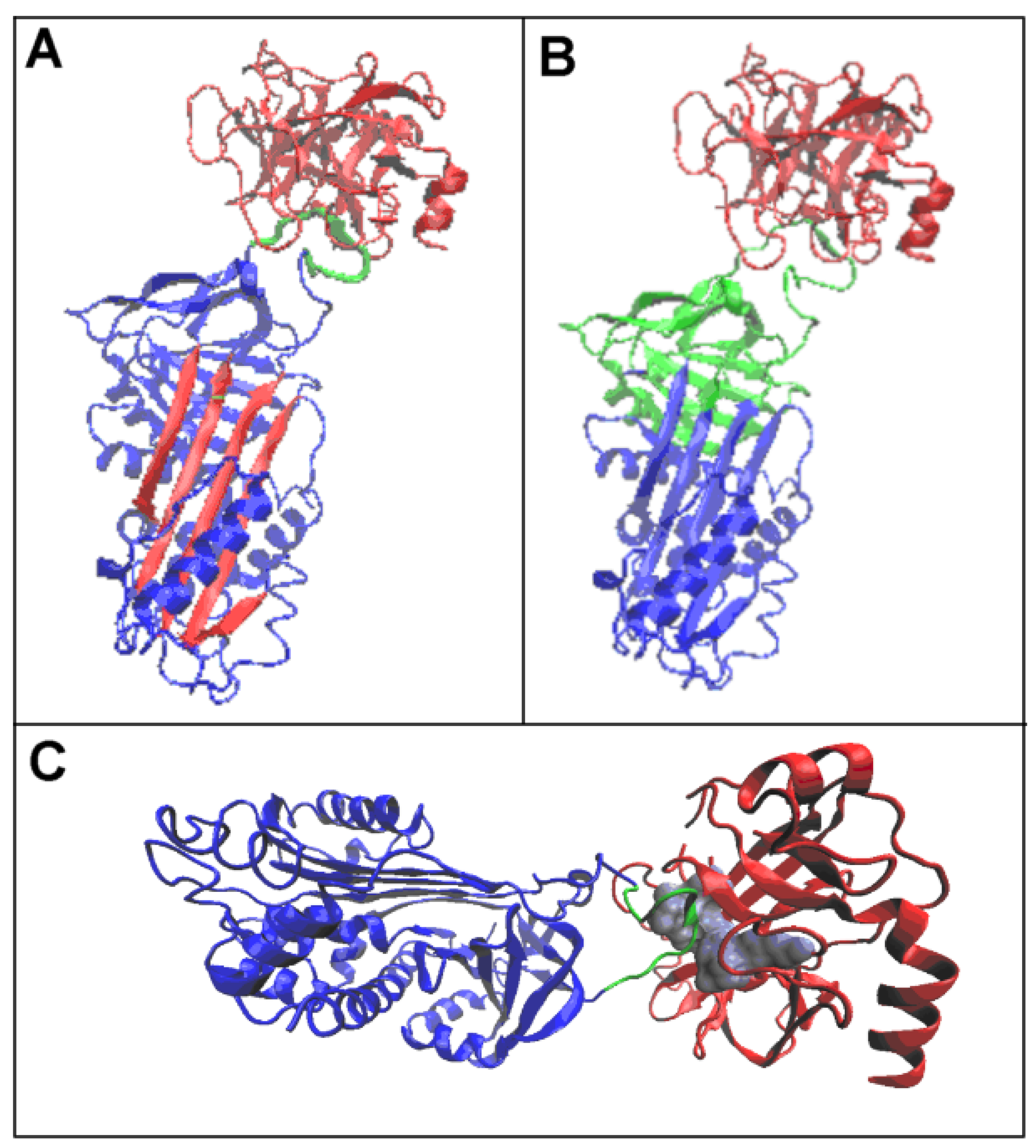
3.4. Status of Beta-Sheets
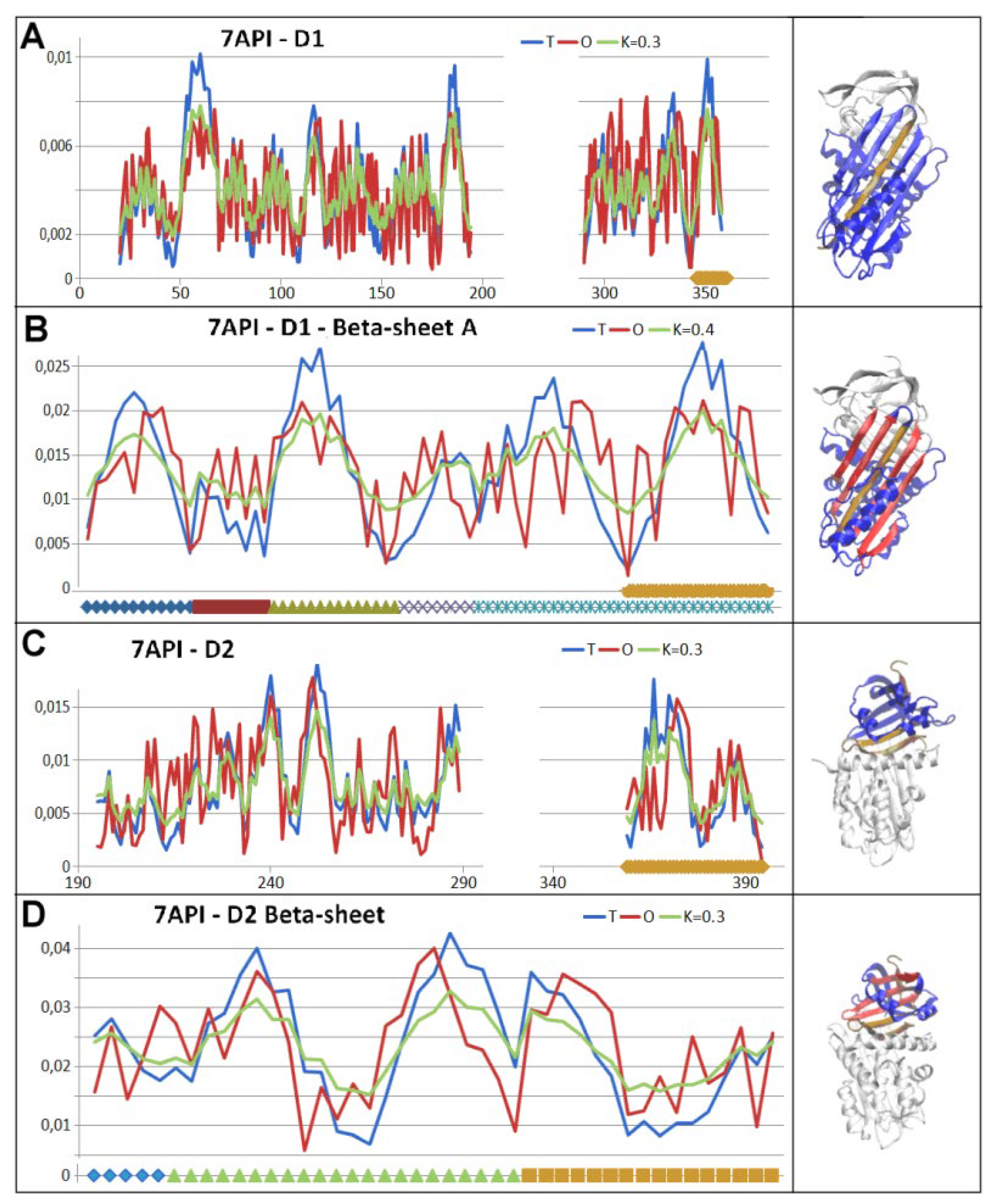
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Cano-Martínez A, Rubio-Ruiz ME, Guarner-Lans V. Homeostasis and evolution in relation to regeneration and repair. J Physiol. 2024; 602(11): 2627-2648. [CrossRef]
- Bechtel W, Bich L. Situating homeostasis in organisms: maintaining organization through time. J Physiol. 2024; 602(22): 6003-6020. [CrossRef]
- Castagna D, Gourdet B, Hjerpe R, MacFaul P, Novak A, Revol G, Rochette E, Jordan A. To homeostasis and beyond! Recent advances in the medicinal chemistry of heterobifunctional derivatives. Prog Med Chem. 2024; 63(1): 61-160. [CrossRef]
- Giordano G, Cuba Samaniego C, Franco E, Blanchini F. Computing the structural influence matrix for biological systems. J Math Biol. 2016; 72(7): 1927-58. [CrossRef]
- Antoneli F, Golubitsky M, Jin J, Stewart I. Homeostasis in input-output networks: Structure, Classification and Applications. Math Biosci. 2025; 384: 109435. [CrossRef]
- Recordati G, Bellini TG. A definition of internal constancy and homeostasis in the context of non-equilibrium thermodynamics. Exp Physiol. 2004; 89(1): 27-38. [CrossRef]
- Lucas A, Yaron JR, Zhang L, Macaulay C, McFadden G. Serpins: Development for Therapeutic Applications. Methods Mol Biol. 2018; 1826: 255-265. [CrossRef]
- Wilkinson DJ. Serpins in cartilage and osteoarthritis: what do we know? Biochem Soc Trans. 2021; 49(2): 1013-1026. [CrossRef]
- Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, Whisstock JC. An overview of the serpin superfamily. Genome Biol. 2006; 7(5): 216. [CrossRef]
- Loebermann H, Tokuoka R, Deisenhofer J, R Huber R. Human alpha 1-proteinase inhibitor. Crystal structure analysis of two crystal modifications, molecular model and preliminary analysis of the implications for function. J Mol Biol. 1984; 177(3): 531-57.
- Elliott PR, Pei XY, Dafforn TR, Lomas DA. Topography of a 2.0 A structure of alpha1-antitrypsin reveals targets for rational drug design to prevent conformational disease. Protein Sci. 2000; 9(7): 1274-81. [CrossRef]
- Geiger M, Wahlmüller F, Furtmüller M. The serpin family. Proteins with multiple functions in health and disease. Springer 2015. [CrossRef]
- Whisstock JC, Pike RN, Jin L, Skinner R, Pei XY, Carrell RW, Lesk AM. Conformational changes in serpins: II. The mechanism of activation of antithrombin by heparin. J Mol Biol. 2000; 301(5): 1287-305. [CrossRef]
- Whisstock JC, Skinner R, Carrell RW, Lesk AM. Conformational changes in serpins: I. The native and cleaved conformations of alpha(1)-antitrypsin. J Mol Biol. 2000; 296(2): 685-99. PMID: 10669617. [CrossRef]
- Whisstock JC, Skinner R, Carrell RW, Lesk AM. Conformational changes in serpins: I. The native and cleaved conformations of alpha(1)-antitrypsin. J Mol Biol. 2000; 295(3): 651-65. [CrossRef]
- Sillitoe I, Bordin N, Dawson N, Waman VP, Ashford P, Scholes HM, Pang CSM, Woodridge L, Rauer C, Sen N, Abbasian M, Le Cornu S, Lam SD, Berka K, Hutařová-Varekova I, Svobodova R, Lees J, Orengo CA. CATH: increased structural coverage of functional space. Nucleic Acids Res. 2021; 49(D1): D266-D273. [CrossRef]
- Ye S, Cech AL, Belmares R, Bergstrom RC, Tong Y, Corey DR, Kanost MR, Goldsmith EJ. The structure of a Michaelis serpin-protease complex. Nat Struct Biol. 2001; 8(11): 979-83. [CrossRef]
- Skinner R, Abrahams JP, Whisstock JC, Lesk AM, Carrell RW, Wardell MR. The 2.6 A structure of antithrombin indicates a conformational change at the heparin binding site J Mol Biol. 1997; 266(3): 601-9. [CrossRef]
- Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000; 407(6806): 923-6. [CrossRef]
- Engh R, Löbermann H, Schneider M, Wiegand G, Huber R, Laurell CB. The S variant of human alpha 1-antitrypsin, structure and implications for function and metabolism. Protein Eng. 1989; 2(6): 407-15. [CrossRef]
- Roterman I, Konieczny L. Protein Is an Intelligent Micelle. Entropy (Basel). 2023; 25(6): 850. [CrossRef]
- Levitt MA. A simplified representation of protein conformations for rapid simulation of protein folding. J. Mol. Biol. 1976, 104(1), 59-107. [CrossRef]
- Kalinowska B, Banach M, Konieczny L, Roterman I. Application of Divergence Entropy to Characterize the Structure of the Hydrophobic Core in DNA Interacting Proteins. Entropy 2015, 17(3), 1477-1507. [CrossRef]
- Kullback S.; Leibler RA. On information and sufficiency. Annals Mathemat Statistics 1951, 22(1), 79-86. [CrossRef]
- Banach M, Stapor K, Konieczny L, Fabian P, Roterman I. Downhill, Ultrafast and Fast Folding Proteins Revised. Int J Mol Sci. 2020; 21(20): 7632. [CrossRef]
- Roterman I, Stapor K, Dułak D, Szoniec G, Konieczny L. Aquaporins as Membrane Proteins: The Current Status. Front Biosci (Schol Ed). 2025; 17(1): 27967. [CrossRef]
- .Roterman I, Stapor K, Konieczny L. Transmembrane proteins-Different anchoring systems. Proteins. 2024; 92(5): 593-609. [CrossRef]
- Roterman I, Stapor K, Dułak D, Konieczny L. Heat Shock Protein and Disaggregase Influencing the Casein Structuralisation. Int J Mol Sci. 2025; 26(13): 6360. [CrossRef]
- Roterman I, Stapor K, Dułak D, Konieczny L. External Force Field for Protein Folding in Chaperonins-Potential Application in In Silico Protein Folding. ACS Omega. 2024 ; 9(16): 18412-18428. [CrossRef]
- Roterman I, Stapor K, Konieczny L. Ab initio protein structure prediction: the necessary presence of external force field as it is delivered by Hsp40 chaperone. BMC Bioinformatics. 2023; 24(1): 418. [CrossRef]
- Lomas DA, Mahadeva R. α1-Antitrypsin polymerization and the serpinopathies: pathobiology and prospects for therapy. J Clin Invest. 2002; 110(11): 1585–1590]. [CrossRef]
- Olson ST, Swanson R, Day D, Verhamme I, Kvassman J, Shore JD. Resolution of Michaelis complex, acylation, and conformational change steps in the reactions of the serpin, plasminogen activator inhibitor-1, with tissue plasminogen activator and trypsin. Biochemistry. 2001; 40(39): 11742-56. [CrossRef]
- Huntington JA, Yamasaki M. Serpin polymerization in vitro. Methods Enzymol. 2011; 501: 379-420. doi: . [CrossRef]
- Yamasaki M, Sendall TJ, Harris LE, Lewis GM, Huntington JA. Loop-sheet mechanism of serpin polymerization tested by reactive center loop mutations. J Biol Chem. 2010; 285(40): 30752-8. [CrossRef]
- Johnson DJ, Langdown J, Li W, Luis SA, Baglin TP, Huntington JA. Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation. J Biol Chem. 2006 ; 281(46): 35478-86. [CrossRef]
- Olivieri C, Li GC, Wang Y, VS M, Walker C, Kim J, Camilloni C, De Simone A, Vendruscolo M, Bernlohr DA, Taylor SS, Veglia G. ATP-competitive inhibitors modulate the substrate binding cooperativity of a kinase by altering its conformational entropy. Sci Adv. 2022; 8(30): eabo0696. [CrossRef]
- Weisberg E, Manley PW, Breitenstein W, Brüggen J, Cowan-Jacob SW, Ray A, Huntly B, Fabbro D, Fendrich G, Hall-Meyers E, Kung AL, Mestan J, Daley GQ, Callahan L, Catley L, Cavazza C, Azam M, Neuberg D, Wright RD, Gilliland DG, Griffin JD. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005; V7(2): V129-41,V. [CrossRef]
- Yamasaki M, Li W, Johnson DJD, Huntington JA. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature. 2008; 455(7217): 1255-8. [CrossRef]
- Marijanovic EM, Fodor J, Riley BT, Porebski BT, Costa MGS, Kass I, Hoke DE, McGowan S, Buckle AM. Reactive centre loop dynamics and serpin specificity. Sci Rep. 2019; 9(1): 3870. [CrossRef]
- Roterman I, Stapor K, Dułak D, Konieczny L. Domain swapping: a mathematical model for quantitative assessment of structural effects. FEBS Open Bio. 2024; 14(12): 2006-2025. [CrossRef]
- Roterman I, Stampor K, Dulak D, Zemanek G, Konieczny L. Mechanism of dimerization and polymerization of serpins – in preparation.
- Elliott PR, Lomas DA, Carrell RW, Abrahams JP. Inhibitory conformation of the reactive loop of alpha 1-antitrypsin. Nat Struct Biol. 1996; 3(8): 676-81. [CrossRef]
- Huntington JA. Serpin structure, function and dysfunction. J Thromb Haemost. 2011; 9 Suppl 1:26-34. [CrossRef]
- Engh RA, Huber R, Bode W, Schulze AJ. Divining the serpin inhibition mechanism: a suicide substrate ‘springe’? Trends Biotechnol. 1995; 13(12): 503-10. [CrossRef]
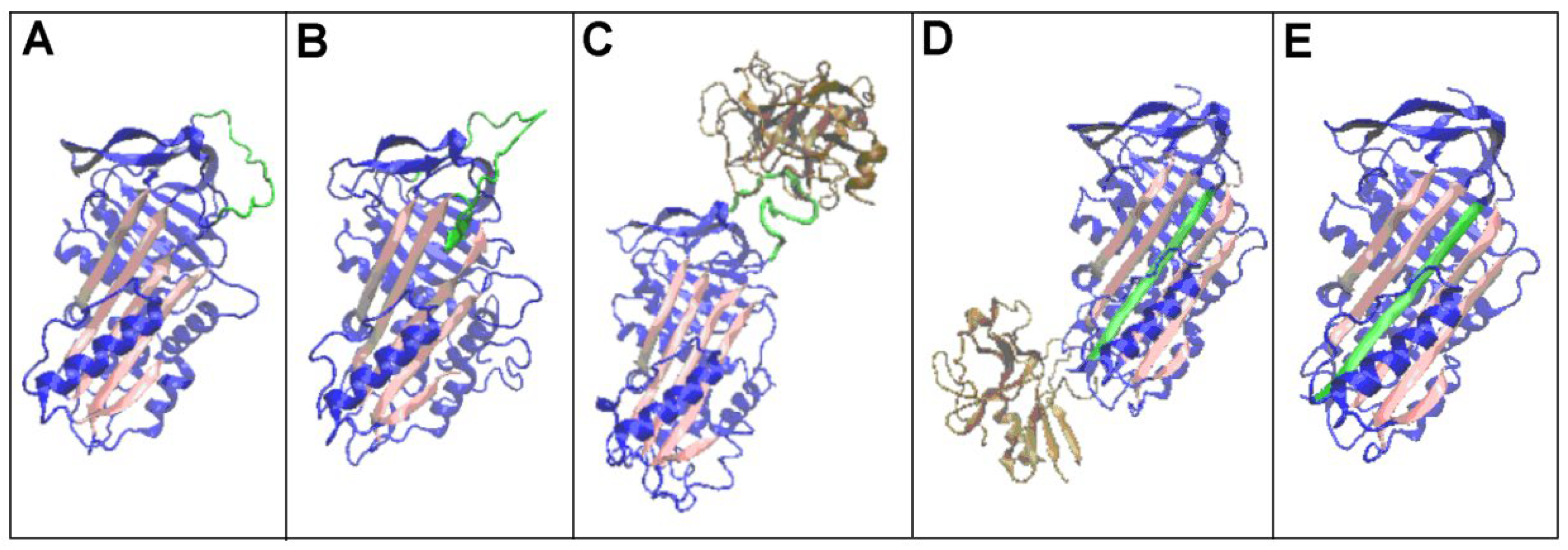
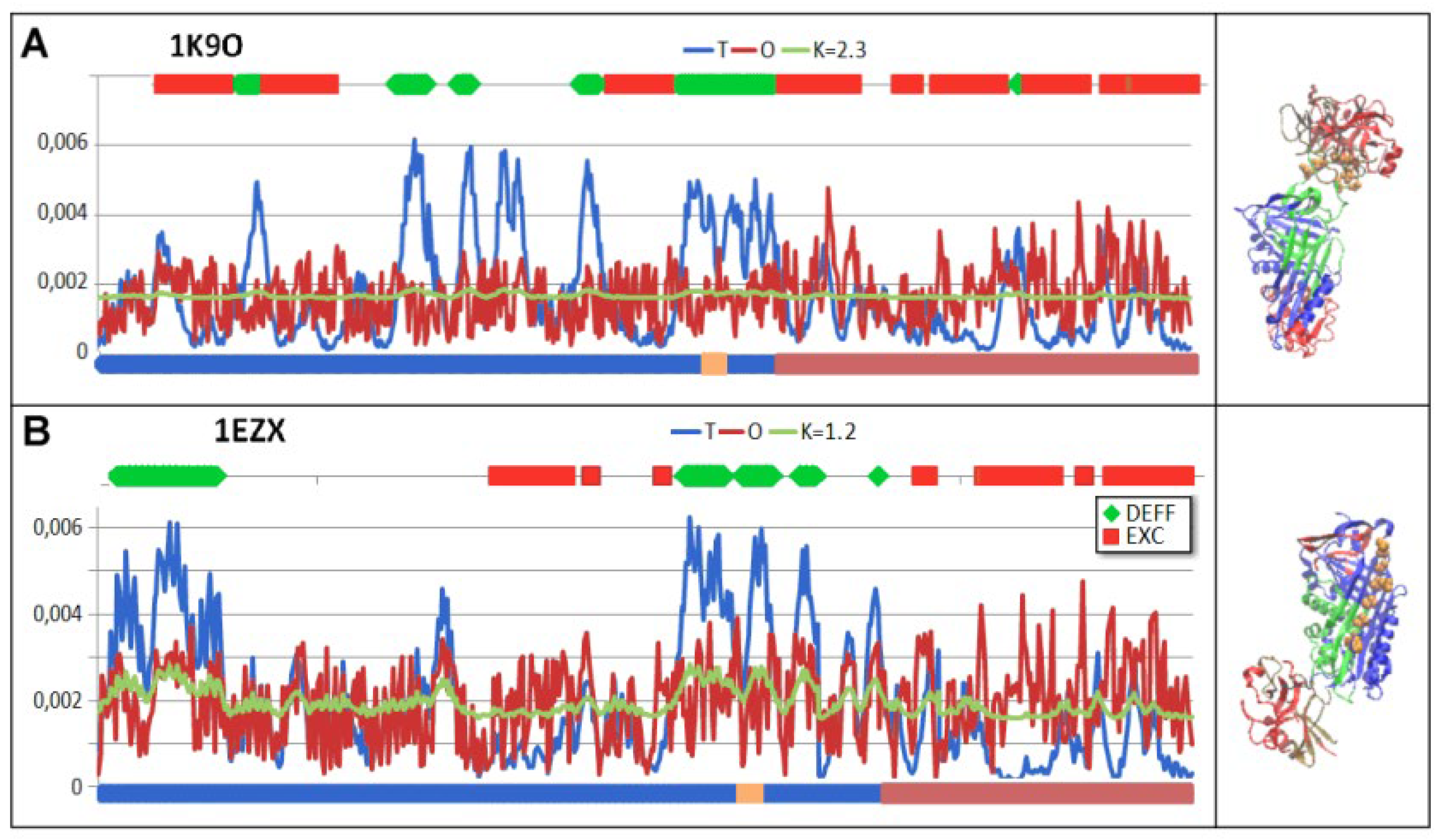
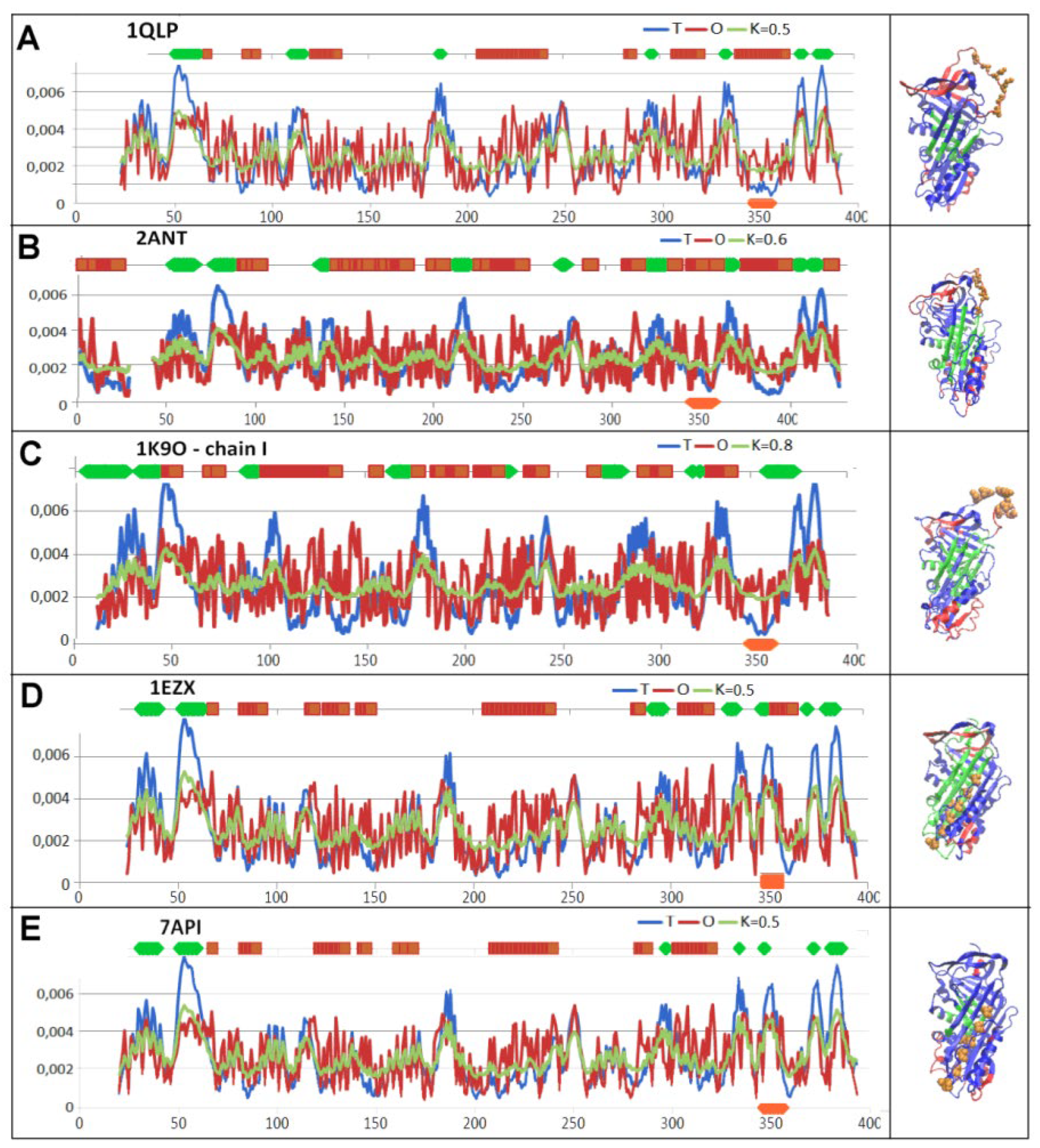
| PDB -ID | Structural form | Ref. |
|---|---|---|
| 1QLP | Human native state α1-antitrypsin | [12] |
| 2ANT | Latent AT III (M*) | [19] |
| 1K9O | Michaelis non-covalent complex | [18] |
| 1EZX | Covalent serpin-protease complex | [20] |
| 7API | Cleaved | [21] |
| FORM | PDB ID | CHAIN IN COMPLEX | CHAIN - INDIVIDUAL | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SERPIN | PROTEASE | SERPIN | PROTEASE | ||||||||
| RD | K | RD | K | RD | K | RD | K | RD | K | ||
| NATIVE LATENT MICHAELIS COVALENT CLEAVED |
1QLP 2ANT 1K9O 1EZX 7API |
0.787 0.726 |
2.3 1.2 |
0.778 0.590 |
1.6 0.6 |
0.711 0.779 |
1.3 1.7 |
0.510 0.572 0.636 0.526 0.519 |
0.5 0.6 0.8 0.5 0.5 |
0.447 0.497 |
0.4 0.4 |
|
FORM |
PDB -ID |
DOMAIN 1 |
DOMAIN2 |
BETA-SHEET A IN D1 |
BETA-SHEET B IN D2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No incorp. | Incorporated | No incorp. | Incorporated | ||||||||||
| RD | K | RD | K | RD | K | RD | K | RD | K | RD | K | ||
| NATIVE LATENT MICHAELIS COVALENT CLEAVED |
1QLP 2ANT 1K9O 1EZX 7API |
0.364 0.489 0.461 0.422 0.425 |
0.2 0.2 0.3 0.3 0.3 |
0.396 0.426 0.416 |
0.2 0.3 0.3 |
0.488 0.578 0.514 0.444 0.446 |
0.4 0.4 0.2 0.3 0.3 |
0.613 0.590 0.483 0.559 0.557 |
0.5 0.4 0.2 0.4 0.4 |
0.584 0.548 0.539 |
0.4 0.4 0.4 |
0.429 0.483 0.598 0.447 0.481 |
0.2 0.2 0.4 0.2 0.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





