Submitted:
07 May 2024
Posted:
07 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
3.1. Self-Assembly and Biological Properties of the Non-Natural Amino Acid 5
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Skwarecki, A. S.; Nowak, M. G.; Milewska, M. J. , Amino Acid and Peptide-Based Antiviral Agents. ChemMedChem 2021, 16, 3106–3135. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, R.; Simonyan, H.; Roviello, G. N., Advances in Amino Acid-Based Chemistry. MDPI: 2023; Vol. 16, p 1490.
- Scognamiglio, P. L.; Riccardi, C.; Palumbo, R.; Gale, T. F.; Musumeci, D.; Roviello, G. N. , Self-assembly of thyminyl l-tryptophanamide (TrpT) building blocks for the potential development of drug delivery nanosystems. Journal of Nanostructure in Chemistry 2023, 1–19. [Google Scholar] [CrossRef]
- Roviello, G. N.; Roviello, V.; Autiero, I.; Saviano, M. , Solid phase synthesis of TyrT, a thymine–tyrosine conjugate with poly (A) RNA-binding ability. RSC advances 2016, 6, 27607–27613. [Google Scholar] [CrossRef] [PubMed]
- Roviello, G. N.; Mottola, A.; Musumeci, D.; Bucci, E. M.; Pedone, C. , Synthesis and aggregation properties of a novel enzymatically resistant nucleoamino acid. Amino Acids 2012, 43, 1465–1470. [Google Scholar] [CrossRef] [PubMed]
- Roviello, G. N. , Novel insights into nucleoamino acids: Biomolecular recognition and aggregation studies of a thymine-conjugated l-phenyl alanine. Amino Acids 2018, 50, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Scognamiglio, P. L.; Vicidomini, C.; Roviello, G. N. , Dancing with Nucleobases: Unveiling the Self-Assembly Properties of DNA and RNA Base-Containing Molecules for Gel Formation. Gels 2023, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Roviello, G. N.; Oliviero, G.; Di Napoli, A.; Borbone, N.; Piccialli, G. , Synthesis, self-assembly-behavior and biomolecular recognition properties of thyminyl dipeptides. Arabian Journal of Chemistry 2020, 13, 1966–1974. [Google Scholar] [CrossRef]
- Musumeci, D.; Ullah, S.; Ikram, A.; Roviello, G. N. , Novel insights on nucleopeptide binding: A spectroscopic and In Silico investigation on the interaction of a thymine-bearing tetrapeptide with a homoadenine DNA. Journal of Molecular Liquids 2022, 347, 117975. [Google Scholar] [CrossRef]
- Ding, Y.; Ting, J. P.; Liu, J.; Al-Azzam, S.; Pandya, P.; Afshar, S. , Impact of non-proteinogenic amino acids in the discovery and development of peptide therapeutics. Amino Acids 2020, 52, 1207–1226. [Google Scholar] [CrossRef]
- Yokoo, H.; Hirano, M.; Misawa, T.; Demizu, Y. , Helical Antimicrobial Peptide Foldamers Containing Non-proteinogenic Amino Acids. ChemMedChem 2021, 16, 1226–1233. [Google Scholar] [CrossRef]
- Pace, A.; Pierro, P. , The new era of 1, 2, 4-oxadiazoles. Organic & biomolecular chemistry 2009, 7, 4337–4348. [Google Scholar]
- James, N. D.; Growcott, J. W. Zibotentan. Drugs of the Future 2009, 34. [Google Scholar] [CrossRef]
- Fizazi, K.; Higano, C. S.; Nelson, J. B.; Gleave, M.; Miller, K.; Morris, T.; Nathan, F. E.; McIntosh, S.; Pemberton, K.; Moul, J. W. , Phase III, randomized, placebo-controlled study of docetaxel in combination with zibotentan in patients with metastatic castration-resistant prostate cancer. Journal of Clinical Oncology 2013, 31, 1740–1747. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, C. S.; Lira, B. F.; Barbosa-Filho, J. M.; Lorenzo, J. G. F.; de Athayde-Filho, P. F. , Synthetic approaches and pharmacological activity of 1, 3, 4-oxadiazoles: a review of the literature from 2000–2012. Molecules 2012, 17, 10192–10231. [Google Scholar] [CrossRef] [PubMed]
- Gour, V. K.; Yahya, S.; Shahar Yar, M. , Unveiling the chemistry of 1, 3, 4-oxadiazoles and thiadiazols: A comprehensive review. Archiv der Pharmazie 2024, 357, 2300328. [Google Scholar] [CrossRef] [PubMed]
- Costa-Almeida, R.; Soares, R.; Granja, P. L. , Fibroblasts as maestros orchestrating tissue regeneration. Journal of tissue engineering and regenerative medicine 2018, 12, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.; McGrath, J.; Navsaria, H. , The role of fibroblasts in tissue engineering and regeneration. British Journal of Dermatology 2007, 156, 1149–1155. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Krishnan, U. M.; Sethuraman, S. , Development of biomaterial scaffold for nerve tissue engineering: Biomaterial mediated neural regeneration. Journal of biomedical science 2009, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hinderer, S.; Brauchle, E.; Schenke-Layland, K. , Generation and assessment of functional biomaterial scaffolds for applications in cardiovascular tissue engineering and regenerative medicine. Advanced healthcare materials 2015, 4, 2326–2341. [Google Scholar] [CrossRef]
- Farag, M. M. , Recent trends on biomaterials for tissue regeneration applications. Journal of Materials Science 2023, 58, 527–558. [Google Scholar] [CrossRef]
- Asl, S. K.; Rahimzadegan, M.; Asl, A. K. , Progress in cardiac tissue engineering and regeneration: Implications of gelatin-based hybrid scaffolds. International Journal of Biological Macromolecules 2024, 129924. [Google Scholar] [CrossRef] [PubMed]
- Arokianathan, J. F.; Ramya, K. A.; Janeena, A.; Deshpande, A. P.; Ayyadurai, N.; Leemarose, A.; Shanmugam, G. , Non-proteinogenic amino acid based supramolecular hydrogel material for enhanced cell proliferation. Colloids and Surfaces B: Biointerfaces 2020, 185, 110581. [Google Scholar] [CrossRef] [PubMed]
- Arezki, N. R.; Williams, A. C.; Cobb, A. J. A.; Brown, M. B. , Design, synthesis and characterization of linear unnatural amino acids for skin moisturization. International Journal of Cosmetic Science 2016, 39, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Ousey, K.; Cutting, K. F.; Rogers, A. A.; Rippon, M. G. , The importance of hydration in wound healing: reinvigorating the clinical perspective. Journal of Wound Care 2016, 25, 122–130. [Google Scholar] [CrossRef]
- Katti, K. S.; Ambre, A. H.; Peterka, N.; Katti, D. R. , Use of unnatural amino acids for design of novel organomodified clays as components of nanocomposite biomaterials. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences 2010, 368, 1963–1980. [Google Scholar] [CrossRef]
- Belokon, Y. N.; Sagyan, A. S.; Djamgaryan, S. A.; Bakhmutov, V. I.; Vitt, S. V.; Batsanov, A. S.; Struchkov, Y. T.; Belikov, V. M. , General method for the asymmetric synthesis of anti-diastereoisomers of β-substituted L-2-aminobutanoic acids via chiral nickel (II) Schiff's base complexes of dehydroaminobutanoic acid. X-Ray crystal and molecular structure of the nickel (II) complex of the Schiff's base from [(benzylprolyl) amino] benzophenone and dehydroaminobutanoic acid. Journal of the Chemical Society, Perkin Transactions 1 1990, 2301–2310. [Google Scholar]
- Abriata, L. A. , A simple spreadsheet program to simulate and analyze the far-UV circular dichroism spectra of proteins. Journal of Chemical Education 2011, 88, 1268–1273. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.-Y. , HDOCK: a web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic acids research 2017, 45, W365–W373. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. , The HDOCK server for integrated protein–protein docking. Nature protocols 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Greco, F.; Falanga, A. P.; Terracciano, M.; D’Ambrosio, C.; Piccialli, G.; Oliviero, G.; Roviello, G. N.; Borbone, N. , CD, UV, and In Silico Insights on the Effect of 1, 3-Bis (1′-uracilyl)-2-propanone on Serum Albumin Structure. Biomolecules 2022, 12, 1071. [Google Scholar] [CrossRef]
- Yin, Y.-w.; Sheng, Y.-j.; Wang, M.; Ma, Y.-q.; Ding, H.-m. , Interaction of serum proteins with SARS-CoV-2 RBD. Nanoscale 2021, 13, 12865–12873. [Google Scholar] [CrossRef]
- Stoddard, S. V.; Wallace, F. E.; Stoddard, S. D.; Cheng, Q.; Acosta, D.; Barzani, S.; Bobay, M.; Briant, J.; Cisneros, C.; Feinstein, S. , In silico design of peptide-based SARS-CoV-2 fusion inhibitors that target wt and mutant versions of SARS-CoV-2 HR1 Domains. Biophysica 2021, 1, 311–327. [Google Scholar] [CrossRef]
- Pawar, S. S.; Rohane, S. H., Review on discovery studio: An important tool for molecular docking. 2021.
- Yang, Z.-P.; Freas, D. J.; Fu, G. C. , Asymmetric synthesis of protected unnatural α-amino acids via enantioconvergent nickel-catalyzed cross-coupling. Journal of the American Chemical Society 2021, 143, 8614–8618. [Google Scholar] [CrossRef] [PubMed]
- Najera, C.; Sansano, J. M. , Catalytic asymmetric synthesis of α-amino acids. Chemical reviews 2007, 107, 4584–4671. [Google Scholar] [CrossRef] [PubMed]
- Fik-Jaskółka, M. A.; Mkrtchyan, A. F.; Saghyan, A. S.; Palumbo, R.; Belter, A.; Hayriyan, L. A.; Simonyan, H.; Roviello, V.; Roviello, G. N. , Spectroscopic and SEM evidences for G4-DNA binding by a synthetic alkyne-containing amino acid with anticancer activity. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 2020, 229, 117884. [Google Scholar] [CrossRef] [PubMed]
- Fik-Jaskółka, M. A.; Mkrtchyan, A. F.; Saghyan, A. S.; Palumbo, R.; Belter, A.; Hayriyan, L. A.; Simonyan, H.; Roviello, V.; Roviello, G. N. , Biological macromolecule binding and anticancer activity of synthetic alkyne-containing l-phenylalanine derivatives. Amino Acids 2020, 52, 755–769. [Google Scholar] [CrossRef]
- Belokon, Y. N.; Sagyan, A. S.; Djamgaryan, S. M.; Bakhmutov, V. I.; Belikov, V. M. , Asymmetric synthesis of β-substituted α-amino acids via a chiral nlii complex of dehydroalanine. Tetrahedron 1988, 44, 5507–5514. [Google Scholar] [CrossRef]
- Bera, S.; Xue, B.; Rehak, P.; Jacoby, G.; Ji, W.; Shimon, L. J.; Beck, R.; Kral, P.; Cao, Y.; Gazit, E. , Self-assembly of aromatic amino acid enantiomers into supramolecular materials of high rigidity. ACS nano 2020, 14, 1694–1706. [Google Scholar] [CrossRef] [PubMed]
- Haskins, N.; Spease, L.; Ucheena Woodfolk, A.; Davenport, J.; Rhinehardt PhD, K., Molecular Docking of Nanoscale Collagen Mimetic Peptides. 2023.
- Chen, S.; Zhang, G.; Liu, Y.; Yang, C.; He, Y.; Guo, Q.; Du, Y.; Gao, F. , Anchoring of hyaluronan glycocalyx to CD44 reduces sensitivity of HER2-positive gastric cancer cells to trastuzumab. The FEBS Journal 2024. [Google Scholar] [CrossRef]
- Suleman, M.; Khattak, A.; Akbar, F.; Rizwan, M.; Tayyab, M.; Yousaf, M.; Khan, A.; Albekairi, N. A.; Agouni, A.; Crovella, S. , Analysis of E2F1 single-nucleotide polymorphisms reveals deleterious non-synonymous substitutions that disrupt E2F1-RB protein interaction in cancer. International Journal of Biological Macromolecules 2024, 260, 129559. [Google Scholar] [CrossRef]
- Jitaru, S.-C.; Enache, A.-C.; Cojocaru, C.; Drochioiu, G.; Petre, B.-A.; Gradinaru, V.-R. , Self-Assembly of a Novel Pentapeptide into Hydrogelated Dendritic Architecture: Synthesis, Properties, Molecular Docking and Prospective Applications. Gels 2024, 10. [Google Scholar] [CrossRef] [PubMed]
- Mosseri, A.; Sancho-Albero, M.; Mercurio, F. A.; Leone, M.; De Cola, L.; Romanelli, A. , Tryptophan-PNA gc Conjugates Self-Assemble to Form Fibers. Bioconjugate Chemistry 2023, 34, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Hanafiah, A.; Abd Aziz, S. N. A.; Nesran, Z. N. M.; Wezen, X. C.; Ahmad, M. F. , Molecular investigation of antimicrobial peptides against Helicobacter pylori proteins using a peptide-protein docking approach. Heliyon 2024, 10. [Google Scholar] [CrossRef] [PubMed]
- Suganthi, M.; Sowmya, H.; Manjunathan, J.; Ramasamy, P.; Thiruvengadam, M.; Varadharajan, V.; Venkidasamy, B.; Senthilkumar, P. , Homology modeling and protein-protein interaction studies of GAPDH from Helopeltis theivora and chitinase from Pseudomonas fluorescens to control infection in tea [Camellia sinensis (L.) O. Kuntze] plants. Plant Stress 2024, 11, 100377. [Google Scholar] [CrossRef]
- Chakraborty, C.; Mallick, B.; Bhattacharya, M.; Byrareddy, S. N. , SARS-CoV-2 Omicron Spike shows strong binding affinity and favourable interaction landscape with the TLR4/MD2 compared to other variants. Journal of Genetic Engineering and Biotechnology 2024, 22, 100347. [Google Scholar] [CrossRef] [PubMed]
- De Vries, S. J.; Van Dijk, M.; Bonvin, A. M. , The HADDOCK web server for data-driven biomolecular docking. Nature protocols 2010, 5, 883–897. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.; Boelens, R.; Bonvin, A. M. , HADDOCK: a protein− protein docking approach based on biochemical or biophysical information. Journal of the American Chemical Society 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liang, Y.; Guo, B.; Yin, Z.; Zhu, D.; Han, Y. , Injectable dry cryogels with excellent blood-sucking expansion and blood clotting to cease hemorrhage for lethal deep-wounds, coagulopathy and tissue regeneration. Chemical Engineering Journal 2021, 403, 126329. [Google Scholar] [CrossRef]
- Solanki, R.; Rostamabadi, H.; Patel, S.; Jafari, S. M. , Anticancer nano-delivery systems based on bovine serum albumin nanoparticles: A critical review. International Journal of Biological Macromolecules 2021, 193, 528–540. [Google Scholar] [CrossRef]
- Zarrilli, F.; Amato, F.; Morgillo, C. M.; Pinto, B.; Santarpia, G.; Borbone, N.; D’Errico, S.; Catalanotti, B.; Piccialli, G.; Castaldo, G.; Oliviero, G. , Peptide Nucleic Acids as miRNA Target Protectors for the Treatment of Cystic Fibrosis. Molecules 2017, 22, 1144. [Google Scholar] [CrossRef]
- Formen, J. S.; Howard, J. R.; Anslyn, E. V.; Wolf, C. , Circular Dichroism Sensing: Strategies and Applications. Angewandte Chemie International Edition 2024, e202400767. [Google Scholar]
- Pirota, V.; Platella, C.; Musumeci, D.; Benassi, A.; Amato, J.; Pagano, B.; Colombo, G.; Freccero, M.; Doria, F.; Montesarchio, D. , On the binding of naphthalene diimides to a human telomeric G-quadruplex multimer model. International Journal of Biological Macromolecules 2021, 166, 1320–1334. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, Y.; Zhao, D. , Structural analysis of biomacromolecules using circular dichroism spectroscopy. Advanced Spectroscopic Methods to Study Biomolecular Structure and Dynamics 2023, 77–103. [Google Scholar]
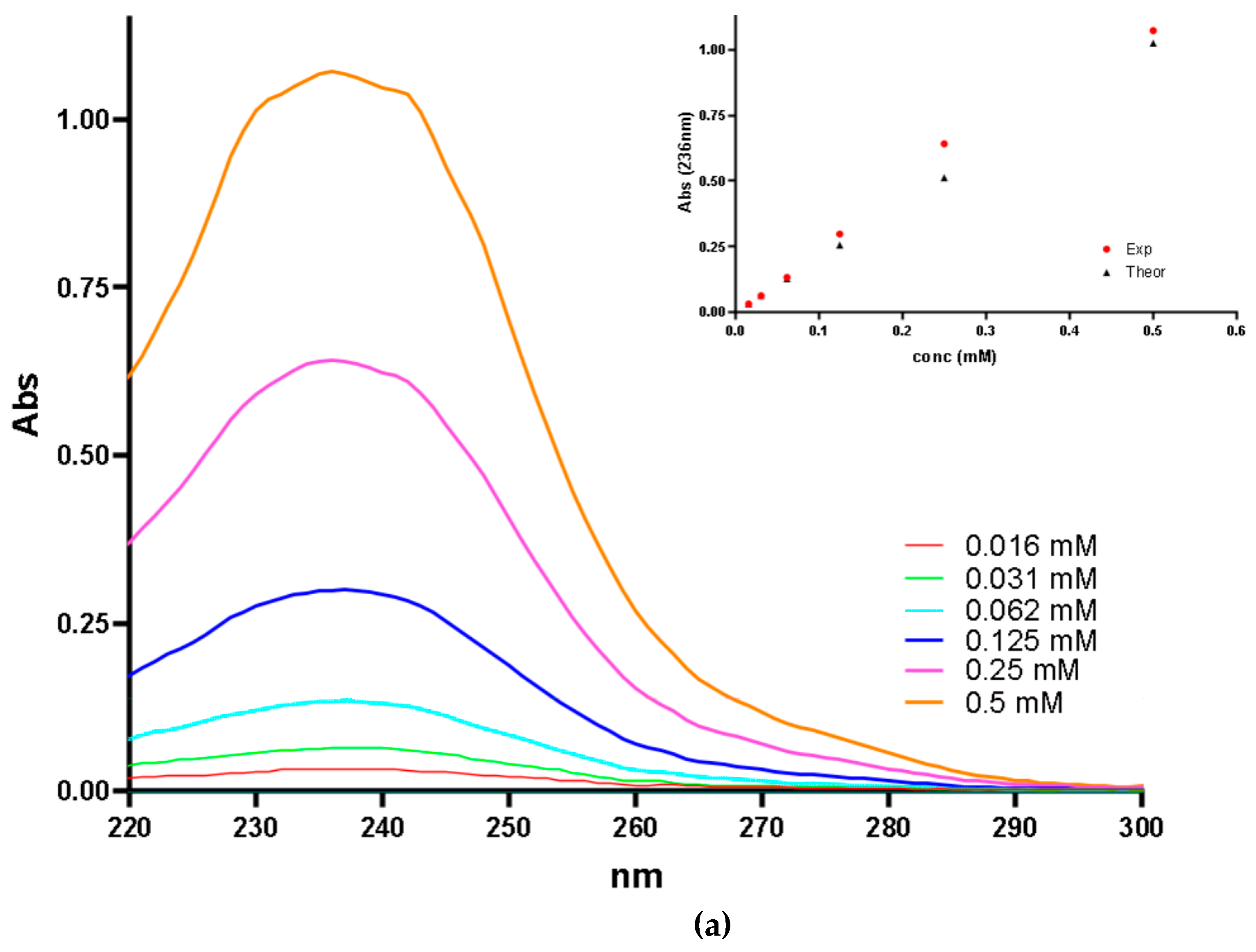
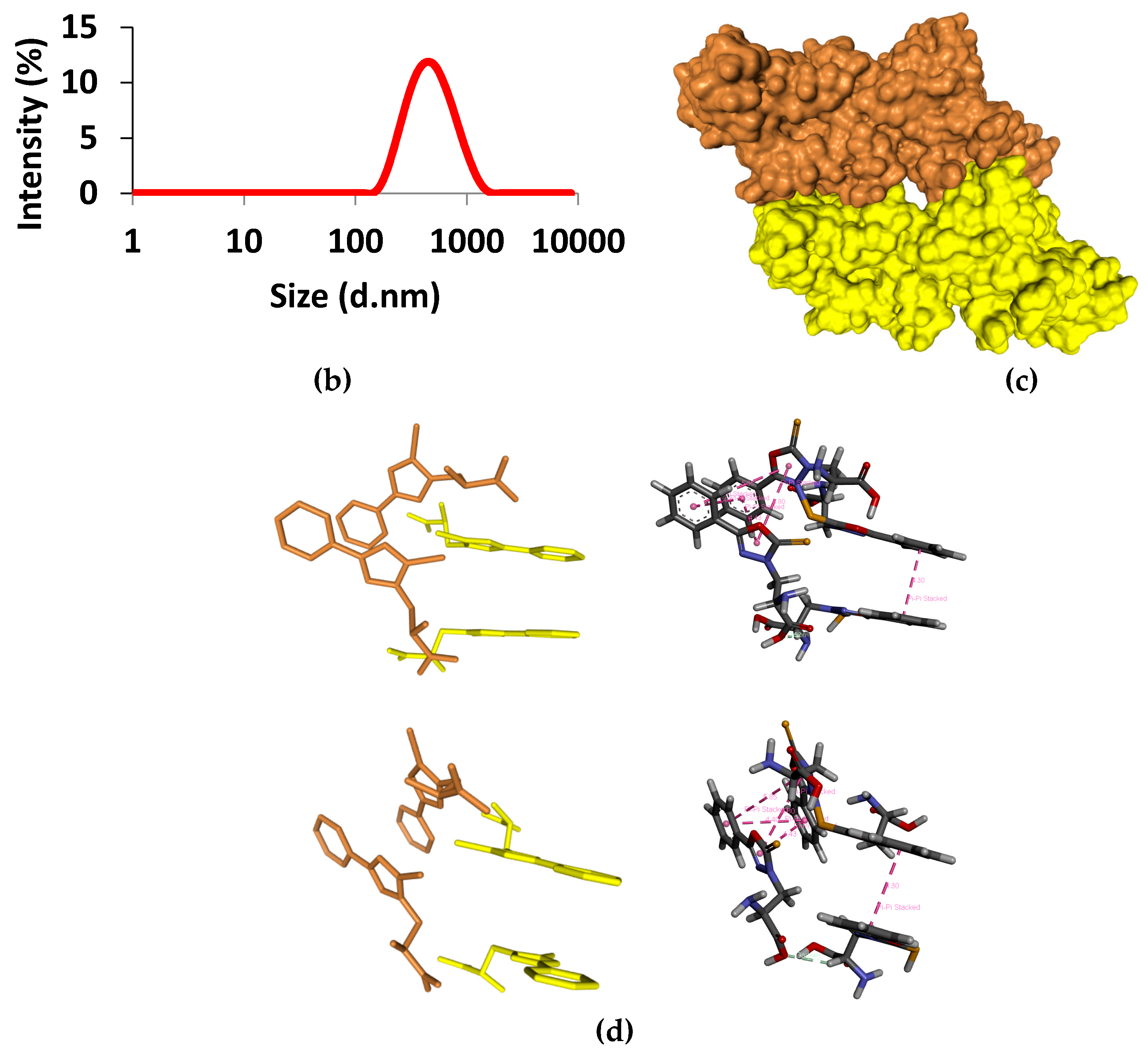
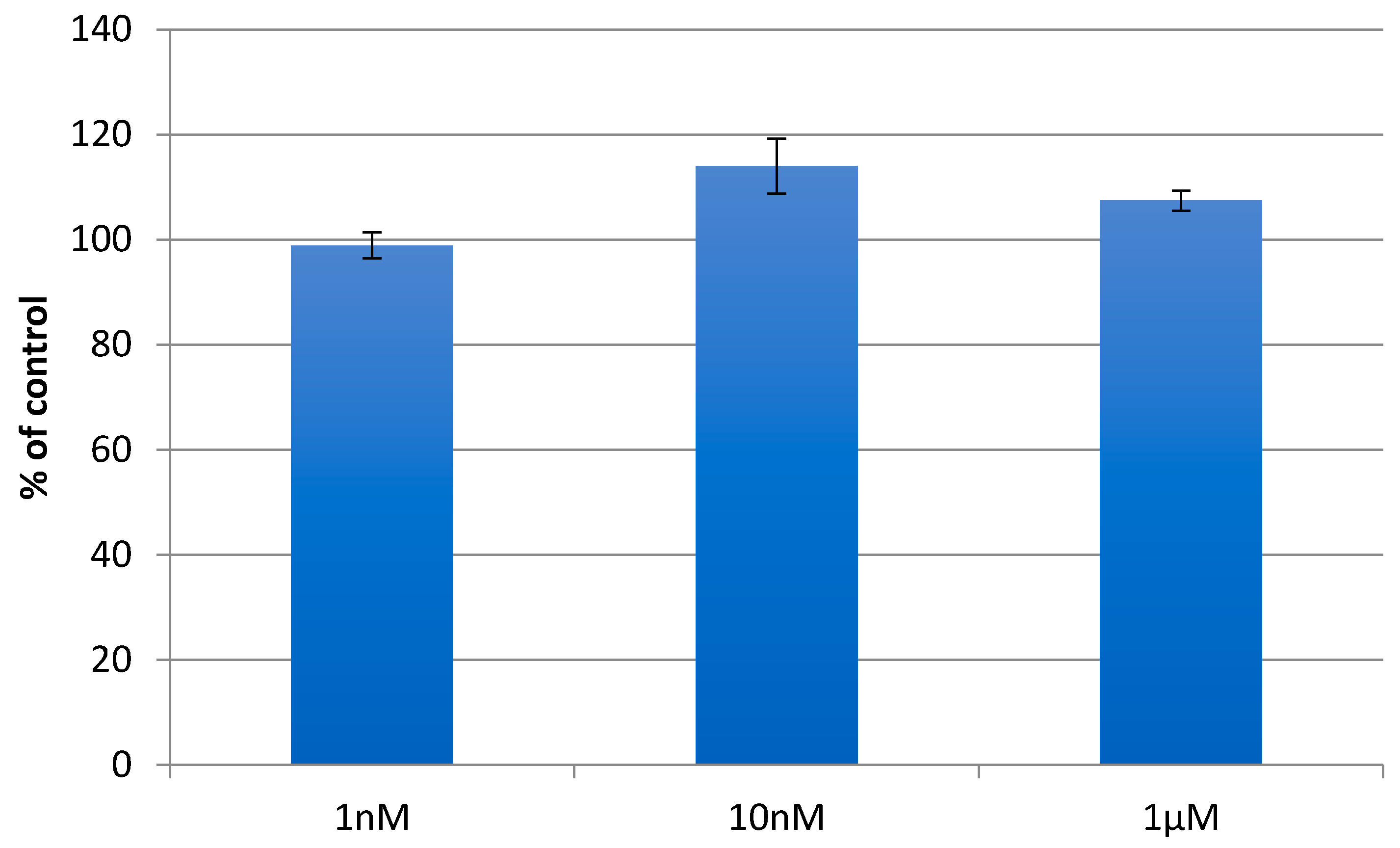
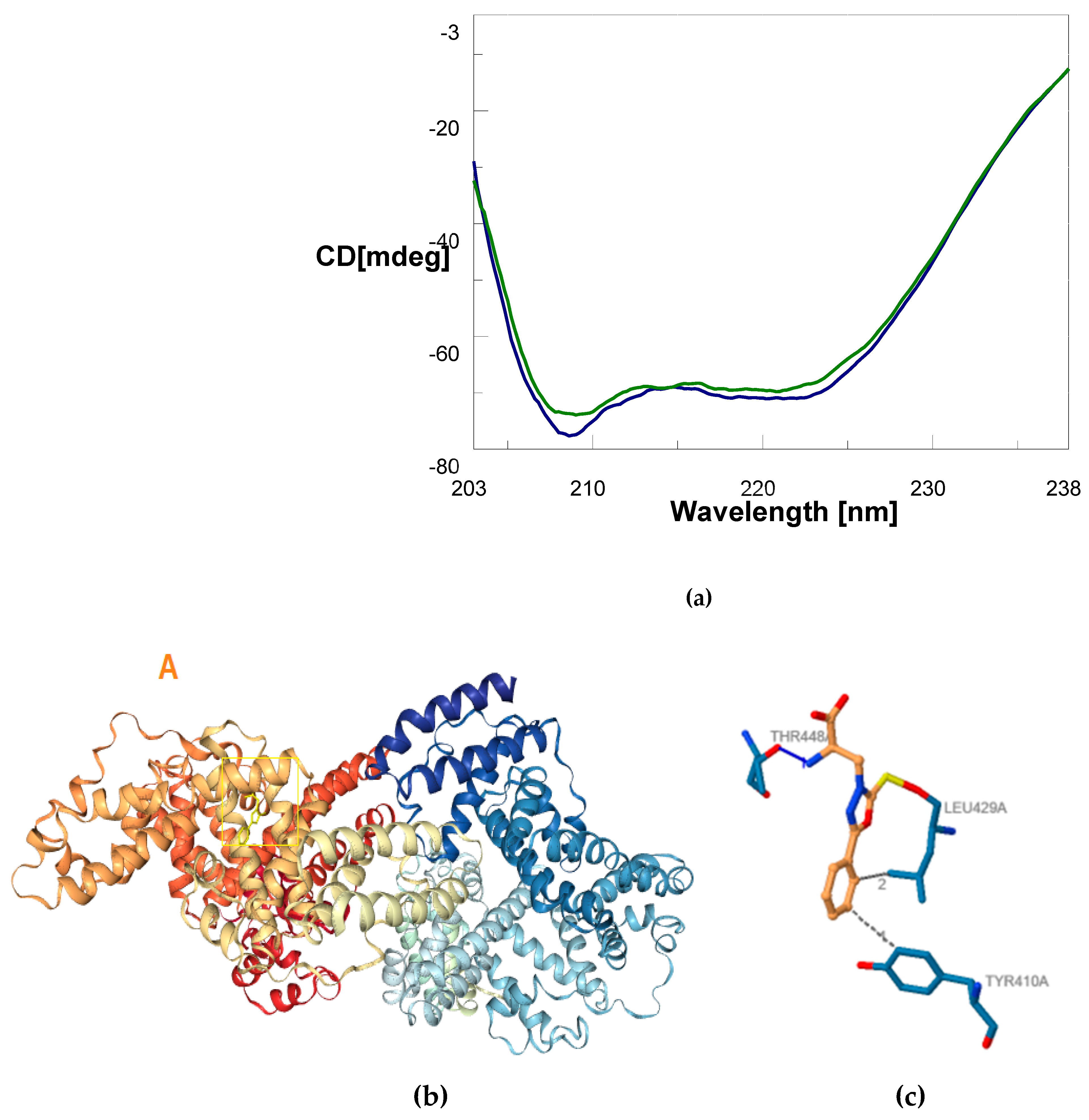
| Secondary Structure | Δ(x-BSA) 1 (%) | change (%) |
|---|---|---|
| α-helix | 62.2 - 69.1 | -6.9 |
| β-sheet | 10.5 - 5.8 | +4.7 |
| Random coil | 27.3 - 25.1 | +2.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





