Submitted:
18 October 2023
Posted:
20 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1 Plant material and sample preparation
2.2 Determination of methylxanthines
2.3 Determination of total phenolic compounds
2.4 Determination of moisture and proteins
2.5 Statistical analysis
3. Results
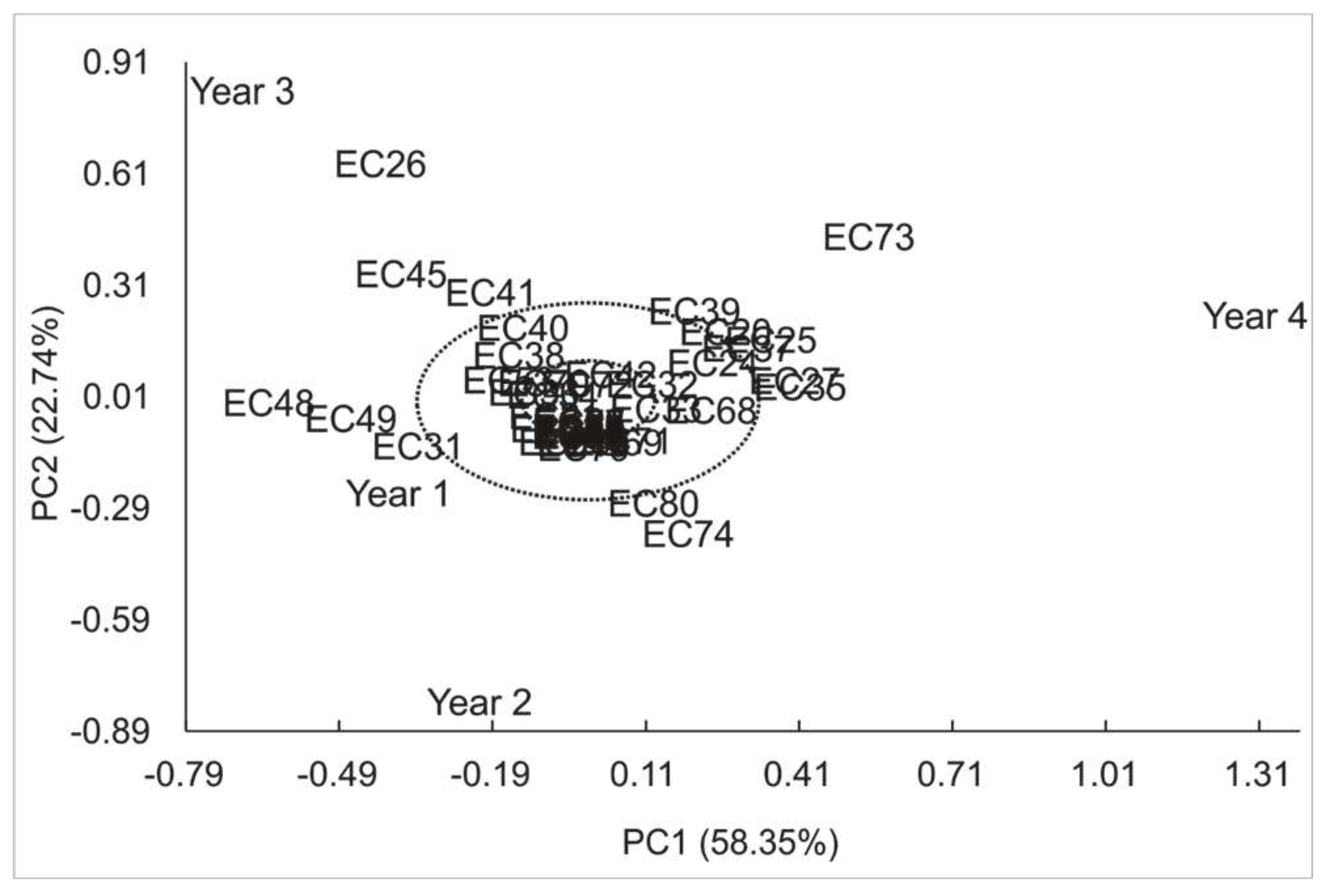
3.1 Caffeine
| Variation sources | GL | SQ | F (Pr>F) | |
| Model | 56 | 119.84 | ||
| Genotype (G) | (53) | (119.03) | 37.87 (<0.0001) | |
| Year (A) | (3) | (0.81) | 4.57 (0.0042) | |
| High Variance (%) Axes Value Explained Cumulative |
GxA | 159 | 9.43 | |
| 1 5.50 58.35 58.35 | IPCA1 | (55) | (5.50) | 3.04(<0.0001) |
| 2 2.14 22.74 81.10 | IPCA2 | (53) | (2.15) | 1.23 (0.1491) |
| 3 1.78 18.90 100.00 | IPCA3 | (51) | (1.78) | 1.06 (0.3731) |
| Mean Error | 290 | 9.57 | ||
| Adjusted Total | 215 | 129.27 |
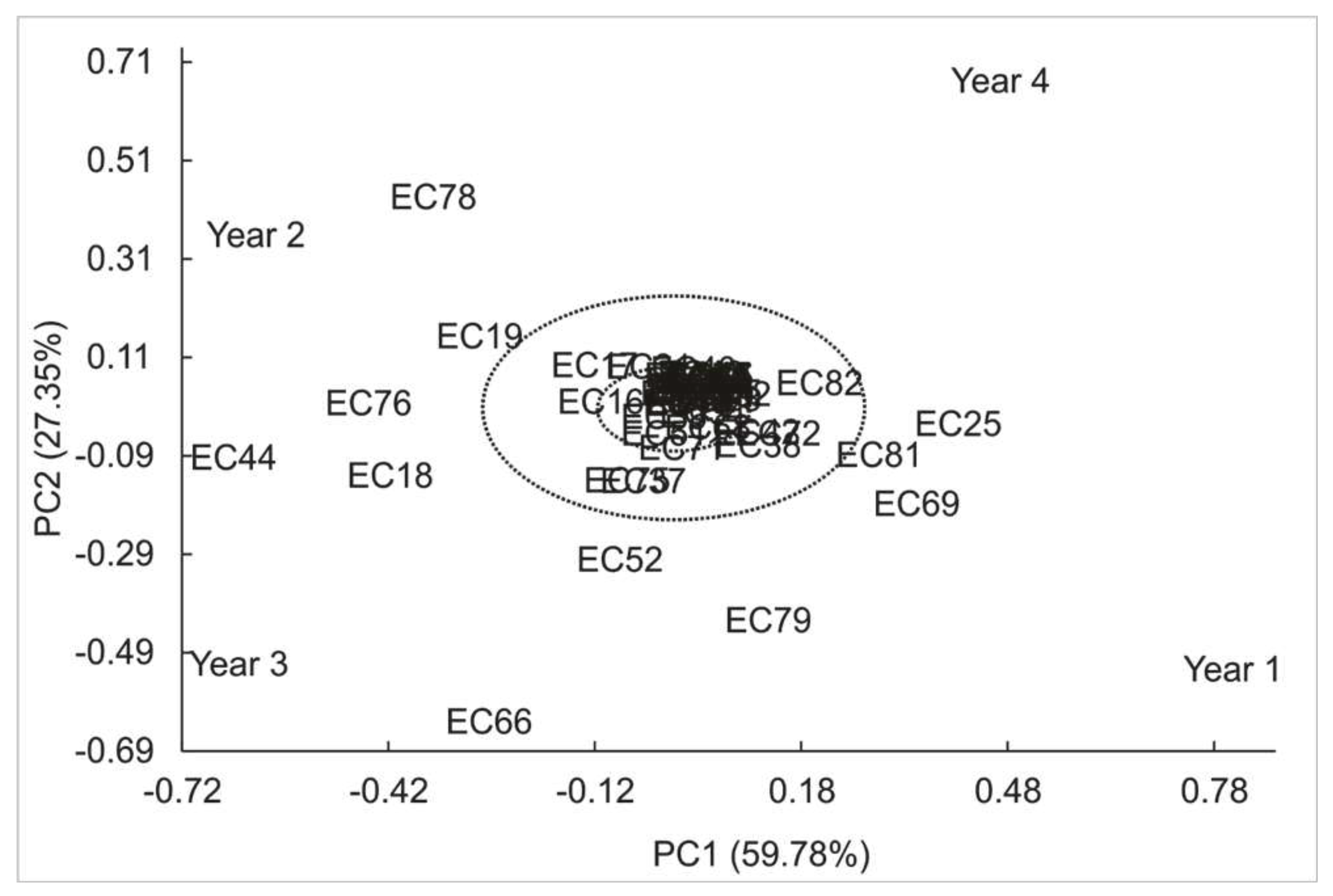
3.2 Theobromine
| Variation sources | GL | SQ | F (Pr>F) | |
| Model | 57 | 19.86 | ||
| Genotype (G) | (54) | (19.38) | 12.71 (<0.0001) | |
| Year (A) | (3) | (0.48) | 5.70 (0.0010) | |
| High Variance (%) Axes Value Explained Cumulative |
GxA | 162 | 4.57 | |
| 1 2.73 59.78 59.78 | IPCA1 | (56) | (2.73) | 3.11 (<0.0001) |
| 2 1.25 27.35 87.13 | IPCA2 | (54) | (1.25) | 1.47 (0.0238) |
| 3 0.59 12.87 100.00 | IPCA3 | (52) | (0.59) | 0.72 (0.9237) |
| Mean Error | 295 | 4.63 | ||
| Adjusted Total | 219 | 24.43 |
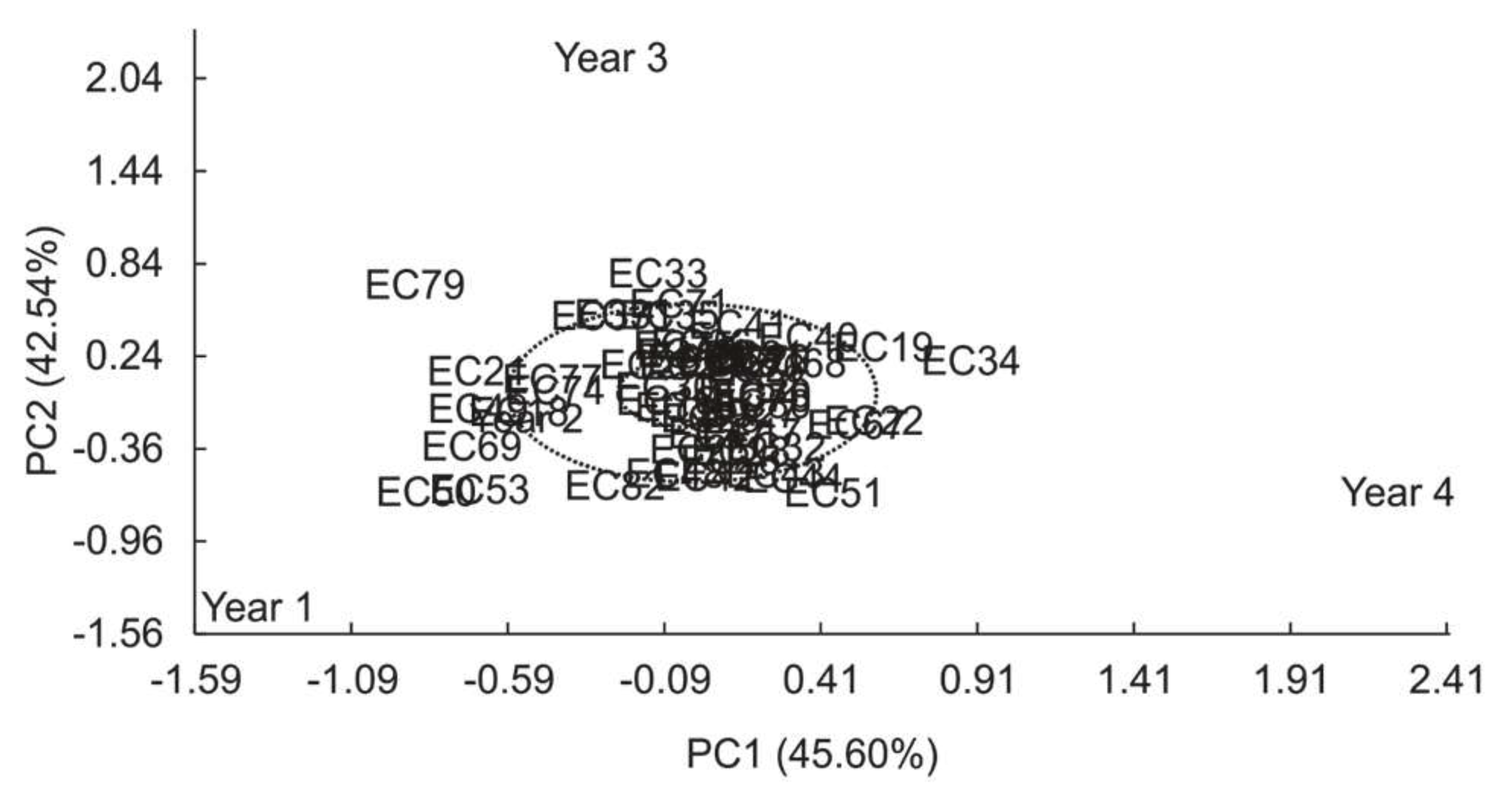
3.3 Total phenolic compounds
| Variation sources | GL | SQ | F (Pr>F) | |
| Model | 57 | 95.58 | ||
| Genotype (G) | (54) | (54.76) | 1.38 (0.0642) | |
| Year (A) | (3) | (40.82) | 18.50 (<0.0001) | |
| High Variance (%) Axes Value Explained Cumulative |
GxA | 162 | 118.50 | |
| 1 54.03 45.60 45.60 | IPCA1 | (56) | (54.03) | 2.37 (<0.0001) |
| 2 50.41 42.54 88.13 | IPCA2 | (54) | (50.41) | 2.29 (<0.0001) |
| 3 14,06 11.87 100.00 | IPCA3 | (52) | (14.06) | 0.66 (0.9625) |
| Mean Error | 295 | 120.12 | ||
| Adjusted Total | 219 | 214.08 |
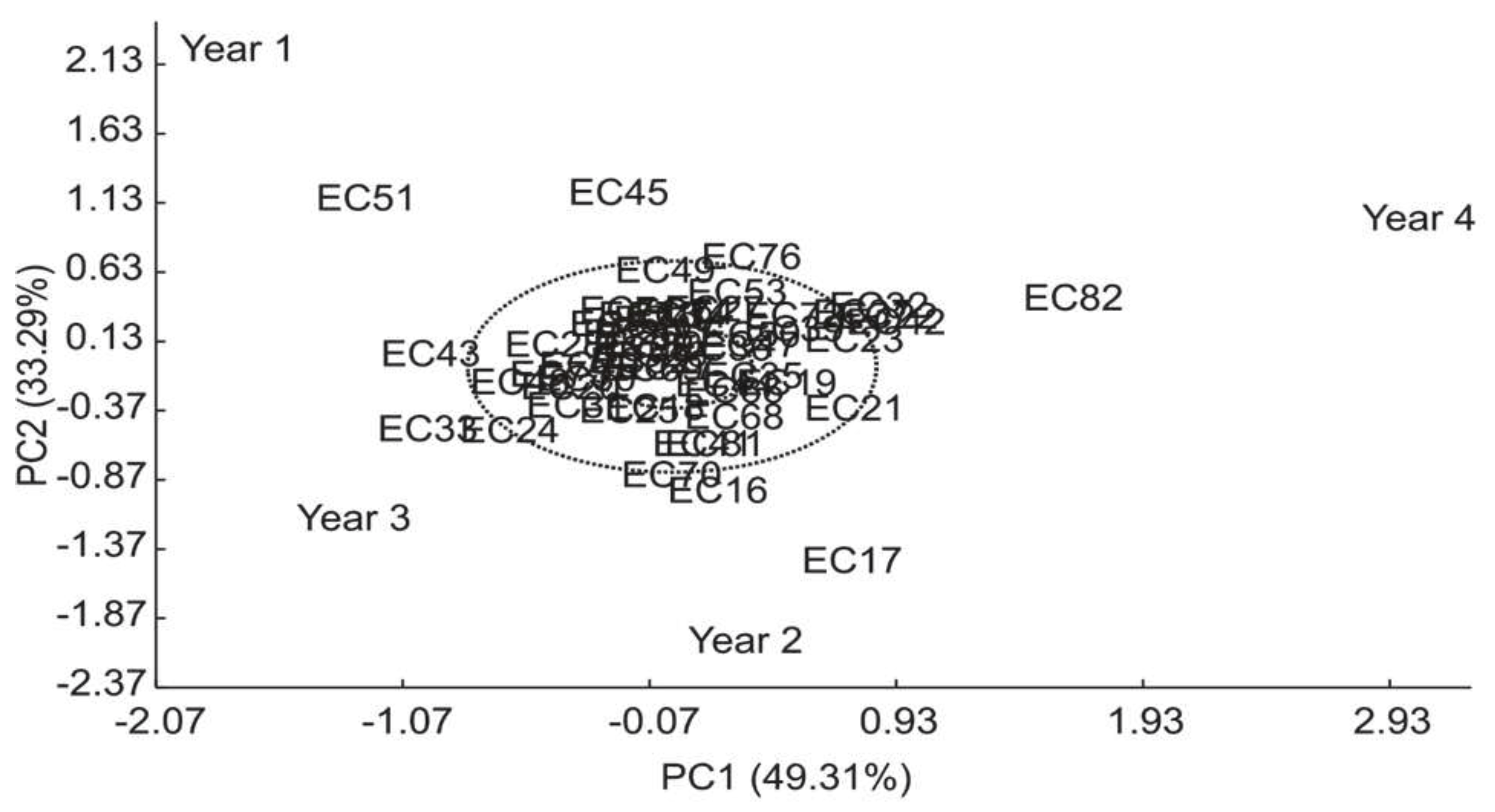
3.4. Proteins
| Variation sources | GL | SQ | F (Pr>F) | |
| Model | 56 | 776.84 | ||
| Genotype (G) | (53) | (354.64) | 2.68 (<0.0001) | |
| Year (A) | (3) | (422.20) | 56.31 (<0.0001) | |
| High Variance (%) Axes Value Explained Cumulative |
GxA | 159 | 397.41 | |
| 1 195.96 49.31 49.31 | IPCA1 | (55) | (195.96) | 3.04(<0.0001) |
| 2 132.31 33.29 82.60 | IPCA2 | (53) | (132.31) | 1.23 (0.1491) |
| 3 69.14 17.40 100.00 | IPCA3 | (51) | (69.14) | 1.06 (0.3731) |
| Mean Error | 290 | 402.90 | ||
| Adjusted Total | 215 | 1174.25 |
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics approval
Consent for publication
References
- Heck CI, de Mejia EG (2007) Yerba mate tea (Ilex paraguariensis): A comprehensive review on chemistry, health implications, and technological considerations. J Food Sci 72(9):138-51. [CrossRef]
- Gan RY, Zhang D, Wang M, Corke H (2018) Health benefits of bioactive compounds from the genus ilex, a source of traditional caffeinated beverages. Nutrients 10(11):1682. [CrossRef]
- De Godoy RCB, Chambers E, Yang G (2020) Development of a preliminary sensory lexicon for mate tea. J Sens Stud 35(3). [CrossRef]
- Cardozo Junior EL, Morand C (2016a) Interest of mate (Ilex paraguariensis A. St.-Hil.) as a new natural functional food to preserve human cardiovascular health - A review. J Funct Foods 21:440–454. [CrossRef]
- Wendling I, Sturion JA, Stuepp CA, et al (2018) Early selection and classification of yerba mate progenies. Pesq agropec bras 53(03). [CrossRef]
- Sturion JA, De Resende MDV (2010) Melhoramento genético da erva-mate, 1a. Embrapa Florestas, Colombo.
- Anesini C, Turner S, Cogoi L, Filip R (2012) Study of the participation of caffeine and polyphenols on the overall antioxidant activity of mate (Ilex paraguariensis). LWT - Food Sci. Technol 45:299–304. [CrossRef]
- Debat HJ, Grabiele M, Aguilera PM et al (2014) Exploring the genes of yerba mate (Ilex paraguariensis A. St.-Hil.) by NGS and de novo transcriptome assembly. PLOS ONE 9(10): e109835. [CrossRef]
- González Arbeláez LF, Fantinelli JC, Ciocci Pardo A et al (2016) Effect of an Ilex paraguariensis (yerba mate) extract on infarct size in isolated rat hearts: the mechanisms involved. Food & Function 7:816–824. [CrossRef]
- De Lima GG, Ruiz HZ, Matos M et al (2019) Prediction of yerba mate caffeine content using near infrared spectroscopy. Spectrosc Lett 52:282–287. [CrossRef]
- Poswal FS, Russell G, Mackonochie M et al (2019) Herbal Teas and their Health Benefits: A Scoping Review. Plant Foods Hum Nutr 74(3):266-276. [CrossRef]
- Oellig C, Schunck J, Schwack W (2018) Determination of caffeine, theobromine and theophylline in Mate beer and Mate soft drinks by high-performance thin-layer chromatography. J Chromatogr A 1533:208–212. [CrossRef]
- Gullón B, Eibes G, Moreira MT et al (2018) Yerba mate waste: A sustainable resource of antioxidant compounds. Ind Crop Prod 113:398–405. [CrossRef]
- Butiuk AP, Martos MA, Adachi O, Hours RA (2016) Study of the chlorogenic acid content in yerba mate (Ilex paraguariensis St. Hil.): Effect of plant fraction, processing step and harvesting season. J Appl Res Med Aromat Plants 3:27–33. [CrossRef]
- Gil M, Wianowska D (2017) Chlorogenic acids – their properties, occurrence and analysis. Annales Universitatis Mariae Curie-Sklodowska, sectio AA – Chemia 72:61. [CrossRef]
- Kaltbach P, Ballert S, Kabrodt K, Schellenberg I (2020) New HPTLC methods for analysis of major bioactive compounds in mate (Ilex paraguariensis) tea. J Food Compos Anal 92:103568. [CrossRef]
- Duarte MM, Gabira MM. Tomasi JC, Amano E, Nogueira AC, Wendling I. Bioactive compounds and leaf anatomy of yerba mate morphotypes. Pesquisa Agropecuária Brasileira, v.57, e02441, 2022.
- Helm CV, Ruiz HZ, Hansel FA et al (2015) Efeito do solvente na extração de teobromina e cafeína em progênies de erva-mate. Embrapa Florestas. https://www.embrapa.br/busca-de-publicacoes/-/publicacao/1038782/efeito-do-solvente-na-extracao-de-teobromina-e-cafeina-em-progenies-de-erva-mate. Accessed. 26 May.
- Mandel J (1971) American Society for Quality A New Analysis of Variance Model for Non-Additive Data A New Analysis of Variance Model for Non-additive Data. Technometrics 13(1):1-18.
- Gabriel KR (1971) Biometrika Trust The Biplot Graphic Display of Matrices with Application to Principal Component Analysis. Biometrika 58(3):453–467.
- Durte, J.B.; Vencovsky, R. Interação genótipo x ambiente: uma introdução a análise “AMMI”. Ribeirão Preto: Sociedade Brasileira de Genética, 199. 60p. (Série Monograficas, 9).
- Kassambara, A. Practical Guide to Cluster Analysis in R. 1 ed. USA: STHDA. 2017.
- Cornrlius, P.L.; Seyedsadr, M.; Crossa, J. Using the shifted multiplicative model to search for “separability” in crop cultivar trials. Theoretical and Applied Genetics, v.84, n.1-2, p.161-172, 1992.
- Brasil (2005) Resolução de Diretoria Colegiada-RDC No. 277, de 22 de Setembro de 2005.
- Schuhli GS, Penteado Junior JF, Wendling I (2019) Descritores mínimos em cultivares de espécies florestais: uma contribuição para erva-mate. Embrapa Florestas. http://www.infoteca.cnptia.embrapa.br/infoteca/handle/doc/1117327. Accessed 26 2. 20 May.
- Heckman MA, Weil J, de Mejia EG (2010) Caffeine (1, 3, 7-trimethylxanthine) in foods: A comprehensive review on consumption, functionality, safety, and regulatory matters. J Food Sci 75(3):77-87. [CrossRef]
- Athayde ML, Coelho GC, Schenkel P (2000) Caffeine and theobromine in epicuticular wax of Ilex paraguariensis A. St.-Hil. Phytochemistry 55(7):853-7. [CrossRef]
- Mohanpuria P, Kumar V, Yadav SK (2010) Tea Caffeine Metabolism, Functions, and Reduction Strategies. Food Sci Biotechnol 19:275–287. [CrossRef]
- Scherer R, Urfer P, Mayol MR et al (2002) Inheritance studies of caffeine and theobromine content of Mate (Ilex paraguariensis) in Misiones, Argentina. Euphytica 126, 203–210. [CrossRef]
- Cardozo Junior EL, Morand C (2016b) Interest of mate (Ilex paraguariensis A. St.-Hil.) as a new natural functional food to preserve human cardiovascular health - A review. J Funct Foods 21:440–454. [CrossRef]
- Cardozo EL, Ferrarese-Filho O, Filho LC et al (2007) Methylxanthines and phenolic compounds in mate (Ilex paraguariensis St. Hil.) progenies grown in Brazil. J Food Compos Anal 20:553–558. [CrossRef]
- Tfouni SAV, Camara MM, Kamikata K et al (2018) Caffeine in teas: Levels, transference to infusion and estimated intake. Food Sci. Technol 38:661–666. [CrossRef]
- Nakamura KL, Cardozo Junior L, Donaduzzi CM, Schuster I (2009) Brazilian Society of Plant Breeding. Printed in Brazil Genetic variation of phytochemical compounds in progenies of Ilex paraguariensis St. Hil. CBAB 9(2). [CrossRef]
- Cardozo Junior EL, Maria Donaduzzi C, Ferrarese-Filho O et al (2010) Quantitative genetic analysis of methylxanthines and phenolic compounds in mate progenies. Pesq agropec bras 45 (2). [CrossRef]
- Zhu B, Chen LB, Lu M, et al (2019) Caffeine Content and Related Gene Expression: Novel Insight into Caffeine Metabolism in Camellia Plants Containing Low, Normal, and High Caffeine Concentrations. J Agric Food Chem 67:3400–3411. [CrossRef]
- Ashihara H, Monteiro AM, Gillies FM et al (1996) Biosynthesis of Caffeine in Leaves of Coffee. Plant Physiol 111(3): 747–753. [CrossRef]
- Da Croce DM (2002) Características físico-químicas de extratos de erva-mate (Ilex paraguariensis St. Hil) no estado de Santa Catarina. Ciênc Florest 12 (2). [CrossRef]
- Duarte MM, de Cássia Tomasi J, Helm CV et al (2020) Caffeinated and decaffeinated mate tea: Effect of toasting on bioactive compounds and consumer acceptance. Rev Bras Ciênc Agrár 15(3):e8513. [CrossRef]
- Gómez-Juaristi M, Martínez-López S, Sarria B et al (2018) Absorption and metabolism of yerba mate phenolic compounds in humans. Food Chemistry 240:1028–1038. [CrossRef]
- Gugliucci A, Bastos DHM (2009) Chlorogenic acid protects paraoxonase 1 activity in high density lipoprotein from inactivation caused by physiological concentrations of hypochlorite. Fitoterapia 80:138–142. [CrossRef]
- Valerga J, Reta M, Lanari MC (2012) Polyphenol input to the antioxidant activity of yerba mate (Ilex paraguariensis) extracts. Food Sci Technol 45:28–35. [CrossRef]
- Sturion JA, Correa G, Resende MDV et al (2004) Controle genético dos teores de polifenóis totais, taninos e cafeína em progênies de erva-mate (Ilex paraguariensis St. Hil.) cultivadas em três classes de solos. Boletim de Pesquisa e Desenvolvimento (Embrapa Florestas). https://www.embrapa.br/busca-de-publicacoes/-/publicacao/287300/controle-genetico-dos-teores-de-polifenois-totais-taninos-e-cafeina-em-progenies-de-erva-mate-ilex-paraguariensis-st-hil-cultivadas-em-tres-classes-de-solos. 26 May.
- Da Silveira TFF, Meinhart AD, de Souza TCL et al (2016) Phenolic compounds from yerba mate based beverages - A multivariate optimization. Food Chemistry 190:1159–1167. [CrossRef]
- Berté KAS, Beux MR, Spada PKWDS et al (2011) Chemical composition and antioxidant activity of yerba-mate (Ilex paraguariensis A.St.-Hil., Aquifoliaceae) extract as obtained by spray drying. J Agric Food Chem 59:5523–5527. [CrossRef]
- Frizon C, Perussello C, Sturion J, Hoffmann-Ribani R (2018) Novel Beverages of Yerba-Mate and Soy: Bioactive Compounds and Functional Properties. Beverages 4:21. [CrossRef]
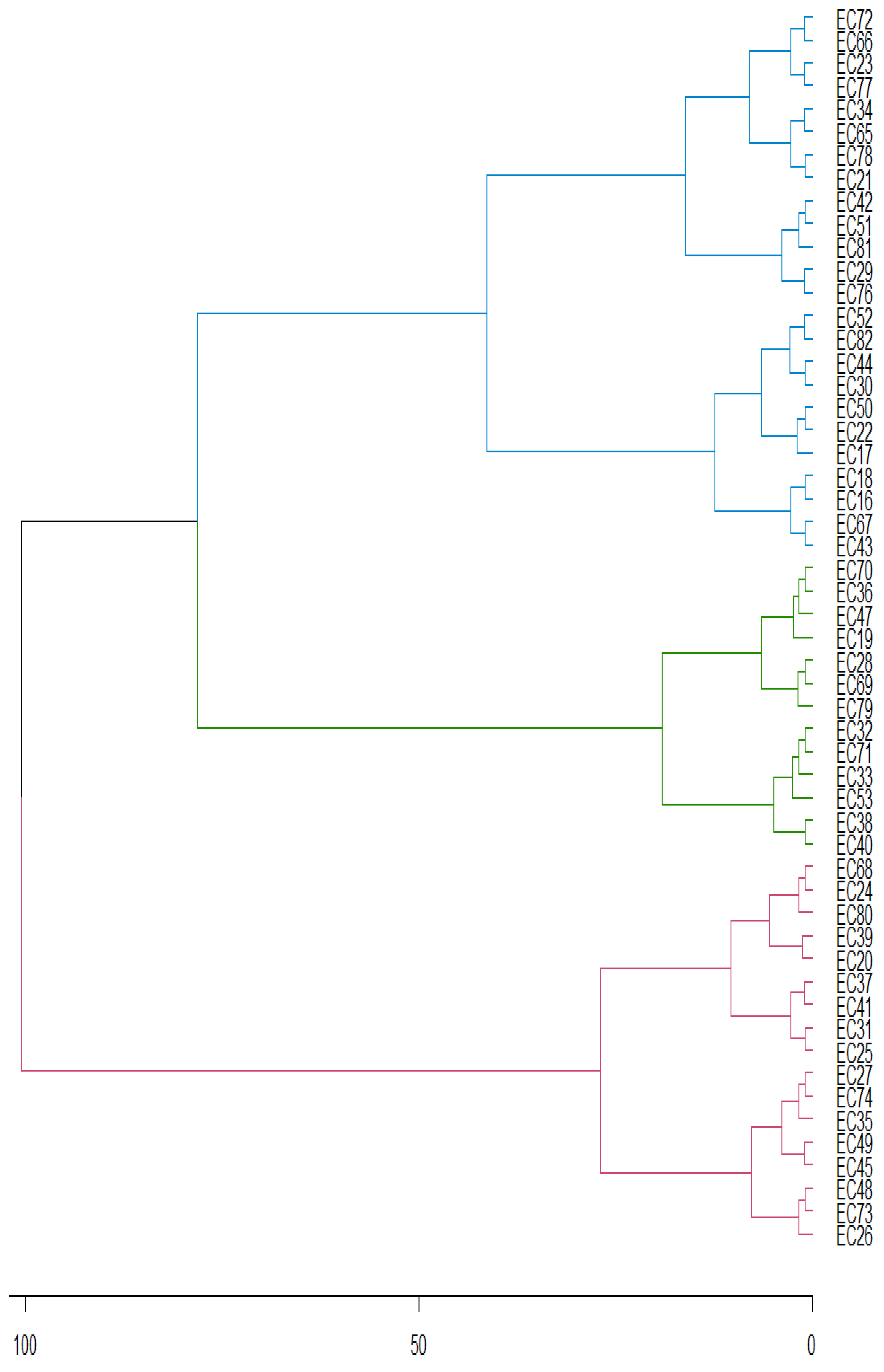
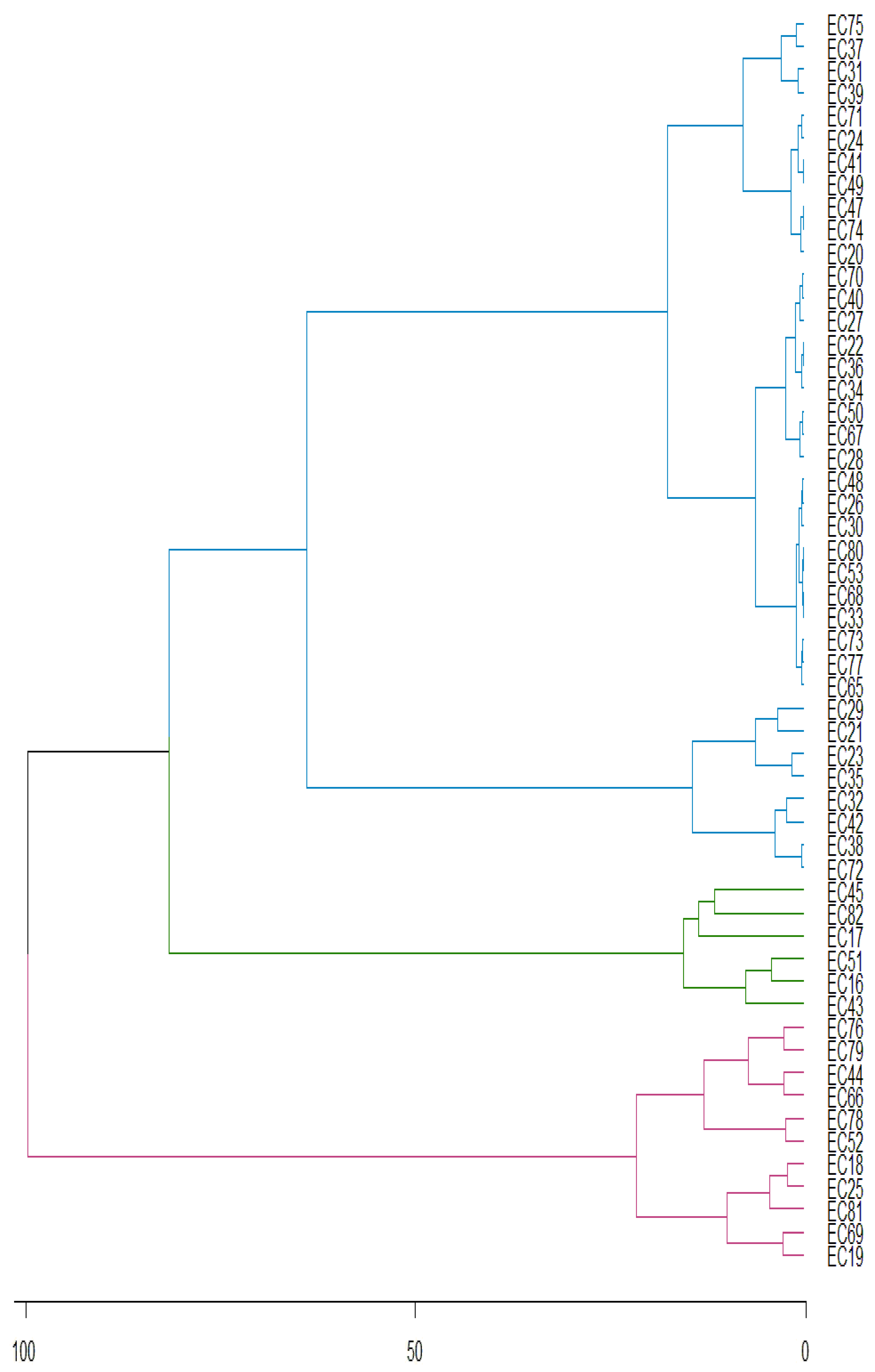
| Genotypes | Means | Chi- | Pr > ChiSq | Genotypes | Means | Chi- | Pr > ChiSq | |
| Square | Square | |||||||
| EC16 | 0.0396 | 0.17 | 0.6780 | EC44 | 0.0413 | 0.05 | 0.8203 | |
| EC17 | 0.0542 | 0.79 | 0.3752 | EC16 | 0.0396 | 0.17 | 0.6780 | |
| EC18 | 0.0674 | 2.71 | 0.0996 | EC44 | 0.0413 | 0.79 | 0.3752 | |
| EC19 | 0.0348 | 1.32 | 0.2501 | EC45 | 2.3290 | 30.50 | <.0001 | |
| EC20 | 0.5961 | 27.19 | <.0001 | EC47 | 0.1208 | 11.35 | 0.0008 | |
| EC21 | 0.4373 | 25.61 | <.0001 | EC48 | 1.7543 | 30.13 | <.0001 | |
| EC22 | 0.0374 | 0.43 | 0.5128 | EC49 | 2.2004 | 30.44 | <.0001 | |
| EC23 | 0.3752 | 24.52 | <.0001 | EC50 | 0.0605 | 1.70 | 0.1925 | |
| EC24 | 1.0741 | 29.17 | <.0001 | EC51 | 0.0839 | 5.47 | 0.0193 | |
| EC25 | 1.5091 | 29.88 | <.0001 | EC52 | 0.0889 | 6.80 | 0.0091 | |
| EC26 | 1.5221 | 29.90 | <.0001 | EC53 | 2.3579 | 30.52 | <.0001 | |
| EC27 | 1.5769 | 29.96 | <.0001 | EC65 | 0.3324 | 23.33 | <.0001 | |
| EC28 | 1.0096 | 27.96 | <.0001 | EC66 | 0.4116 | 25.15 | <.0001 | |
| EC29 | 0.0850 | 5.89 | 0.0152 | EC67 | 0.0479 | 0.15 | 0.7018 | |
| EC30 | 0.0374 | 0.38 | 0.5362 | EC68 | 0.7601 | 28.15 | <.0001 | |
| EC31 | 1.1940 | 29.42 | <.0001 | EC69 | 0.8194 | 28.29 | <.0001 | |
| EC32 | 0.8286 | 28.44 | <.0001 | EC70 | 1.5931 | 29.98 | <.0001 | |
| EC33 | 1.0823 | 29.19 | <.0001 | EC71 | 2.1910 | 30.45 | <.0001 | |
| EC34 | 1.1340 | 29.30 | <.0001 | EC72 | 1.2243 | 29.47 | <.0001 | |
| EC35 | 0.2751 | 22.21 | <.0001 | EC73 | 1.6523 | 30.04 | <.0001 | |
| EC36 | 1.1766 | 29.38 | <.0001 | EC74 | 1.1718 | 29.01 | <.0001 | |
| EC37 | 1.9442 | 30.28 | <.0001 | EC76 | 0.0906 | 7.09 | 0.0078 | |
| EC38 | 1.6107 | 29.99 | <.0001 | EC77 | 0.6200 | 27.19 | <.0001 | |
| EC39 | 0.7686 | 28.19 | <.0001 | EC78 | 0.0799 | 5.00 | 0.0253 | |
| EC40 | 1.7908 | 30.16 | <.0001 | EC79 | 2.3846 | 30.51 | <.0001 | |
| EC41 | 1.4447 | 29.80 | <.0001 | EC80 | 0.7048 | 27.29 | <.0001 | |
| EC42 | 1.1004 | 29.23 | <.0001 | EC81 | 0.0576 | 1.18 | 0.2770 | |
| EC43 | 1.3957 | 29.74 | <.0001 | EC82 | 0.0438 | 2.80 | 0.0946 |
| Genotypes | Means | Chi- | Pr > ChiSq | Genotypes | Means | Chi- | Pr > ChiSq | |
| Square | Square | |||||||
| EC16 | 0.3026 | 3.01 | 0.0829 | EC44 | 0.4760 | 5.52 | 0.0188 | |
| EC17 | 0.2586 | 2.41 | 0.1204 | EC45 | 0.0161 | 0.26 | 0.6073 | |
| EC18 | 0.4678 | 5.00 | 0.0253 | EC47 | 0.0627 | 0.48 | 0.4885 | |
| EC19 | 0.3431 | 3.62 | 0.0570 | EC48 | 0.0076 | 2.37 | 0.1240 | |
| EC20 | 0.0447 | 5.73 | 0.0167 | EC49 | 0.0898 | 1.44 | 0.2300 | |
| EC21 | 0.0862 | 0.13 | 0.7216 | EC50 | 0.0543 | 0.35 | 0.5519 | |
| EC22 | 0.0294 | 0.66 | 0.4177 | EC51 | 0.1211 | 4.58 | 0.0323 | |
| EC23 | 0.0688 | 5.76 | 0.0164 | EC52 | 0.6187 | 6.47 | 0.0110 | |
| EC24 | 0.1058 | 6.02 | 0.0141 | EC53 | 0.0173 | 0.00 | 0.9627 | |
| EC25 | 0.3101 | 0.02 | 0.8859 | EC65 | 0.0237 | 1.81 | 0.1786 | |
| EC26 | 0.0200 | 1.61 | 0.2045 | EC66 | 0.5809 | 3.59 | 0.0582 | |
| EC27 | 0.0303 | 0.09 | 0.7673 | EC67 | 0.0045 | 4.99 | 0.0255 | |
| EC28 | 0.0112 | 3.38 | 0.0661 | EC68 | 0.1182 | 8.03 | 0.0046 | |
| EC29 | 0.0373 | 2.14 | 0.1431 | EC69 | 0.2487 | 1.92 | 0.1657 | |
| EC30 | 0.0004 | 7.20 | 0.0073 | EC70 | 0.0162 | 6.18 | 0.0129 | |
| EC31 | 1.7719 | 5.65 | 0.0174 | EC71 | 0.1529 | 5.22 | 0.0224 | |
| EC32 | 0.0493 | 6.62 | 0.0101 | EC72 | 0.0796 | 2.19 | 0.1390 | |
| EC33 | 0.0081 | 4.61 | 0.0317 | EC73 | 0.0581 | 0.47 | 0.4911 | |
| EC34 | 0.0739 | 0.00 | 1.0000 | EC74 | 0.1612 | 1.15 | 0.2838 | |
| EC35 | 0.0199 | 7.56 | 0.0060 | EC75 | 0.4280 | 0.01 | 0.9351 | |
| EC36 | 0.0230 | 3.14 | 0.0766 | EC76 | 0.3440 | 5.87 | 0.0154 | |
| EC37 | 0.5567 | 6.63 | 0.0101 | EC77 | 0.0090 | 3.85 | 0.0499 | |
| EC38 | 0.0997 | 0.81 | 0.3695 | EC78 | 0.5312 | 5.58 | 0.0181 | |
| EC39 | 0.1649 | 1.28 | 0.2572 | EC79 | 0.5640 | 0.29 | 0.5904 | |
| EC40 | 0.0709 | 0.24 | 0.6236 | EC80 | 0.1005 | 3.04 | 0.0814 | |
| EC41 | 0.9768 | 5.01 | 0.0251 | EC81 | 0.2100 | 1.31 | 0.2528 | |
| EC42 | 0.1173 | 6.80 | 0.0091 | EC82 | 0.1230 | 4.58 | 0.0323 | |
| EC43 | 0.0602 | 0.88 | 0.3491 |
| Genotypes | Means | Chi- | Pr > ChiSq | Genotypes | Means | Chi- | Pr > ChiSq | |
| Square | Square | |||||||
| EC16 | 9.0128 | 0.00 | 0.9479 | EC44 | 8.1318 | 0.46 | 0.4999 | |
| EC17 | 8.1033 | 1.05 | 0.3049 | EC45 | 8.6207 | 0.04 | 0.8496 | |
| EC18 | 7.5834 | 2.91 | 0.0882 | EC47 | 8.1529 | 0.41 | 0.5245 | |
| EC19 | 9.2024 | 1.33 | 0.2481 | EC48 | 8.2046 | 0.29 | 0.5871 | |
| EC20 | 8.6407 | 0.28 | 0.5950 | EC49 | 8.5733 | 0.01 | 0.9140 | |
| EC21 | 7.5239 | 3.31 | 0.0687 | EC50 | 8.6158 | 0.03 | 0.8563 | |
| EC22 | 8.7405 | 0.16 | 0.6936 | EC51 | 8.3449 | 0.09 | 0.7704 | |
| EC23 | 8.7693 | 0.20 | 0.6580 | EC52 | 8.8932 | 0.08 | 0.7752 | |
| EC24 | 9.1320 | 0.41 | 0.5245 | EC53 | 8.5883 | 0.02 | 0.8935 | |
| EC25 | 8.4304 | 0.26 | 0.6081 | EC65 | 8.2999 | 0.00 | 0.9828 | |
| EC26 | 8.0816 | 0.59 | 0.4435 | EC66 | 8.2536 | 0.16 | 0.6896 | |
| EC27 | 8.2580 | 0.55 | 0.4582 | EC67 | 8.7437 | 0.22 | 0.6425 | |
| EC28 | 8.1072 | 1.07 | 0.3010 | EC68 | 8.7821 | 1.85 | 0.1739 | |
| EC29 | 7.8740 | 1.32 | 0.2498 | EC69 | 7.7633 | 2.43 | 0.1194 | |
| EC30 | 8.0060 | 0.82 | 0.3653 | EC70 | 7.4366 | 0.68 | 0.4084 | |
| EC31 | 8.0287 | 0.75 | 0.3879 | EC71 | 9.0001 | 0.00 | 0.9637 | |
| EC32 | 8.2664 | 0.19 | 0.6659 | EC72 | 8.7127 | 0.01 | 0.9114 | |
| EC33 | 8.7675 | 0.19 | 0.6603 | EC73 | 8.6298 | 3.58 | 0.0586 | |
| EC34 | 9.4237 | 2.27 | 0.1323 | EC74 | 7.4875 | 0.00 | 0.9658 | |
| EC35 | 8.6304 | 0.04 | 0.8365 | EC75 | 8.2633 | 0.02 | 0.8873 | |
| EC36 | 8.4143 | 0.03 | 0.8656 | EC76 | 8.3533 | 2.67 | 0.1023 | |
| EC37 | 8.5419 | 0.00 | 0.9572 | EC77 | 7.6202 | 0.25 | 0.6157 | |
| EC38 | 8.3793 | 0.05 | 0.8172 | EC78 | 8.1388 | 1.79 | 0.1806 | |
| EC39 | 8.4205 | 0.03 | 0.8742 | EC79 | 7.7742 | 0.87 | 0.3509 | |
| EC40 | 8.3520 | 0.08 | 0.7800 | EC80 | 7.8665 | 2.45 | 0.1177 | |
| EC41 | 7.9771 | 0.92 | 0.3377 | EC81 | 7.0278 | 0.66 | 0.4170 | |
| EC42 | 7.4232 | 4.07 | 0.0436 | EC82 | 8.9908 | 0.00 | 0.9828 | |
| EC43 | 7.5776 | 2.50 | 0.1138 |
| Genotypes | Means | Chi- | Pr > ChiSq | Genotypes | Means | Chi- | Pr > ChiSq | |
| Square | Square | |||||||
| EC16 | 13.3475 | 0.03 | 0.8544 | EC43 | 14.7075 | 2.78 | 0.0952 | |
| EC17 | 10.3950 | 8.91 | 0.0028 | EC44 | 12.1425 | 1.05 | 0.3046 | |
| EC18 | 14.0125 | 0.65 | 0.4209 | EC45 | 16.2075 | 6.97 | 0.0083 | |
| EC19 | 11.6125 | 2.54 | 0.1108 | EC47 | 11.3775 | 3.44 | 0.0638 | |
| EC20 | 12.9750 | 0.24 | 0.6277 | EC48 | 13.8200 | 0.39 | 0.5298 | |
| EC21 | 12.8350 | 0.10 | 0.7508 | EC49 | 15.2000 | 3.38 | 0.0662 | |
| EC22 | 11.7525 | 2.08 | 0.1492 | EC50 | 11.1350 | 4.52 | 0.0335 | |
| EC23 | 11.3125 | 3.71 | 0.0541 | EC51 | 14.4250 | 1.38 | 0.2402 | |
| EC24 | 13.0875 | 0.55 | 0.4578 | EC52 | 13.5150 | 0.41 | 0.5238 | |
| EC25 | 12.8925 | 0.01 | 0.9131 | EC53 | 16.5800 | 8.54 | 0.0035 | |
| EC26 | 14.0875 | 0.76 | 0.3827 | EC65 | 12.7725 | 0.34 | 0.5608 | |
| EC27 | 14.6025 | 1.77 | 0.1836 | EC66 | 12.6300 | 0.27 | 0.6005 | |
| EC28 | 12.5550 | 0.34 | 0.5599 | EC67 | 11.6725 | 1.24 | 0.2663 | |
| EC29 | 13.7550 | 0.74 | 0.3882 | EC68 | 14.1300 | 0.83 | 0.3621 | |
| EC30 | 12.6575 | 0.47 | 0.4944 | EC69 | 14.1000 | 0.78 | 0.3766 | |
| EC31 | 14.5175 | 1.58 | 0.2092 | EC70 | 12.9625 | 0.02 | 0.8908 | |
| EC32 | 13.8325 | 0.41 | 0.5223 | EC71 | 13.9100 | 0.51 | 0.4770 | |
| EC33 | 13.3525 | 0.04 | 0.8507 | EC72 | 13.7300 | 0.30 | 0.5859 | |
| EC34 | 12.7325 | 0.05 | 0.8258 | EC73 | 16.0675 | 4.76 | 0.0290 | |
| EC35 | 14.3075 | 1.15 | 0.2844 | EC74 | 14.0200 | 0.27 | 0.6009 | |
| EC36 | 14.2100 | 0.97 | 0.3255 | EC76 | 13.0775 | 0.01 | 0.9378 | |
| EC37 | 14.2125 | 0.97 | 0.3244 | EC77 | 12.7725 | 0.09 | 0.7675 | |
| EC38 | 14.7925 | 2.23 | 0.1352 | EC78 | 13.3575 | 0.04 | 0.8469 | |
| EC39 | 13.5625 | 0.15 | 0.6981 | EC79 | 12.6975 | 0.01 | 0.9114 | |
| EC40 | 15.4900 | 4.31 | 0.0380 | EC80 | 12.5275 | 0.67 | 0.4140 | |
| EC41 | 13.0575 | 0.01 | 0.9222 | EC81 | 11.5050 | 2.93 | 0.0868 | |
| EC42 | 14.3575 | 1.24 | 0.2649 | EC82 | 13.1575 | 0.24 | 0.6277 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





