Submitted:
09 September 2023
Posted:
12 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
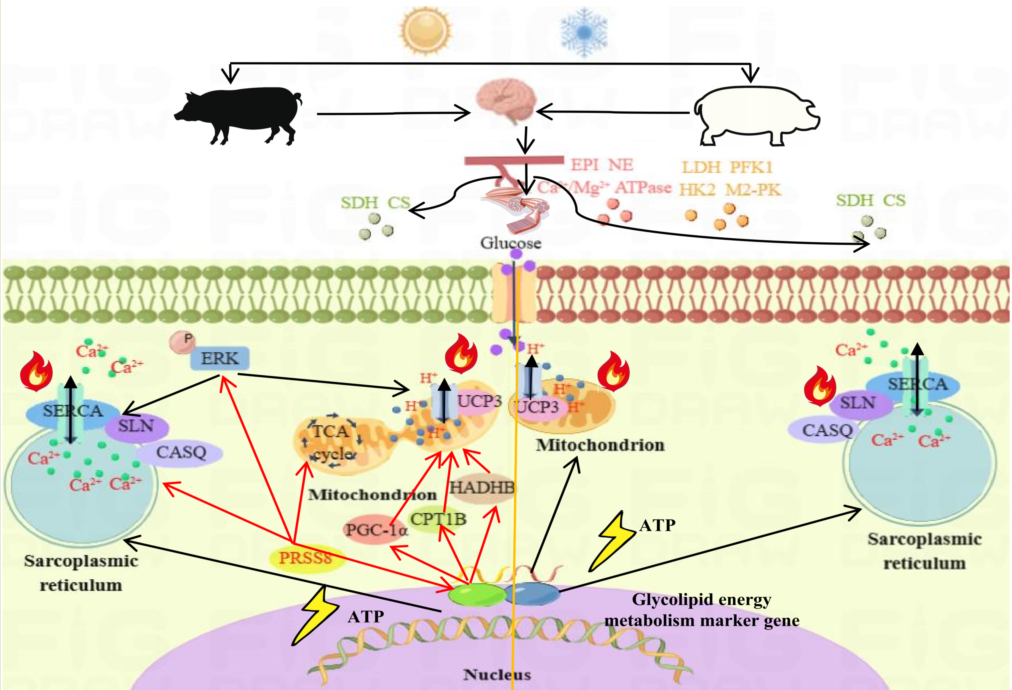
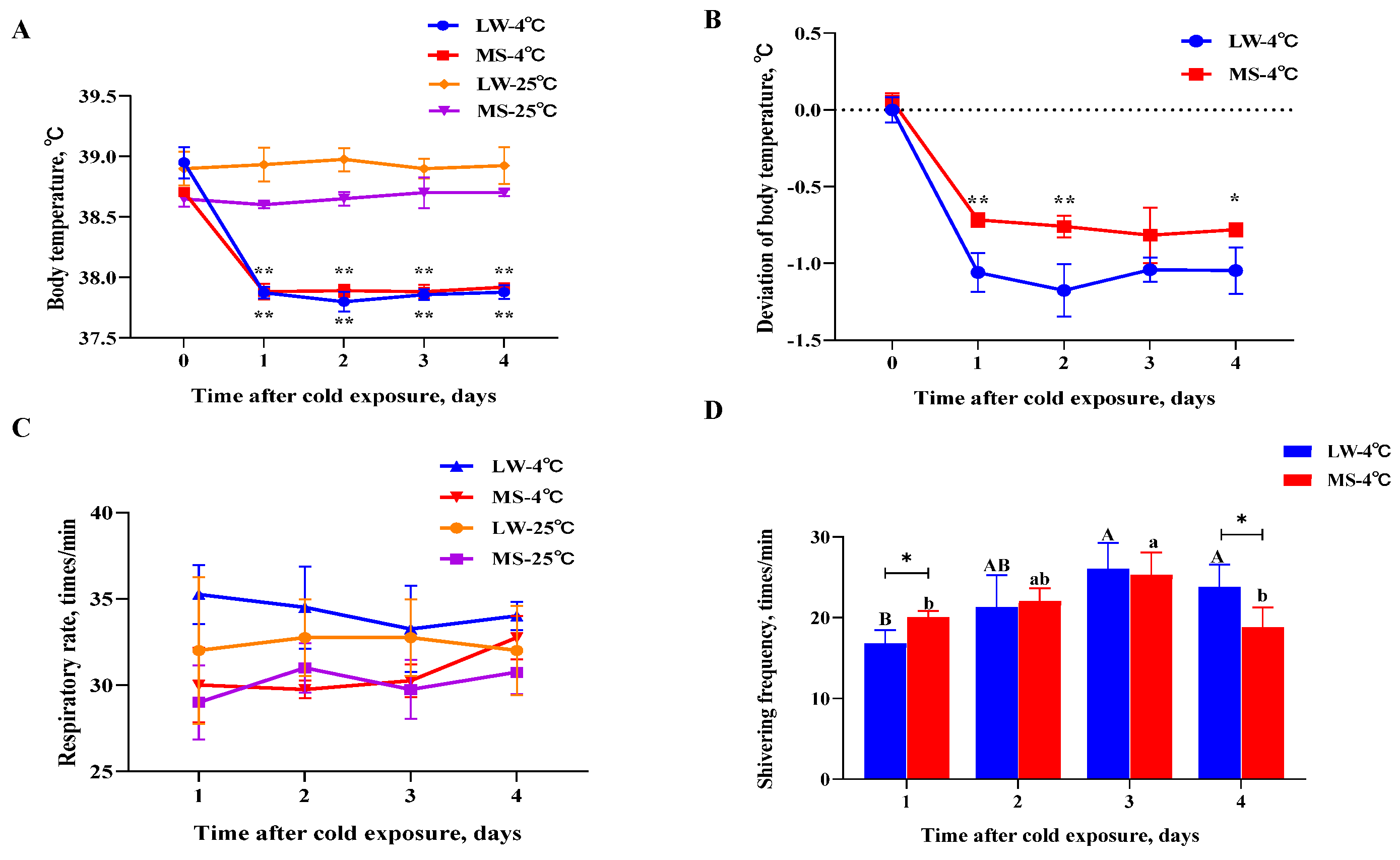
2.1. Different Hysiological Response Patterns of Large White Pigs and Mashen Pigs under Cold Stress
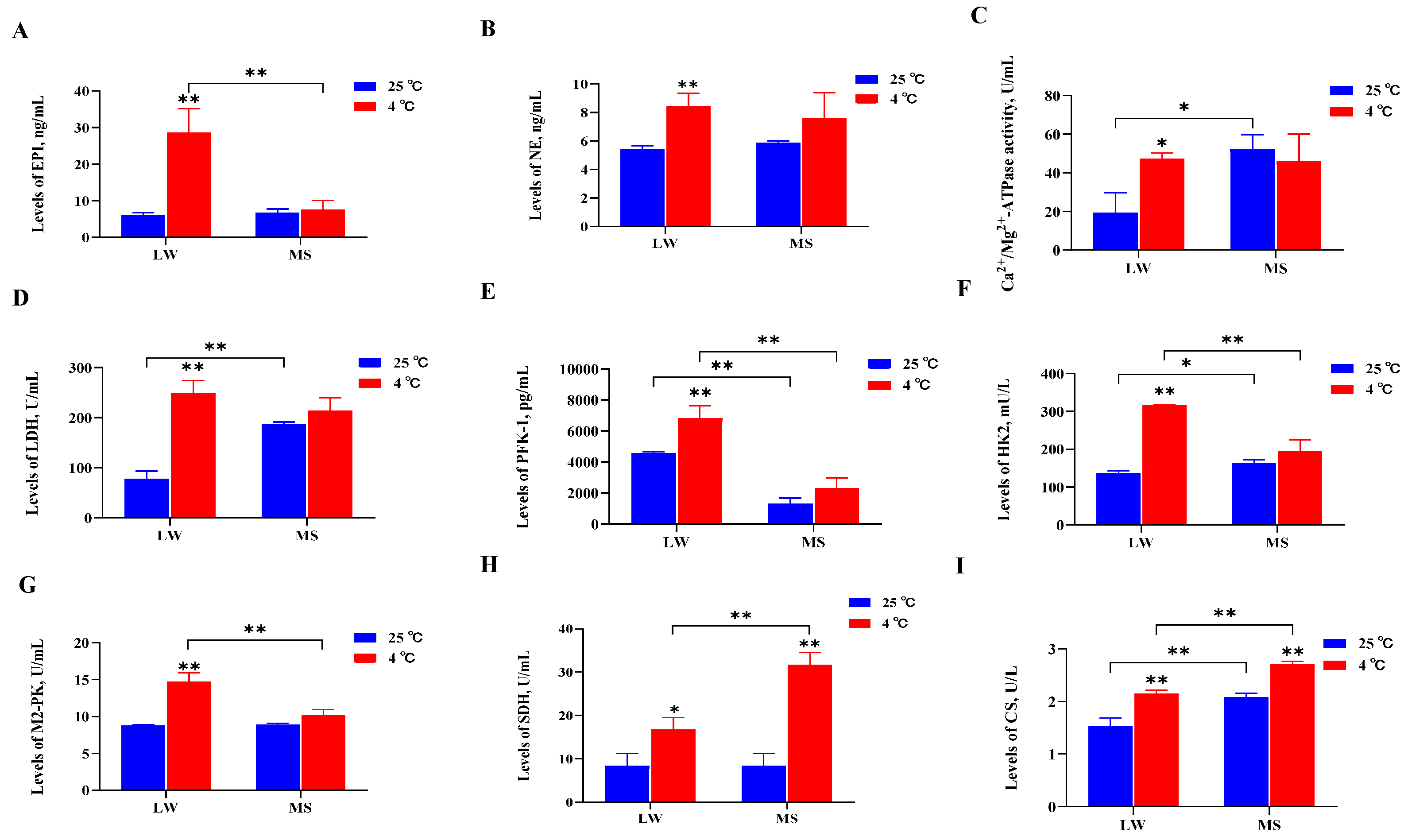
2.2. Serum Biochemistry and Hormone Analyses of Large White Pigs and Mashen Pigs under Cold Stress
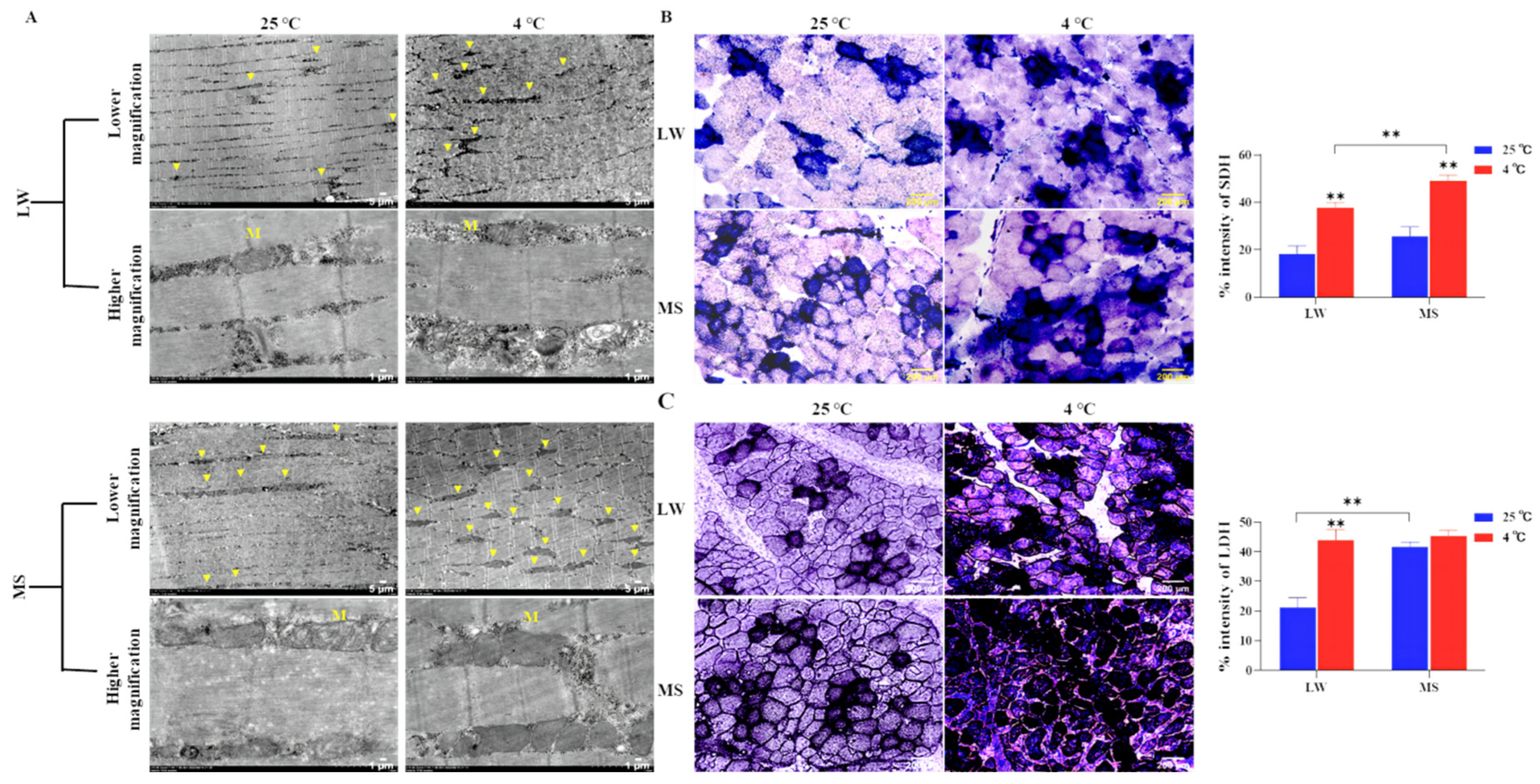
2.3. Effects of Cold Stress on Ultrastructure and Enzyme Activity of Longissimus Dorsi Muscle in Large White Pigs and Mashen Pigs under Cold Stress
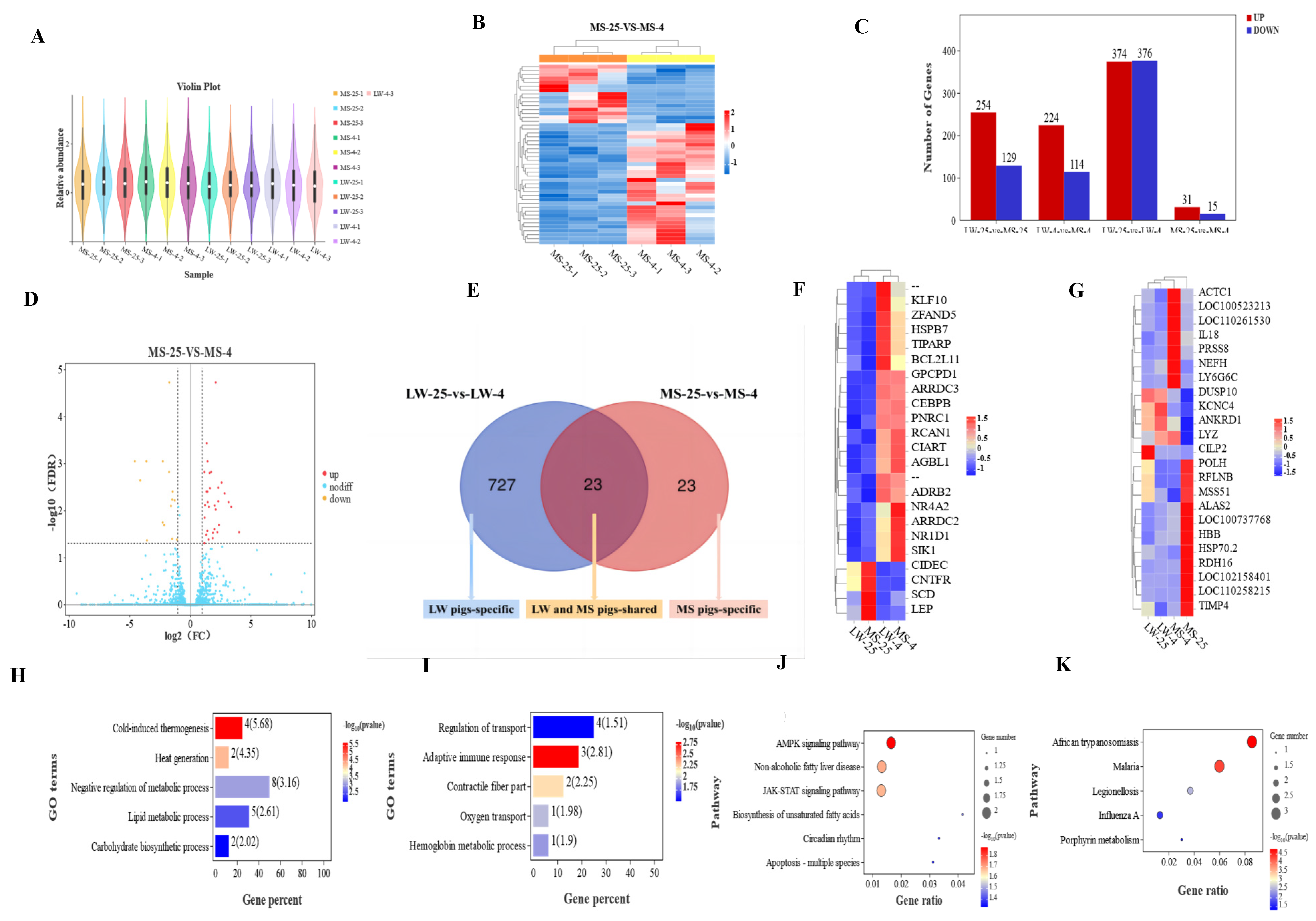
2.4. Differential Expression Genes Analysis of Longissimus Dorsi Muscle in Large White Pigs and Mashen Pigs under Cold Stress
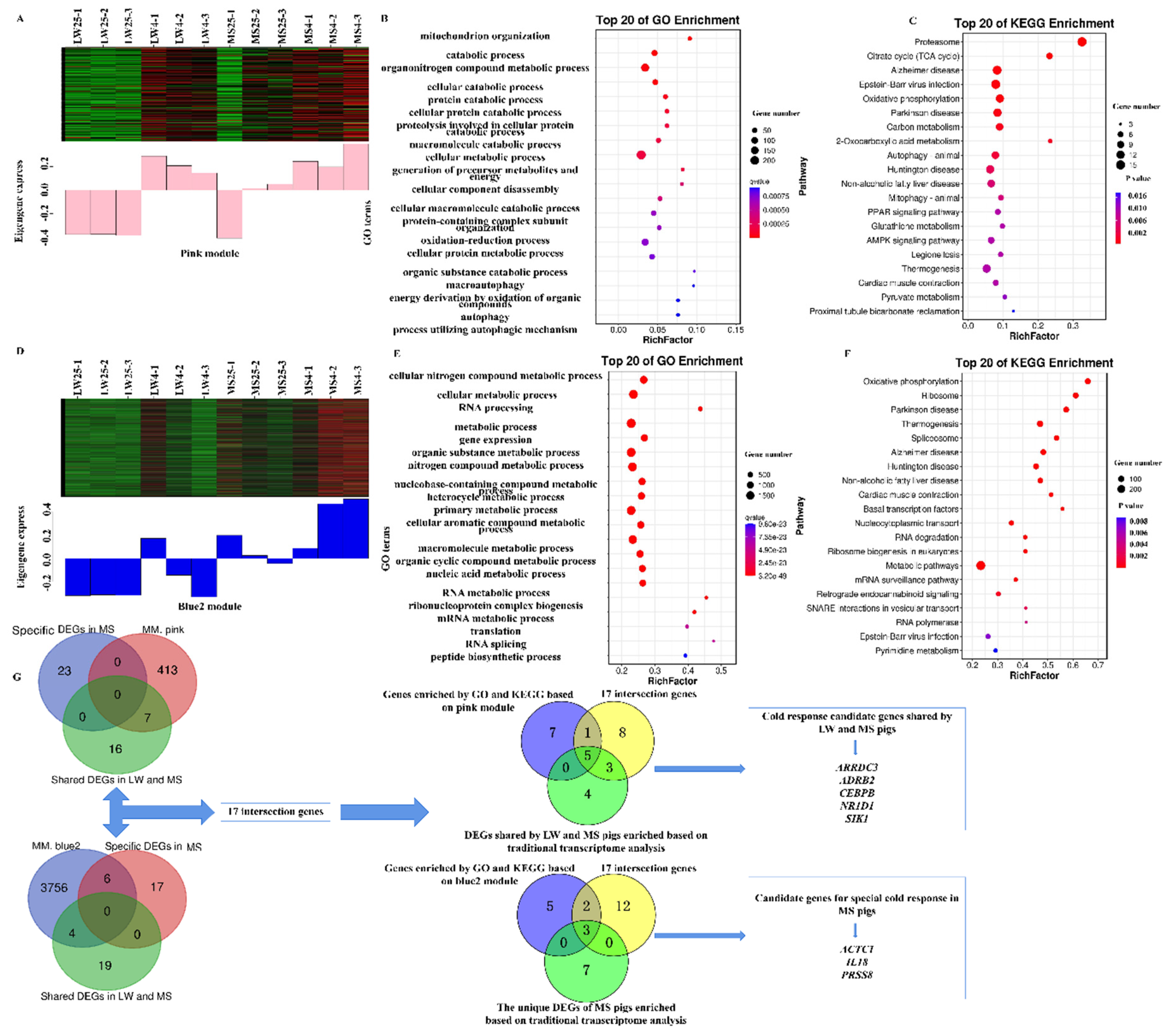
2.5. Functional Annotation and Pathway Enrichment Analysis of Common and Specific Cold-Responsive Differentially Expressed Genes in Large White Pigs and Mashen Pigs under Cold Stress
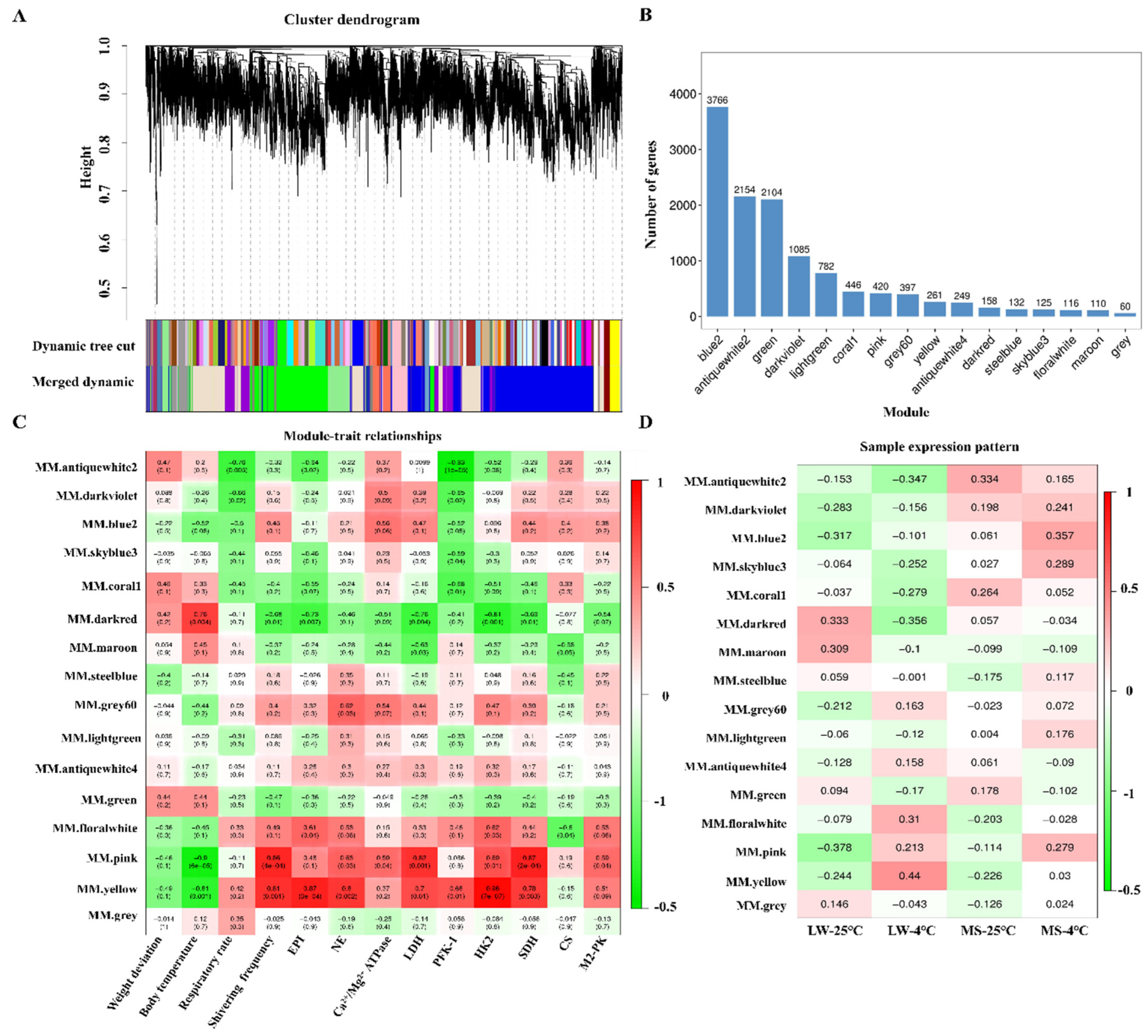
2.6. Weighted Co-Expression Network Analysis of Longissimus Dorsi Muscle in Large White Pigs and Mashen Pigs under Cold Stress
2.7. Functional Annotation and Pathway Enrichment Analysis of the Target Modules
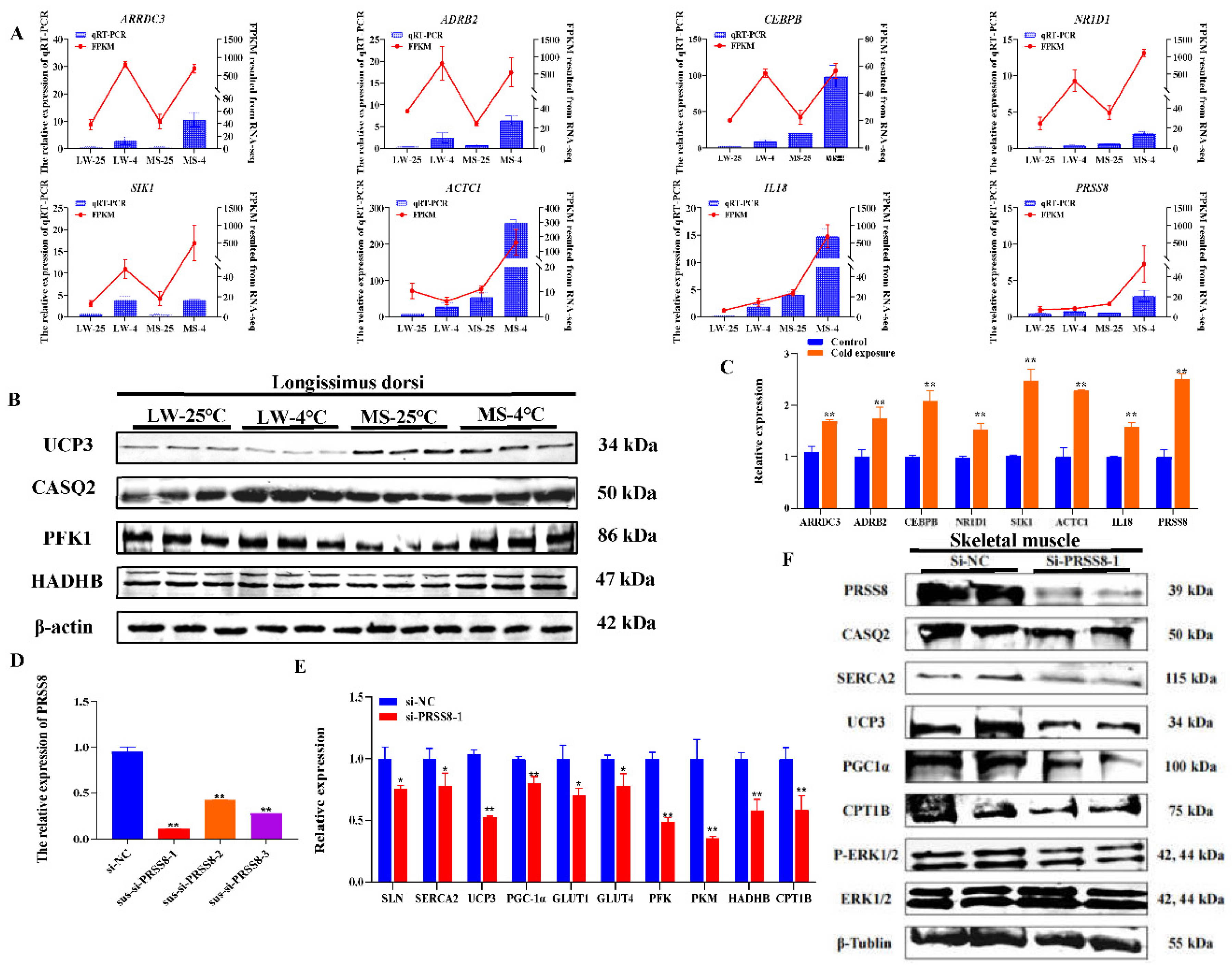
2.8. Validation Results of Common and Specific Cold Response Candidate Genes in Pig Longissimus Dorsi Muscle and Satellite Cells
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Phenotypic Detection and Sample Collection
4.3. Serum Biochemistry and Hormone Analyses
4.4. Electron Microscopy and Immunohistochemistry
4.5. RNA-seq
4.6. Weighted Gene Co-Expression Network Analysis
4.7. Cell Culture
4.8. Quantitative Real-Time Polymerase Chain Reaction
4.9. Western Blotting
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Toghiani, S.; Hay, E.; Fragomeni, B.; Rekaya, R.; Roberts, AJ. Genotype by environment interaction in response to cold stress in a composite beef cattle breed. Animal. 2020, 31, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Cheng, X.; Fang, H.; Ren, Y.; Li, X.; Ren, W.; Xue, B.; Yang, C. Effect of cold stress on ovarian & uterine microcirculation in rats and the role of endothelin system. Reprod. Biol. Endocrin. 2020, 18, 29. [Google Scholar] [CrossRef]
- Le Dividich, J.; Noblet, J. Colostrum intake and thermoregulation in the neonatal pig in relation to environmental temperature. Biol. Neonate. 1981, 40, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Iida, R.; Koketsu, Y. Climatic factors associated with peripartum pig deaths during hot and humid or cold seasons. Prev. Vet. Med. 2014, 115, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Čobanović, N.; Stajković, S.; Blagojević, B.; Betić, N.; Dimitrijević, M.; Vasilev, D.; Karabasil, N. The effects of season on health, welfare, and carcass and meat quality of slaughter pigs. Int. J. Biometeorol. 2020, 64, 1899–1909. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Tian, S.; Jin, L.; Zhou, G.; Li, Y.; Zhang, Y.; Wang, T.; Yeung, C.; Chen, L.; Ma, J.; et al. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet. 2013, 45, 1431–1438. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, X.; Jing, X.; He, X.; Wang, L.; Liu, Y.; Liu, D. Transcriptomics Analysis on Excellent Meat Quality Traits of Skeletal Muscles of the Chinese Indigenous Min Pig Compared with the Large White Breed. Int. J. Mol. Sci. 2017, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Lin, J.; Huang, J.; Zhang, H.; Zhang, R.; Zhang, X.; Cao, C.; Hambly, C.; Qin, G.; Yao, J.; et al. UCP1Reconstitution of using CRISPR/Cas9 in the white adipose tissue of pigs decreases fat deposition and improves thermogenic capacity. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E9474–E9482. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Cao, C.; Tao, C.; Ye, R.; Dong, M.; Zheng, Q.; Wang, C.; Jiang, X.; Qin, G.; Yan, C.; et al. Cold adaptation in pigs depends on UCP3 in beige adipocytes. J. Mol. Cell. Biol. 2017, 9, 364–375. [Google Scholar] [CrossRef]
- Sun, G.; Song, X.; Zou, Y.; Teng, T.; Jiang, L.; Shi, B. Dietary Glucose Ameliorates Impaired Intestinal Development and Immune Homeostasis Disorders Induced by Chronic Cold Stress in Pig Model. Int. J. Mol. Sci. 2022, 23, 7730. [Google Scholar] [CrossRef]
- Lu, C.; Liu, Y.; Ma, Y.; Wang, S.; Cai, C.; Yang, Y.; Zhao, Y.; Liang, G.; Cao, G.; Li, B.; et al. Comparative Evaluation of the Ileum Microbiota Composition in Piglets at Different Growth Stages. Front. Microbiol. 2021, 12, 765691. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Li, M.; Zhang, Y.; Meng, S.; Yang, Y.; Gao, P.; Guo, X.; Cao, G.; Li, B. Comparative Transcriptome Analyses of Between Pig Breeds Differing in Muscle Characteristics. Front. Genet. 2020, 11, 526309. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Guo, Y.; Lu, C.; Cai, C.; Gao, P.; Cao, G.; Li, B.; Guo, X.; Yang, Y. Effect of Different Pig Fecal Microbiota Transplantation on Mice Intestinal Function and Microbiota Changes During Cold Exposure. Front. Vet. Sci. 2022, 9, 805815. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, T.; Li, W.; Zhang, W.; Cai, C.; Lu, C.; Gao, P.; Cao, G.; Li, B.; Guo, X.; et al. Effects of Low-Ambient-Temperature Stimulation on Modifying the Intestinal Structure and Function of Different Pig Breeds. Animals (Basel). 2022, 12, 2740. [Google Scholar] [CrossRef]
- Fuller-Jackson, J.; Henry, B. Adipose and skeletal muscle thermogenesis: studies from large animals. J. Endocrinol. 2018, 237, R99–R115. [Google Scholar] [CrossRef] [PubMed]
- Cheshire, W.P. Thermoregulatory disorders and illness related to heat and cold stress. Auton. Neurosci. 2016, 196, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Halvorson, I.; Thornhill, J. Posterior hypothalamic stimulation of anesthetized normothermic and hypothermic rats evokes shivering thermogenesis. Brain. Res. 1993, 610, 208–215. [Google Scholar] [CrossRef]
- Nabi, G.; Xing, D.; Sun, Y.; Zhang, Q.; Li, M.; Jiang, C.; Ahmad, I.M.; Wingfield, J.C.; Wu, Y.; Li, D. Coping with extremes: High-altitude sparrows enhance metabolic and thermogenic capacities in the pectoralis muscle and suppress in the liver relative to their lowland counterparts. Gen. Comp. Endocrinol. 2021, 313, 113890. [Google Scholar] [CrossRef]
- Wang, S.; Gopinath, T.; Larsen, E.K.; Weber, D.K.; Walker, C.; Uddigiri, V.R.; Mote, K.R.; Sahoo, S.K.; Periasamy, M.; Veglia, G. Structural basis for sarcolipin's regulation of muscle thermogenesis by the sarcoplasmic reticulum Ca-ATPase. Sci. Adv. 2021, 7, eabi7154. [Google Scholar] [CrossRef]
- Gorski, P.; Ceholski, D.; Young, H. Structure-Function Relationship of the SERCA Pump and Its Regulation by Phospholamban and Sarcolipin. Adv. Exp. Med. Biol. 2017, 981, 77–119. [Google Scholar] [CrossRef]
- Bal, N.C.; Maurya, S.K.; Singh, S.; Wehrens, X.H.; Periasamy, M. Increased Reliance on Muscle-based Thermogenesis upon Acute Minimization of Brown Adipose Tissue Function. J. Biol. Chem. 2016, 291, 17247–17257. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, X.; Wang, C.; Wang, D. Lasiopodomys brandtiiRecruitment of Muscle Genes as an Effect of Brown Adipose Tissue Ablation in Cold-Acclimated Brandt's Voles. Int. J. Mol. Sci. 2022, 24, 342. [Google Scholar] [CrossRef] [PubMed]
- Nowack, J.; Vetter, S.; Stalder, G.; Painer, J.; Kral, M.; Smith, S.; Le, M.; Jurcevic, P.; Bieber, C.; Arnold, W.; et al. Muscle nonshivering thermogenesis in a feral mammal. Sci. Rep. 2019, 9, 6378. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Li, Y.; Ma, F.; Zhao, R.; Yang, X. Zinc-α2-glycoprotein promotes skeletal muscle lipid metabolism in cold-stressed mice. Endocr. J. 2021, 68, 53–62. [Google Scholar] [CrossRef]
- Codella, R.; Alves, T.; Befroy, D.; Choi, C.; Luzi, L.; Rothman, D.; Kibbey, R.; Shulman, G. Overexpression of UCP3 decreases mitochondrial efficiency in mouse skeletal muscle in vivo. FEBS Lett. 2023, 597, 309–319. [Google Scholar] [CrossRef]
- Hilse, KE.; Kalinovich, AV.; Rupprecht, A.; Smorodchenko, A.; Zeitz, U.; Staniek, K.; Erben, RG.; Pohl, EE. The expression of UCP3 directly correlates to UCP1 abundance in brown adipose tissue. Biochim. Biophys. Acta. 2016, 1857, 72–78. [Google Scholar] [CrossRef]
- Maurya, S.K.; Herrera, J.L.; Sahoo, S.K.; Reis, F.C.G.; Vega, R.B.; Kelly, D.P.; Periasamy, M. Sarcolipin Signaling Promotes Mitochondrial Biogenesis and Oxidative Metabolism in Skeletal Muscle. Cell. Rep. 2018, 24, 2919–2931. [Google Scholar] [CrossRef]
- Hangalapura, B.N.; Kaiser, M.G.; Poel, J.J.; Parmentier, H.K.; Lamont, S.J. Cold stress equally enhances in vivo pro-inflammatory cytokine gene expression in chicken lines divergently selected for antibody responses. Dev. Comp. Immunol. 2006, 30, 503–511. [Google Scholar] [CrossRef]
- Górski, J.; Kuryliszyn, A.; Wereszczyńska, U. Effect of acute cold exposure on the mobilization of intramuscular glycogen and triglycerides in the rat. Acta. Physiol. Pol. 1981, 32, 755–759. [Google Scholar]
- Umehara, T.; Murase, T.; Abe, Y.; Yamashita, H.; Shibaike, Y.; Kagawa, S.; Yamamoto, T.; Ikematsu, K. Identification of potential markers of fatal hypothermia by a body temperature-dependent gene expression assay. Int. J. Legal. Med. 2019, 133, 335–345. [Google Scholar] [CrossRef]
- Hou, L.; Shi, J.; Cao, L.; Xu, G.; Hu, C.; Wang, C. Pig has no uncoupling protein 1. Biochem. Biophys. Res. Commun. 2017, 487, 795–800. [Google Scholar] [CrossRef]
- Brosh, A. Heart rate measurements as an index of energy expenditure and energy balance in ruminants: a review. J. Anim. Sci. 2007, 85, 1213–1227. [Google Scholar] [CrossRef]
- Le Dividich, J.; Mormède, P.; Catheline, M.; Caritez, J.C. Body composition and cold resistance of the neonatal pig from European (Large White) and Chinese (Meishan) breeds. Biol. Neonate. 1991, 59, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Heldmaier, G. Cold adaptation by short daily cold exposures in the young pig. J. Appl. Physiol. 1974, 36, 163–168. [Google Scholar] [CrossRef]
- Herpin, P.; Lossec, G.; Schmidt, I.; Cohen-Adad, F.; Duchamp, C.; Lefaucheur, L.; Goglia, F.; Lanni, A. Effect of age and cold exposure on morphofunctional characteristics of skeletal muscle in neonatal pigs. Pflugers. Arch. 2002, 444, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Le Dividich, J.; Noblet, J. Thermoregulation and energy metabolism in the neonatal pig. Ann. Rech. Vet. 1983, 14, 375–381. [Google Scholar] [PubMed]
- Berthon, D.; Herpin, P.; Bertin, R.; De Marco, F.; le Dividich, J. Metabolic changes associated with sustained 48-hr shivering thermogenesis in the newborn pig. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 1996, 114, 327–335. [Google Scholar] [CrossRef]
- Ren, J.; Long, Y.; Liu, R.; Song, G.; Li, Q.; Cui, Z. Characterization of Biological Pathways Regulating Acute Cold Resistance of Zebrafish. Int. J. Mol. Sci. 2021, 22, 3028. [Google Scholar] [CrossRef]
- Hangalapura, B.; Nieuwland, M.; Buyse, J.; Kemp, B.; Parmentier, H. Effect of duration of cold stress on plasma adrenal and thyroid hormone levels and immune responses in chicken lines divergently selected for antibody responses. Poult. Sci. 2004, 83, 1644–1649. [Google Scholar] [CrossRef]
- Singh, A.K.; Aryal, B.; Chaube, B.; Rotllan, N.; Varela, L.; Horvath, T.L.; Suárez, Y.; Fernández-Hernando, C. Brown adipose tissue derived ANGPTL4 controls glucose and lipid metabolism and regulates thermogenesis. Mol. Metab. 2018, 11, 59–69. [Google Scholar] [CrossRef]
- Faure, J.; Lebret, B.; Bonhomme, N.; Ecolan, P.; Kouba, M.; Lefaucheur, L. Metabolic adaptation of two pig muscles to cold rearing conditions. J. Anim. Sci. 2013, 91, 1893–1906. [Google Scholar] [CrossRef] [PubMed]
- Cheviron, Z.; Bachman, G.; Connaty, A.; McClelland, G.; Storz, J. Regulatory changes contribute to the adaptive enhancement of thermogenic capacity in high-altitude deer mice. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 8635–8640. [Google Scholar] [CrossRef]
- Xie, S.; Yang, X.; Wang, D.; Zhu, F.; Yang, N.; Hou, Z.; Ning, Z. Thyroid transcriptome analysis reveals different adaptive responses to cold environmental conditions between two chicken breeds. PLoS One. 2018, 13, e0191096. [Google Scholar] [CrossRef]
- Ge, G.; Long, Y.; Shi, L.; Ren, J.; Yan, J.; Li, C.; Li, Q.; Cui, Z. Transcriptomic profiling revealed key signaling pathways for cold tolerance and acclimation of two carp species. BMC Genomics. 2020, 21, 539. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; You, W.; Zhou, Y.; Chen, W.; Wang, Y.; Shan, T. Cold-induced lipid dynamics and transcriptional programs in white adipose tissue. BMC Biol. 2019, 17, 74. [Google Scholar] [CrossRef]
- Smith, S.D.; Kawash, J.K.; Karaiskos, S.; Biluck, I.; Grigoriev, A. Evolutionary adaptation revealed by comparative genome analysis of woolly mammoths and elephants. DNA Res. 2017, 24, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Jansen, H.T.; Trojahn, S.; Saxton, M.W.; Quackenbush, C.R.; Evans Hutzenbiler, B.D.; Nelson, O.L.; Cornejo, O.E.; Robbins, C.T.; Kelley, J.L. Hibernation induces widespread transcriptional remodeling in metabolic tissues of the grizzly bear. Commun. Biol. 2019, 2, 336. [Google Scholar] [CrossRef]
- Zhu, C.; Xu, X.; Zhou, S.; Zhou, B.; Liu, Y.; Xu, H.; Tian, Y.; Zhu, X. Apis melliferaWGCNA based identification of hub genes associated with cold response and development in metamorphic pupae. Front. Physiol. 2023, 14, 1169301. [Google Scholar] [CrossRef]
- Wang, J.; Guan, J.; Yixi, K.; Shu, T.; Chai, Z.; Wang, J.; Wang, H.; Wu, Z.; Cai, X.; Zhong, J.; et al. Comparative transcriptome analysis of winter yaks in plateau and plain. Reprod. Domest. Anim. 2022, 57, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, W.; An, J.; Yang, P.; Guo, L.; Li, Y.; Lv, J.; Yu, S. Transcriptome analyses and weighted gene coexpression network analysis reveal key pathways and genes involved in the rapid cold resistance of the Chinese white wax scale insect. Arch. Insect. Biochem. Physiol. 2021, 107, e21781. [Google Scholar] [CrossRef]
- Periasamy, M.; Maurya, S.K.; Sahoo, S.K.; Singh, S.; Sahoo, S.K.; Reis, F.C.G.; Bal, N.C. Role of SERCA Pump in Muscle Thermogenesis and Metabolism. Comp. Physiol. 2017, 7, 879–890. [Google Scholar] [CrossRef]
- Rowland, L.A.; Bal, N.C.; Periasamy, M. The role of skeletal-muscle-based thermogenic mechanisms in vertebrate endothermy. Biol. Rev. Camb. Philos. Soc. 2015, 90, 1279–1297. [Google Scholar] [CrossRef] [PubMed]
- Iwen, K.A.; Backhaus, J.; Cassens, M.; Waltl, M.; Hedesan, O.C.; Merkel, M.; Heeren, J.; Sina, C.; Rademacher, L.; Windjäger, A.; et al. Cold-Induced Brown Adipose Tissue Activity Alters Plasma Fatty Acids and Improves Glucose Metabolism in Men. J. Clin. Endocr. Metab. 2017, 102, 4226–4234. [Google Scholar] [CrossRef] [PubMed]
- Grefhorst, A.; van den Beukel, J.C.; Dijk, W.; Steenbergen, J.; Voortman, G.J.; Leeuwenburgh, S.; Visser, T.J.; Kersten, S.; Friesema, E.C.H.; Themmen, A.P.N.; et al. Multiple effects of cold exposure on livers of male mice. J. Endocrinol. 2018, 238, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Shi, Q.; Li, W.; Mu, X.; Peng, J.; Li, M.; Chen, M.; Huang, H.; Wang, C.; Gao, K.; et al. ARRDC1 and ARRDC3 act as tumor suppressors in renal cell carcinoma by facilitating YAP1 degradation. Am. J. Cancer. Res. 2018, 8, 132–143. [Google Scholar]
- Lin, C.; MacGurn, J.; Chu, T.; Stefan, C.; Emr, S. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008, 135, 714–725. [Google Scholar] [CrossRef]
- He, R.; He, Y.; Tang, Y.; Zhou, H.; McLeod, H.; Liu, J. The potential anticancer effect of beta-blockers and the genetic variations involved in the interindividual difference. Pharmacogenomics. 2016, 17, 74–79. [Google Scholar] [CrossRef]
- Smas, C.; Sul, H. Control of adipocyte differentiation. Biochem. J. 1995, 697–710. [Google Scholar] [CrossRef]
- Tang, W.; Weng, L.; Wang, X.; Liu, C.; Hu, G.; Yin, S.; Tao, Y.; Hong, N.; Guo, H.; Liu, W.; et al. The Mediator subunit MED20 organizes the early adipogenic complex to promote development of adipose tissues and diet-induced obesity. Cell Rep. 2021, 36, 109314. [Google Scholar] [CrossRef]
- Lee, J.; Chae, S.; Nan, J.; Koo, Y.; Lee, S.; Park, Y.; Hwang, D.; Han, W.; Lee, D.; Kim, Y.; et al. SENP2 suppresses browning of white adipose tissues by de-conjugating SUMO from C/EBPβ. Cell Rep. 2022, 38, 110408. [Google Scholar] [CrossRef]
- Everett, L.; Lazar, M. Nuclear receptor Rev-erbα: up, down, and all around. Trends. Endocrinol. Metab. 2014, 25, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Altarejos, J.; Montminy, M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 2011, 12, 141–151. [Google Scholar] [CrossRef]
- Sakamaki, J.; Fu, A.; Reeks, C.; Baird, S.; Depatie, C.; Al Azzabi, M.; Bardeesy, N.; Gingras, A.; Yee, S.; Screaton, R. Role of the SIK2-p35-PJA2 complex in pancreatic β-cell functional compensation. Nat. Cell Biol. 2014, 16, 234–244. [Google Scholar] [CrossRef]
- Koo, S.; Flechner, L.; Qi, L.; Zhang, X.; Screaton, R.; Jeffries, S.; Hedrick, S.; Xu, W.; Boussouar, F.; Brindle, P.; et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005, 437, 1109–1111. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Seo, W.; Lee, M.; Kim, S.; Koo, S. Salt-inducible kinase regulates hepatic lipogenesis by controlling SREBP-1c phosphorylation. J. Biol. Chem. 2009, 284, 10446–10452. [Google Scholar] [CrossRef]
- Pourcel, L.; Buron, F.; Arib, G.; Le Fourn, V.; Regamey, A.; Bodenmann, I.; Girod, P.; Mermod, N. Influence of cytoskeleton organization on recombinant protein expression by CHO cells. Biotechnol. Bioeng. 2020, 117, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Boutilier, J.; Taylor, R.; Ram, R.; McNamara, E.; Nguyen, Q.; Goullée, H.; Chandler, D.; Mehta, M.; Balmer, L.; Laing, N.; et al. Variable cardiac α-actin (Actc1) expression in early adult skeletal muscle correlates with promoter methylation. Biochim. Biophys. Acta. Gene Regul. Mech. 2017, 1860, 1025–1036. [Google Scholar] [CrossRef]
- Bahadoran, S.; Hassanpour, H.; Arab, S.; Abbasnia, S.; Kiani, A. Changes in the expression of cardiac genes responsive to thyroid hormones in the chickens with cold-induced pulmonary hypertension. Poult. Sci. 2021, 100, 101263. [Google Scholar] [CrossRef]
- Kahaly, GJ.; Dillmann, WH. Thyroid hormone action in the heart. Endocr. Rev. 2005, 26, 704–728. [Google Scholar] [CrossRef]
- Ji, K.; Jiao, D.; Yang, G.; Degen, A.; Zhou, J.; Liu, H.; Wang, W.; Cong, H. Transcriptome analysis revealed potential genes involved in thermogenesis in muscle tissue in cold-exposed lambs. Front. Genet. 2022, 13, 1017458. [Google Scholar] [CrossRef]
- Nakanishi, K.; Yoshimoto, T.; Tsutsui, H.; Okamura, H. Interleukin-18 regulates both Th1 and Th2 responses. Annu. Rev. Immunol. 2001, 19, 423–474. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, K. Unique Action of Interleukin-18 on T Cells and Other Immune Cells. Front. Immunol. 2018, 9, 763. [Google Scholar] [CrossRef]
- El-Darawish, Y.; Li, W.; Yamanishi, K.; Pencheva, M.; Oka, N.; Yamanishi, H.; Matsuyama, T.; Tanaka, Y.; Minato, N.; Okamura, H. Frontline Science: IL-18 primes murine NK cells for proliferation by promoting protein synthesis, survival, and autophagy. J. Leukocyte. Biol. 2018, 104, 253–264. [Google Scholar] [CrossRef]
- Deason, K.; Troutman, T.; Jain, A.; Challa, D.; Mandraju, R.; Brewer, T.; Ward, E.; Pasare, C. BCAP links IL-1R to the PI3K-mTOR pathway and regulates pathogenic Th17 cell differentiation. J. Exp. Med. 2018, 215, 2413–2428. [Google Scholar] [CrossRef]
- Uchimura, K.; Hayata, M.; Mizumoto, T.; Miyasato, Y.; Kakizoe, Y.; Morinaga, J.; Onoue, T.; Yamazoe, R.; Ueda, M.; Adachi, M.; et al. The serine protease prostasin regulates hepatic insulin sensitivity by modulating TLR4 signalling. Nat. Commun. 2014, 5, 3428. [Google Scholar] [CrossRef] [PubMed]
- Sekine, T.; Takizawa, S.; Uchimura, K.; Miyazaki, A.; Tsuchiya, K. Liver-Specific Overexpression of Prostasin Attenuates High-Fat Diet-Induced Metabolic Dysregulation in Mice. Int. J. Mol. Sci. 2021, 22, 8314. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




