Submitted:
12 May 2025
Posted:
13 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. RNA Extraction, Library Construction, and Sequencing
2.3. Read Mapping and Differential Expression Analysis
2.4. Functional Enrichment and Pathway Analysis
2.5. Protein–Protein Interaction (PPI) Network Analysis
2.6. Validation by Quantitative Real-Time PCR (qRT-PCR)
3. Results
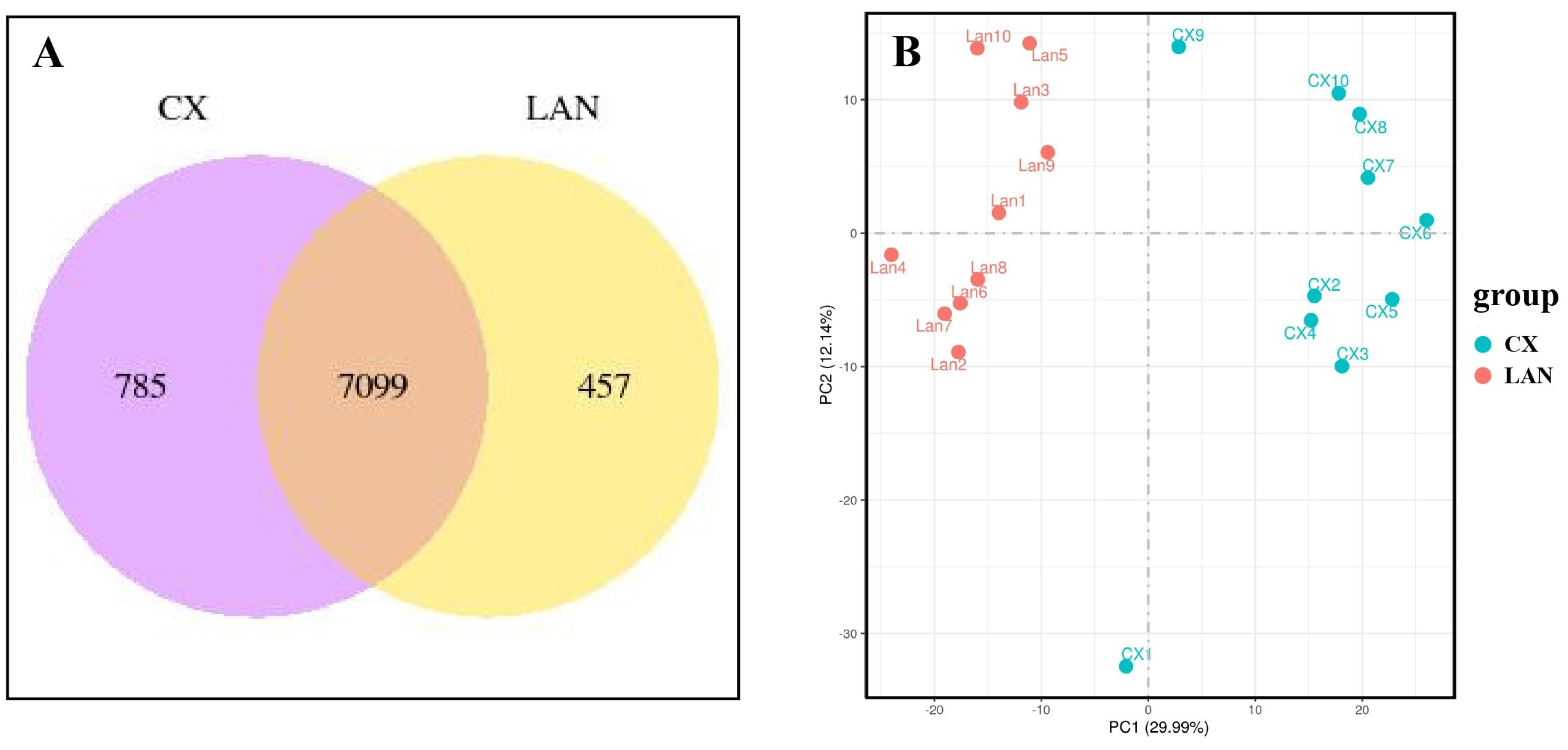
3.1. Overview of RNA Sequencing Data
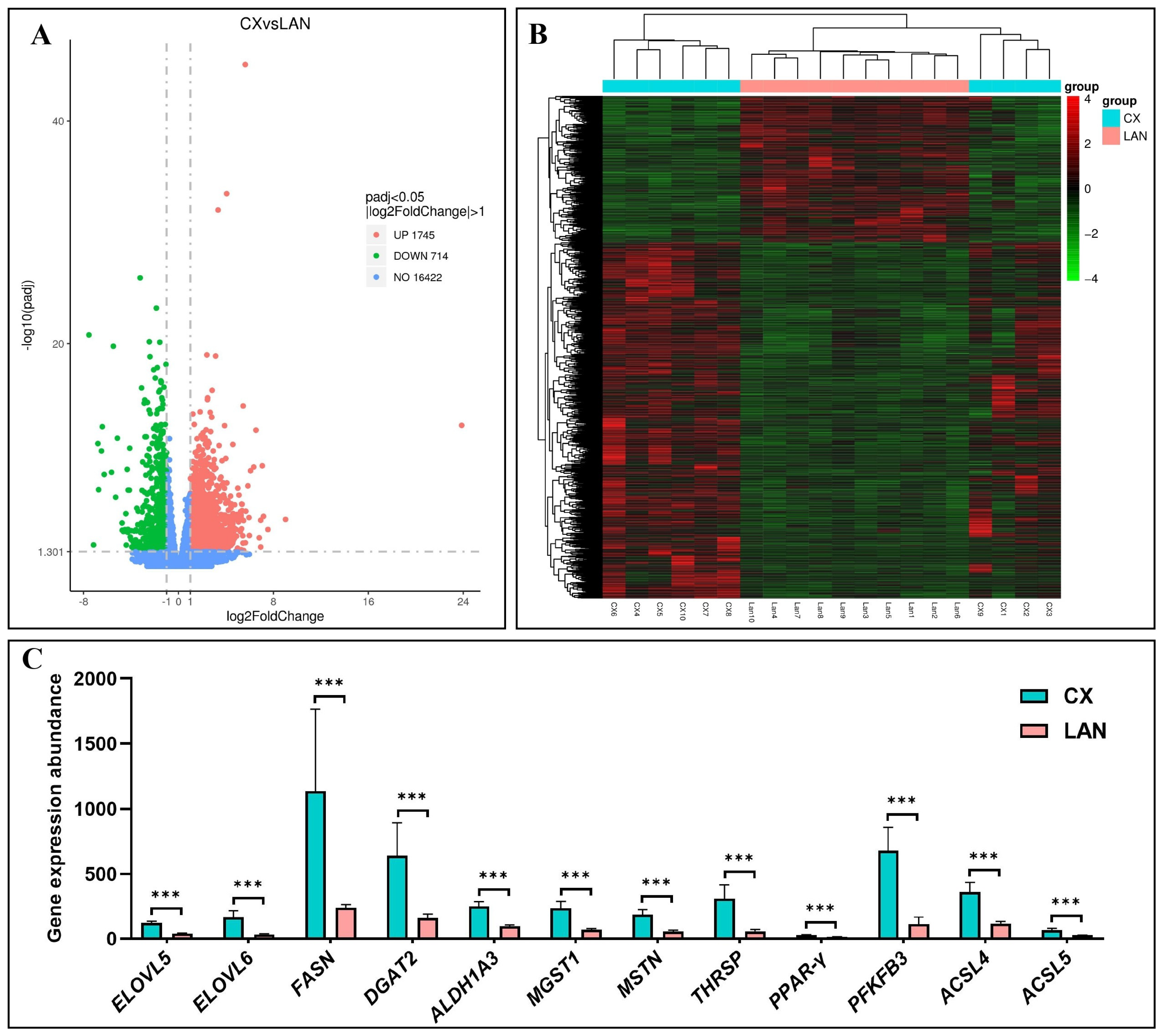
3.2. Identification of Differentially Expressed Genes
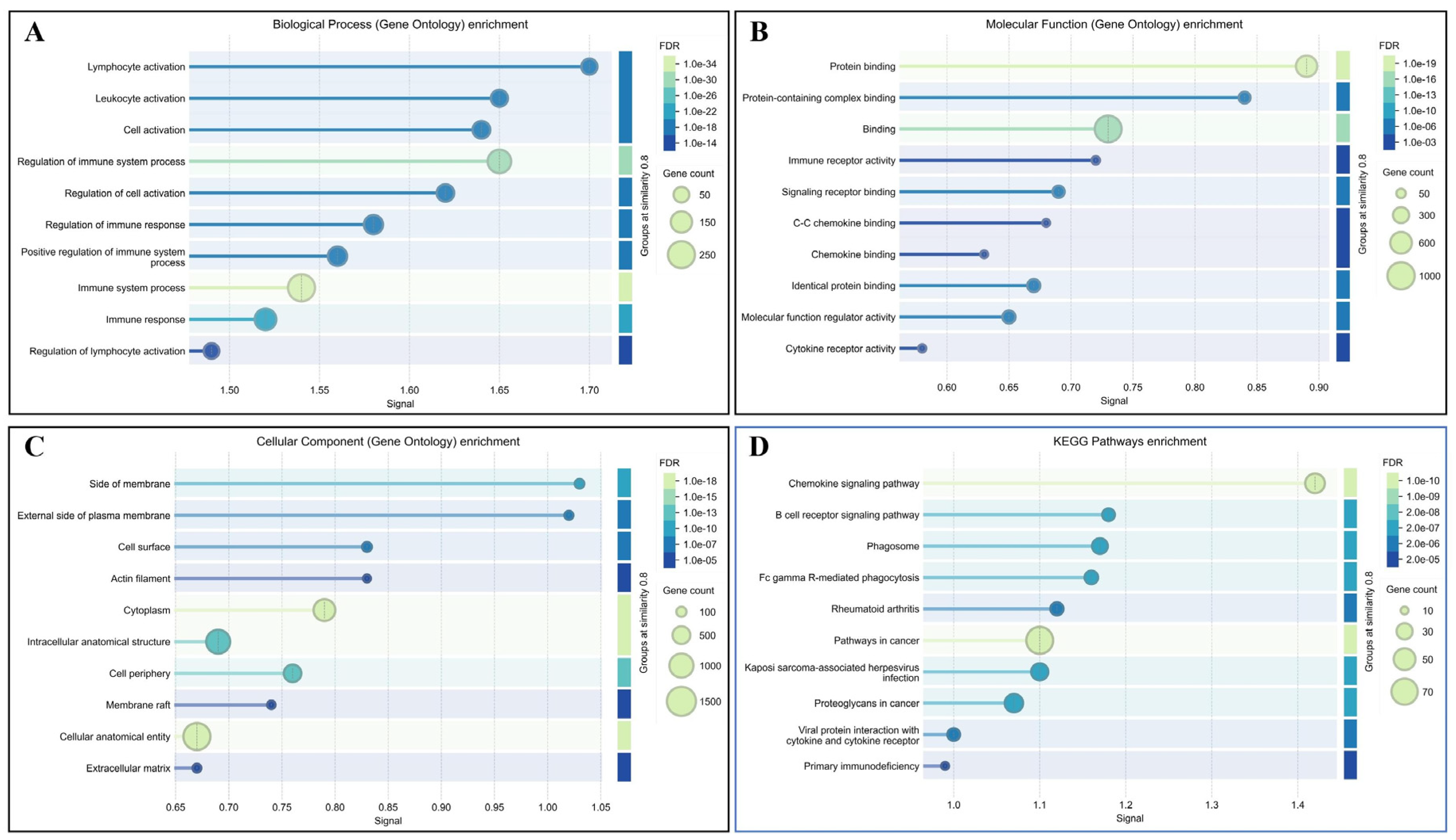
3.3. Functional Enrichment Analysis of DEGs
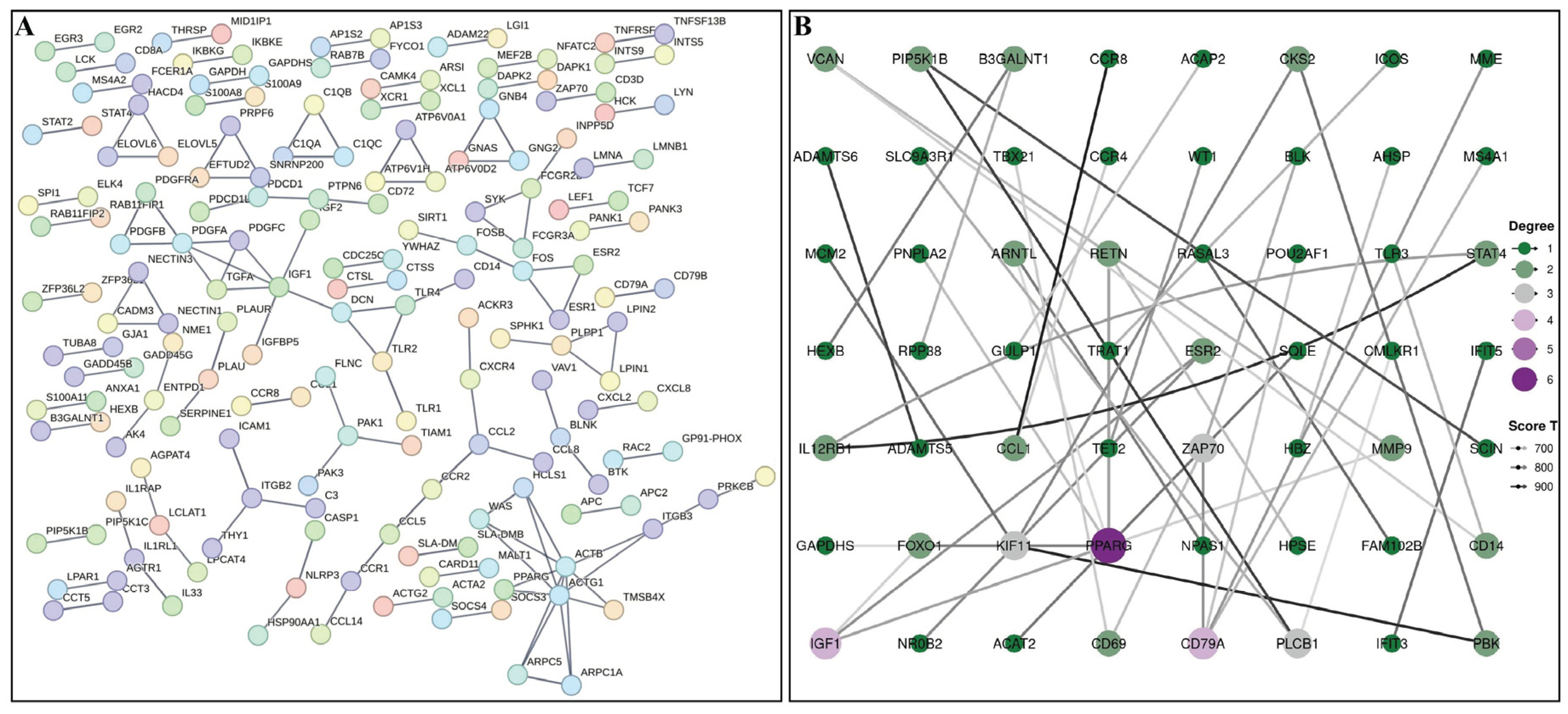
3.4. PPI Network Analysis of DEGs
3.5. Validation of DEGs by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Kim, S.W.; Gormley, A.; Jang, K.; Duarte, M. Current status of global pig production: an overview and research trends. Animal Bioscience 2023, 37, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Zhang, P.; Yin, J.; Zhang, X. New insights into the effects of dietary amino acid composition on meat quality in pigs: A review. Meat science 2024, 221, 109721. [Google Scholar] [CrossRef] [PubMed]
- Aboah, J.; Lees, N. Consumers use of quality cues for meat purchase: Research trends and future pathways. Meat science 2020, 166, 108142. [Google Scholar] [CrossRef]
- Chen, G.; Cai, Y.; Su, Y.; Wang, D.; Pan, X.; Zhi, X. Study of meat quality and flavour in different cuts of Duroc-Bamei binary hybrid pigs. Veterinary Medicine and Science 2020, 7, 724–734. [Google Scholar] [CrossRef]
- Fu, Q.-Q.; Shi, H.; Hu, D.; Cheng, J.; Chen, S.; Ben, A. Pork longissimus dorsi marinated with edible mushroom powders: Evaluation of quality traits, microstructure, and protein degradation. Food research international 2022, 158, 111503. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.T.; Zheng, Y.; Xu, J.; Zhao, Z.; Zhang, Q.; Zhang, Y.; Li, M.; Zou, H.; Azeem, R.-M.; Sun, W.-S.; et al. Transcriptome and metabolome insights into key genes regulating fat deposition and meat quality in pig breeds. Animals 2024, 14, 3560. [Google Scholar] [CrossRef]
- Sharma, P.; Doultani, S.; Hadiya, K.; George, L.; Highland, H. Overview of marker-assisted selection in animal breeding. Journal of Advances in Biology & Biotechnology 2024, 27, 303–318. [Google Scholar] [CrossRef]
- Meuwissen, T.; Hayes, B.; Goddard, M. Genomic selection: A paradigm shift in animal breeding. Animal Frontiers 2016, 6, 6–14. [Google Scholar] [CrossRef]
- Šimon, M.; Bogićević, S.; Kaić, A.; Luštrek, B.; Potočnik, K. Exploring genetic influences on equine meat quality: A bioinformatics approach. Foods 2025, 14, 533. [Google Scholar] [CrossRef]
- Cesar, A.; Regitano, L.; Reecy, J.; Poleti, M.; Oliveira, P.; De Oliveira, G.; Moreira, G.; Mudadu, M.; Tizioto, P.; Koltes, J.; et al. Identification of putative regulatory regions and transcription factors associated with intramuscular fat content traits. BMC Genomics 2018, 19, 499. [Google Scholar] [CrossRef]
- Hasan, M.; Feugang, J.; Liao, S. A nutrigenomics approach using RNA sequencing technology to study nutrient–gene interactions in agricultural animals. Current Developments in Nutrition 2019, 3, nzz082. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-Z.; Wu, J.; Zhou, J.; Zhang, Y.; Qiao, M.; Sun, H.; Li, Z.; Li, L.; Chen, N.; Oyelami, F.; et al. Integration of ATAC-seq and RNA-seq analysis identifies key genes affecting intramuscular fat content in pigs. Frontiers in Nutrition 2022, 9, 1016956. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, A.; Óvilo, C.; Núñez, Y.; Benítez, R.; López-García, A.; García, F.; Félix, M.; Laranjo, M.; Charneca, R.; Martins, J. Transcriptomic profiling of skeletal muscle reveals candidate genes influencing muscle growth and associated lipid composition in portuguese local pig breeds. Animals 2021, 11, 1423. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Żukowski, K.; Eckert, R.; Gurgul, A.; Piórkowska, K.; Oczkowicz, M. Comprehensive analysis of the whole transcriptomes from two different pig breeds using RNA-Seq method. Animal genetics 2014, 45, 674–684. [Google Scholar] [CrossRef]
- Liu, W.; Gong, T.; Xu, Y. The co-expression of steroidogenic enzymes with T1R3 during testicular development in the Congjiang Xiang pig. Animal reproduction science 2023, 251, 107216. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Chen, Z.; Ruan, Y.; Xu, H. Differential regulatory roles of microRNAs during intramuscular adipogenesis in Chinese Guizhou Congjiang Xiang pigs. Epigenetics 2022, 17, 1800–1819. [Google Scholar] [CrossRef]
- Guo, J.; Shan, T.; Wu, T.; Zhu, L.; Ren, Y.; An, S.; Wang, Y. Comparisons of different muscle metabolic enzymes and muscle fiber types in Jinhua and Landrace pigs. Journal of animal science 2011, 89 1, 185–191. [Google Scholar] [CrossRef]
- Kasprzyk, A.; Tyra, M.; Babicz, M. Fatty acid profile of pork from a local and a commercial breed. Archives Animal Breeding 2015, 58, 379–385. [Google Scholar] [CrossRef]
- Vidal, O.; Noguera, J.; Amills, M.; Varona, L.; Gil, M.; Jiménez, N.; Dávalos, G.; Folch, J.; Sánchez, A. Identification of carcass and meat quality quantitative trait loci in a Landrace pig population selected for growth and leanness. Journal of animal science 2005, 83, 293–300. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, Z.-Y.; Liu, Q.-X. Gene coexpression networks reveal key drivers of phenotypic divergence in porcine muscle. BMC Genomics 2015, 16, 50. [Google Scholar] [CrossRef]
- Tan, L.; Chen, Z.; Teng, M.; Chen, B.; Xu, H. Genome-wide analysis of mRNAs, lncRNAs, and circRNAs during intramuscular adipogenesis in Chinese Guizhou Congjiang pigs. PLoS ONE 2022, 17, e0261293. [Google Scholar] [CrossRef]
- Yang, Y.; Yuan, H.; Yao, B.; Zhao, S.; Wang, X.; Xu, L.; Zhang, L. Genetic adaptations of the Tibetan pig to high-altitude hypoxia on the Qinghai–Tibet plateau. International Journal of Molecular Sciences 2024, 25, 11303. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Yang, Y.; Liu, Y.; Chen, J.; Yang, B.; Tang, Z. High serum reproductive hormone levels at mid-pregnancy support Meishan pig prolificacy. Journal of Integrative Agriculture 2023, 22, 3489–3499. [Google Scholar] [CrossRef]
- Renaville, B.; Bacciu, N.; Lanzoni, M.; Mossa, F.; Piasentier, E. Association of single nucleotide polymorphisms in fat metabolism candidate genes with fatty acid profiles of muscle and subcutaneous fat in heavy pigs. Meat science 2018, 139, 220–227. [Google Scholar] [CrossRef]
- Jakobsson, A.; Westerberg, R.; Jacobsson, A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Progress in lipid research 2006, 45, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Chitraju, C.; Walther, T.; Farese, R. The triglyceride synthesis enzymes DGAT1 and DGAT2 have distinct and overlapping functions in adipocytes[S]. Journal of Lipid Research 2019, 60, 1112–1120. [Google Scholar] [CrossRef]
- Yu, T.-Y.; Tian, X.; Li, D.; He, Y.; Yang, P.; Cheng, Y.; Zhao, X.; Sun, J.; Yang, G. Transcriptome, proteome and metabolome analysis provide insights on fat deposition and meat quality in pig. Food research international 2023, 166, 112550. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Cao, S.; Yang, L.; Li, Z. Flavor formation based on lipid in meat and meat products: A review. Journal of food biochemistry 2022, 46, 14439. [Google Scholar] [CrossRef]
- Wu, F.; Chen, Z.; Zhang, Z.; Wang, Z.; Zhang, Z.; Wang, Q.; Pan, Y. The role of SOCS3 in regulating meat quality in Jinhua pigs. International Journal of Molecular Sciences 2023, 24, 10593. [Google Scholar] [CrossRef]
- Pothakam, N.; Supakankul, P.; Norseeda, W.; Liu, G.; Teltathum, T.; Naraballobh, W.; Khamlor, T.; Sringarm, K.; Mekchay, S. Association of adipocytokine IL-1A and IL-6 genes with intramuscular fat content and fatty acid composition in pigs. Meat science 2021, 179, 108554. [Google Scholar] [CrossRef]
- Gu, X.; Chang, X.; Yang, L.; Chamba, Y.; Geng, F. Quantitative proteomic analysis of Tibetan pig livers at different altitudes. Molecules 2023, 28, 1694. [Google Scholar] [CrossRef]
- Qiu, Y.; Gan, M.; Wang, X.; Liao, T.; Chen, Q.; Lei, Y.; Chen, L.; Wang, J.; Zhao, Y.; Niu, L.; et al. The global perspective on peroxisome proliferator-activated receptor γ (PPARγ) in ectopic fat deposition: A review. International journal of biological macromolecules 2023, 127042. [Google Scholar] [CrossRef]
- Janani, C.; Kumari, R. PPAR gamma gene--A review. Diabetes & metabolic syndrome 2015, 9, 46–50. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.-L.; Wu, L.; Guo, Y.; Yang, G.; Li, X.; Shi, X.e. Rosiglitazone-induced PPARγ activation promotes intramuscular adipocyte adipogenesis of pig. Animal Biotechnology 2023, 34, 3708–3717. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Zhou, Y.; Yang, J.; Li, J.; Peng, Y.; Zhang, X.; Miao, Y.; Jiang, W.; Bu, G.; Hou, L.; et al. Targeted overexpression of PPARγ in skeletal muscle by random insertion and CRISPR/Cas9 transgenic pig cloning enhances oxidative fiber formation and intramuscular fat deposition. The FASEB Journal 2021, 35, e21308. [Google Scholar] [CrossRef] [PubMed]
- Commins, S.; Borish, L.; Steinke, J. Immunologic messenger molecules: cytokines, interferons, and chemokines. The Journal of allergy and clinical immunology 2010, 125, S53–S72. [Google Scholar] [CrossRef] [PubMed]
- Blanc, R.; Kallenbach, J.; Bachman, J.; Mitchell, A.; Paris, N.; Chakkalakal, J. Inhibition of inflammatory CCR2 signaling promotes aged muscle regeneration and strength recovery after injury. Nature Communications 2020, 11, 4167. [Google Scholar] [CrossRef]
- Ren, Y.; Wang, F.; Sun, R.; Zheng, X.; Liu, Y.; Lin, Y.; Hong, L.; Huang, X.; Chao, Z. The genetic selection of HSPD1 and HSPE1 reduce inflammation of liver and spleen while restraining the growth and development of skeletal muscle in Wuzhishan pigs. Animals 2024, 14, 174. [Google Scholar] [CrossRef]
- Nara, H.; Watanabe, R. Anti-inflammatory effect of muscle-derived interleukin-6 and its involvement in lipid metabolism. International Journal of Molecular Sciences 2021, 22, 9889. [Google Scholar] [CrossRef]
- Wang, W.; Li, W.; Liu, W.; Wang, Z.; Xie, B.; Yang, X.; Tang, Z. Exploring multi-tissue alternative splicing and skeletal muscle metabolism regulation in obese- and lean-type pigs. Genes 2024, 15, 196. [Google Scholar] [CrossRef]
| Gene_name | Gene_id | CX | LAN | Log2FC | Adjusted p-value |
| AP1S2 | ENSSSCG00045031746 | 414.59 | 41.03 | 3.33 | 1.00e-32 |
| FCGR3A | ENSSSCG00045036350 | 264.93 | 50.60 | 2.39 | 1.05e-19 |
| SLA-DQA | ENSSSCG00045011704 | 749.70 | 105.10 | 2.83 | 1.59e-16 |
| CD37 | ENSSSCG00045013461 | 47.82 | 8.73 | 2.45 | 7.50e-16 |
| EMP2 | ENSSSCG00045000984 | 477.11 | 134.64 | 1.82 | 1.40e-14 |
| ARMCX2 | ENSSSCG00045011080 | 47.47 | 7.32 | 2.67 | 1.55e-14 |
| CHPT1 | ENSSSCG00045032049 | 2151.92 | 928.29 | 1.21 | 2.16e-14 |
| C5AR1 | ENSSSCG00045034745 | 102.66 | 14.72 | 2.80 | 4.14e-14 |
| DNASE1L3 | ENSSSCG00045035470 | 49.55 | 3.38 | 3.88 | 2.36e-13 |
| CD53 | ENSSSCG00045035517 | 183.79 | 36.60 | 2.33 | 5.59e-13 |
| Gene_name | Gene_id | CX | LAN | Log2FC | Adjusted p-value |
| VPS72 | ENSSSCG00045022741 | 471.19 | 1721.26 | -1.86 | 6.47e-24 |
| UBL5 | ENSSSCG00045039454 | 207.27 | 1137.93 | -2.45 | 6.84e-21 |
| GDE1 | ENSSSCG00045015707 | 616.04 | 1837.79 | -1.57 | 7.50e-21 |
| AQP7 | ENSSSCG00045001378 | 41.03 | 217.30 | -2.40 | 1.49e-19 |
| CUL1 | ENSSSCG00045021962 | 456.96 | 949.08 | -1.05 | 7.38e-19 |
| DUSP27 | ENSSSCG00045009424 | 740.94 | 2549.34 | -1.78 | 1.46e-18 |
| LYSMD2 | ENSSSCG00045031255 | 103.88 | 403.52 | -1.95 | 1.26e-17 |
| DDX24 | ENSSSCG00045022939 | 150.26 | 411.68 | -1.45 | 2.73e-17 |
| LRRC42 | ENSSSCG00045022986 | 321.70 | 889.54 | -1.46 | 3.18e-17 |
| PDZD9 | ENSSSCG00045028818 | 181.87 | 1576.16 | -3.11 | 9.77e-17 |
| Gene name | CX | LAN | Fold change (CX/LAN) | Log2FC | p-value | RNA-Seq Log2FC | Consistency |
| AP1S2 | 9.87±1.45 | 1.06±0.21 | 9.31 | 3.22 | <0.001 | 3.33 | 96.70% |
| FCGR3A | 5.64±0.82 | 1.03±0.18 | 5.48 | 2.45 | <0.001 | 2.39 | 97.50% |
| SLA-DQA | 7.42±1.16 | 1.05±0.22 | 7.07 | 2.82 | <0.001 | 2.83 | 99.60% |
| CD37 | 5.32±0.94 | 1.04±0.19 | 5.12 | 2.35 | <0.001 | 2.45 | 95.90% |
| EMP2 | 3.38±0.56 | 0.97±0.15 | 3.48 | 1.8 | <0.001 | 1.82 | 98.90% |
| VPS72 | 0.26±0.04 | 0.95±0.17 | 0.27 | -1.89 | <0.001 | -1.86 | 98.40% |
| UBL5 | 0.17±0.03 | 0.96±0.18 | 0.18 | -2.47 | <0.001 | -2.45 | 99.20% |
| GDE1 | 0.34±0.05 | 0.98±0.20 | 0.35 | -1.51 | <0.001 | -1.57 | 96.20% |
| AQP7 | 0.19±0.04 | 1.02±0.21 | 0.19 | -2.39 | <0.001 | -2.40 | 99.60% |
| CUL1 | 0.48±0.07 | 0.99±0.16 | 0.48 | -1.06 | <0.001 | -1.05 | 99.00% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




