Submitted:
23 August 2023
Posted:
24 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Identification and physicochemical properties of RRM1 gene family members in rice
2.2. Chromosome location of OsRRM1 gene family and construction of phylogenetic tree
2.3. Analysis of conserved domain, gene structure and motif of OsRRM1 gene family
2.4. Interspecies collinearity analysis of OsRRM1 gene family
2.5. GO and KEGG analysis of OsRRM1 gene family
2.6. Analysis of presumptive cis-regulatory elements in the promoter region of OsRRM1 gene
2.7. Expression pattern analysis of RRM1 gene in rice treated with blast fungus
2.8. Plant materials and rice blast stress treatment
2.9. Analysis of OsRRM1 gene expression by qRT-PCR
3. Results
3.1. Screening and identification of RRM1 gene family members in rice
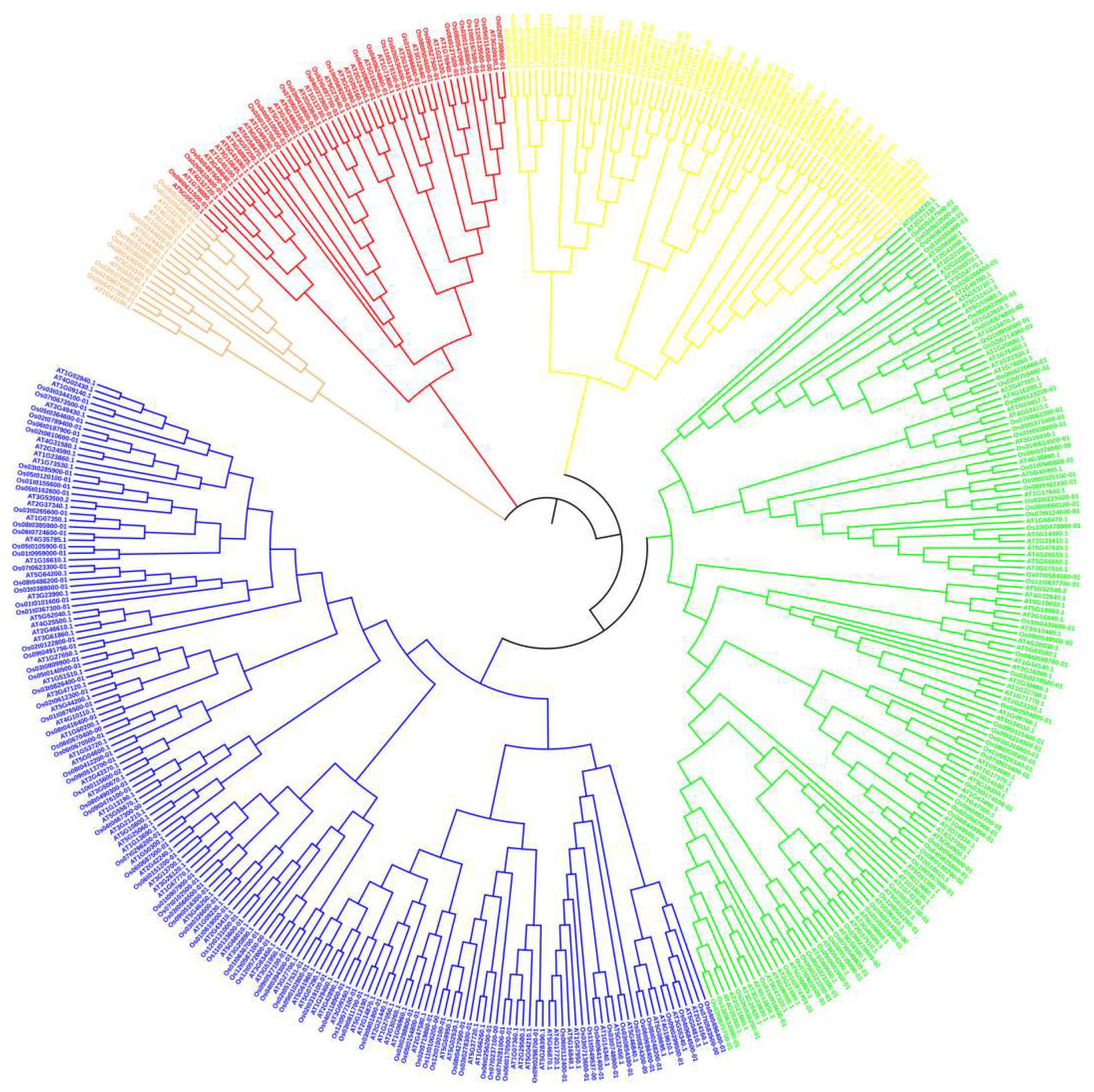
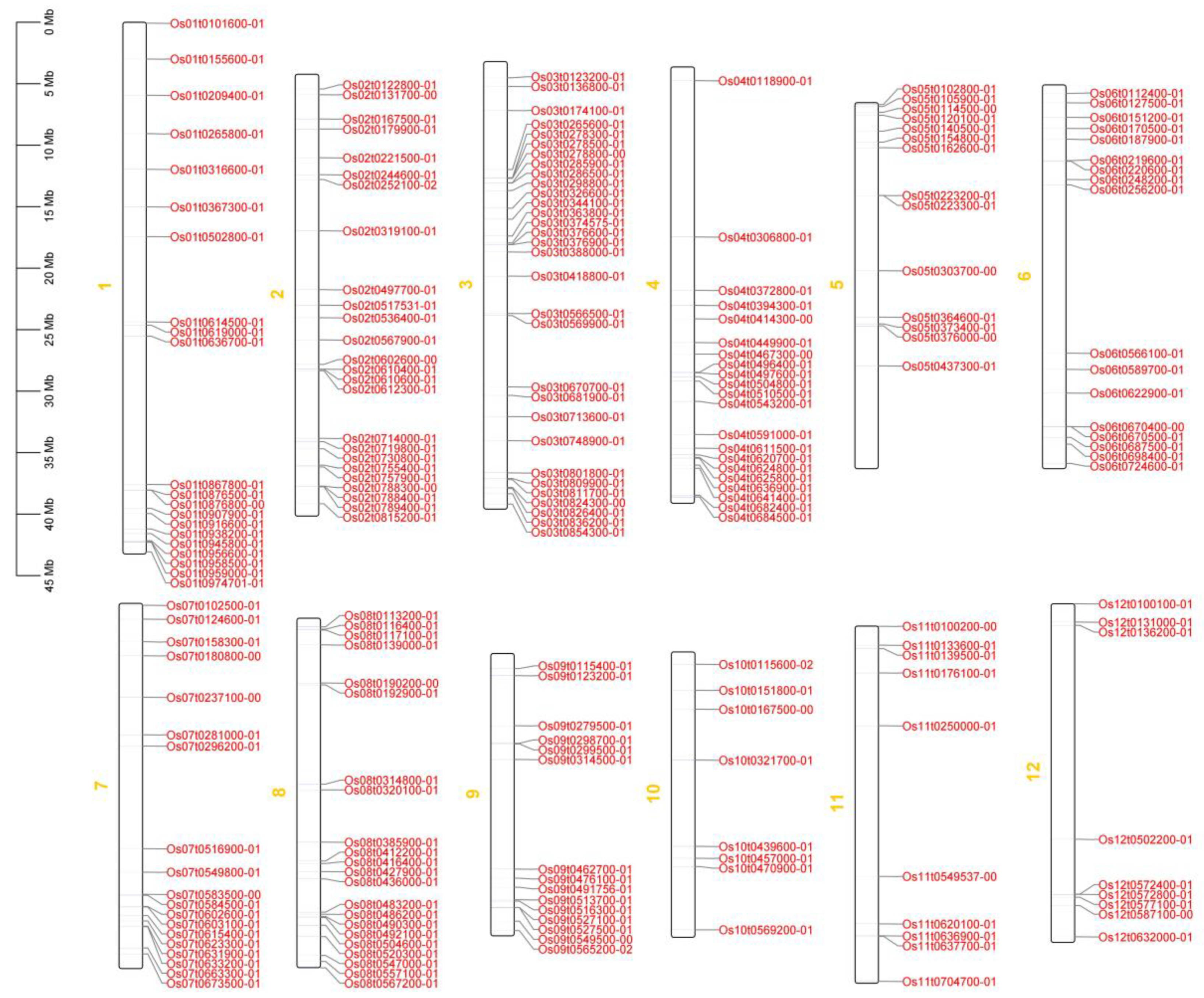
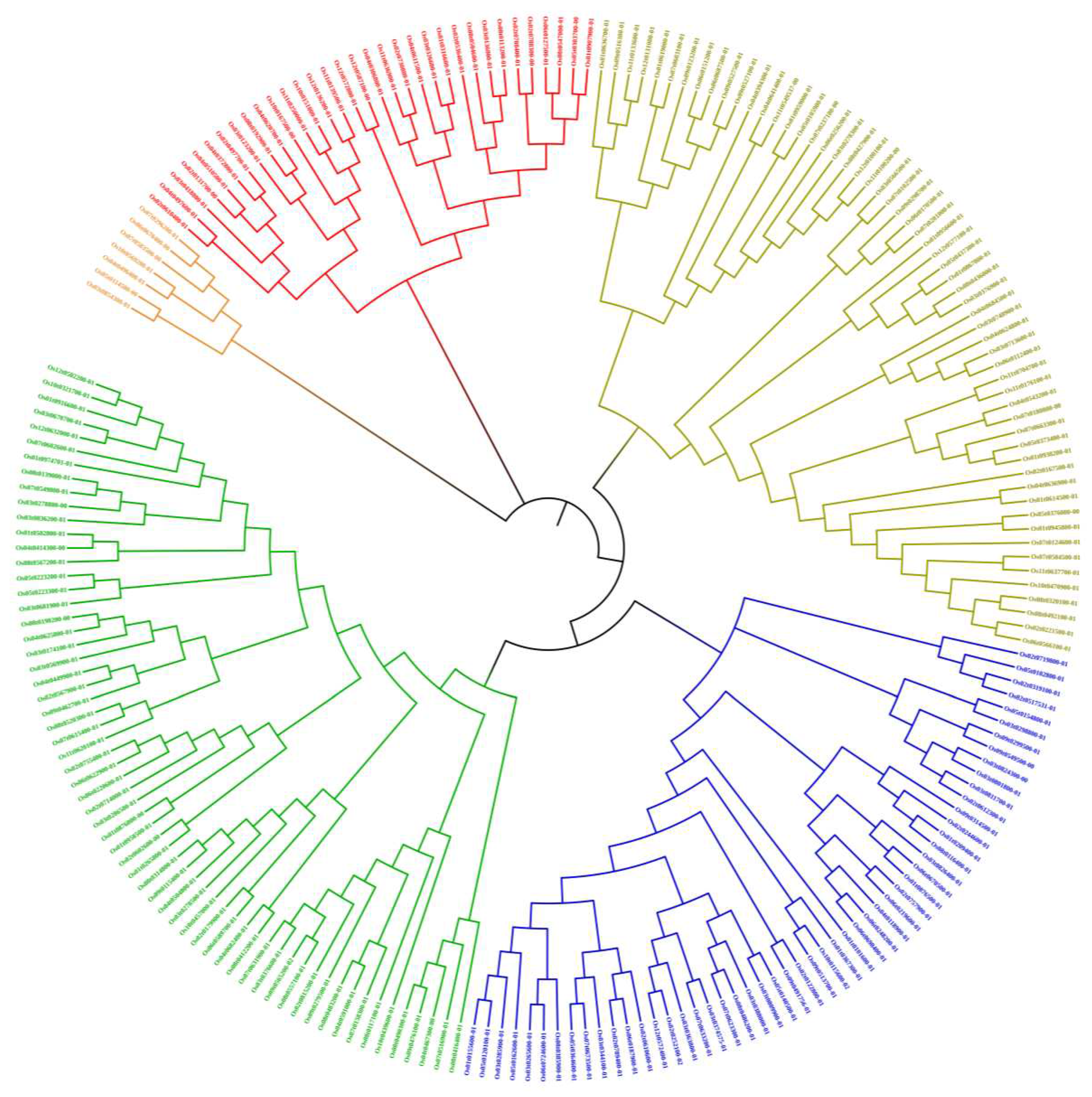
3.2. Chromosome localization and phylogenetic tree analysis of OsRRM1 gene family
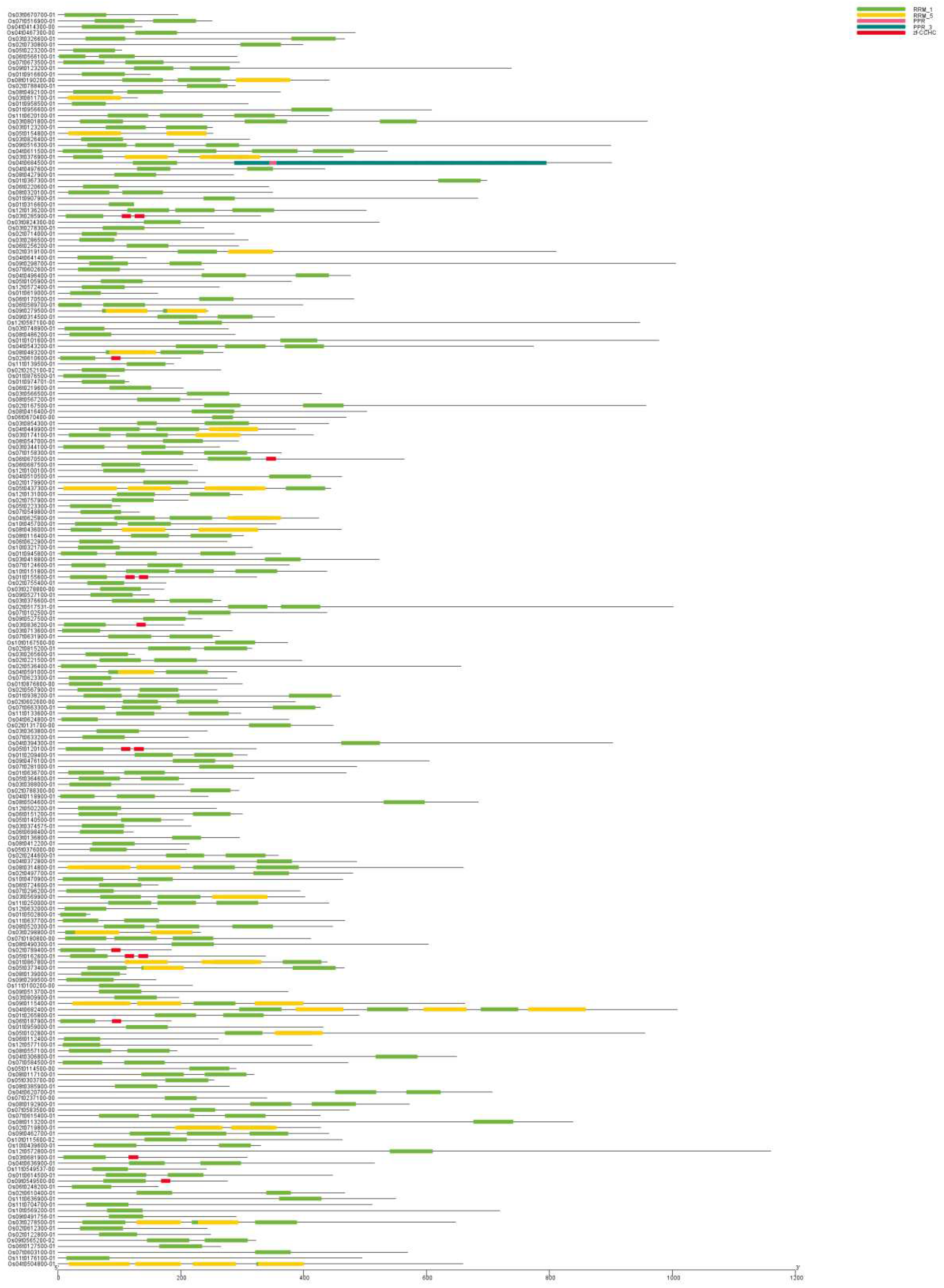
3.3. Motif analysis and gene structure analysis of OsRRM1 gene family
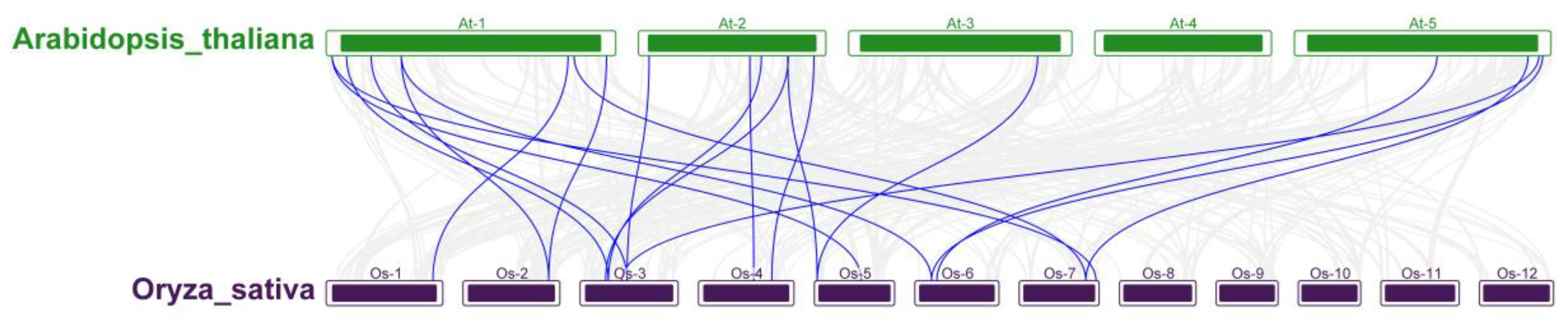
3.4. Evolutionary analysis of OsRRM1 gene family and collinearity analysis of RRM1 gene family between rice and Arabidopsis Thaliana
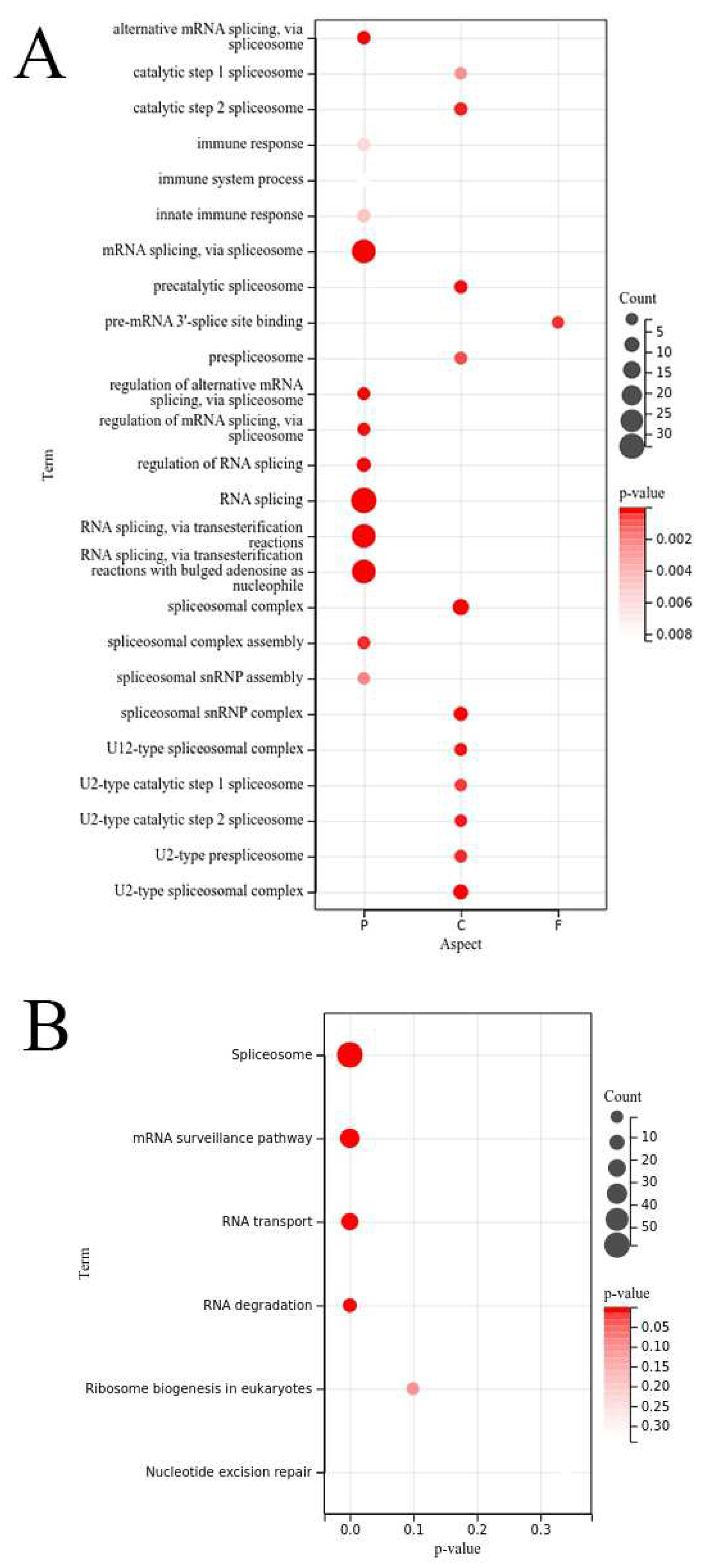
3.5. GO and KEGG analysis of OsRRM1 gene family
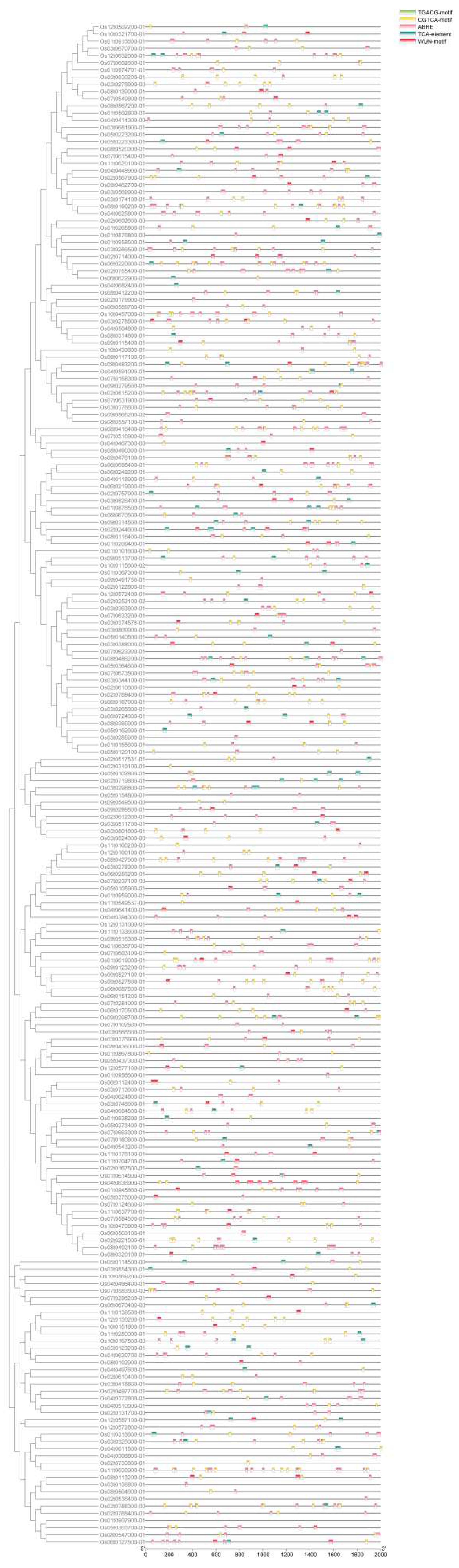
3.6. Characterization of presumptive cis-regulatory elements in the promoter region of OsRRM1 gene
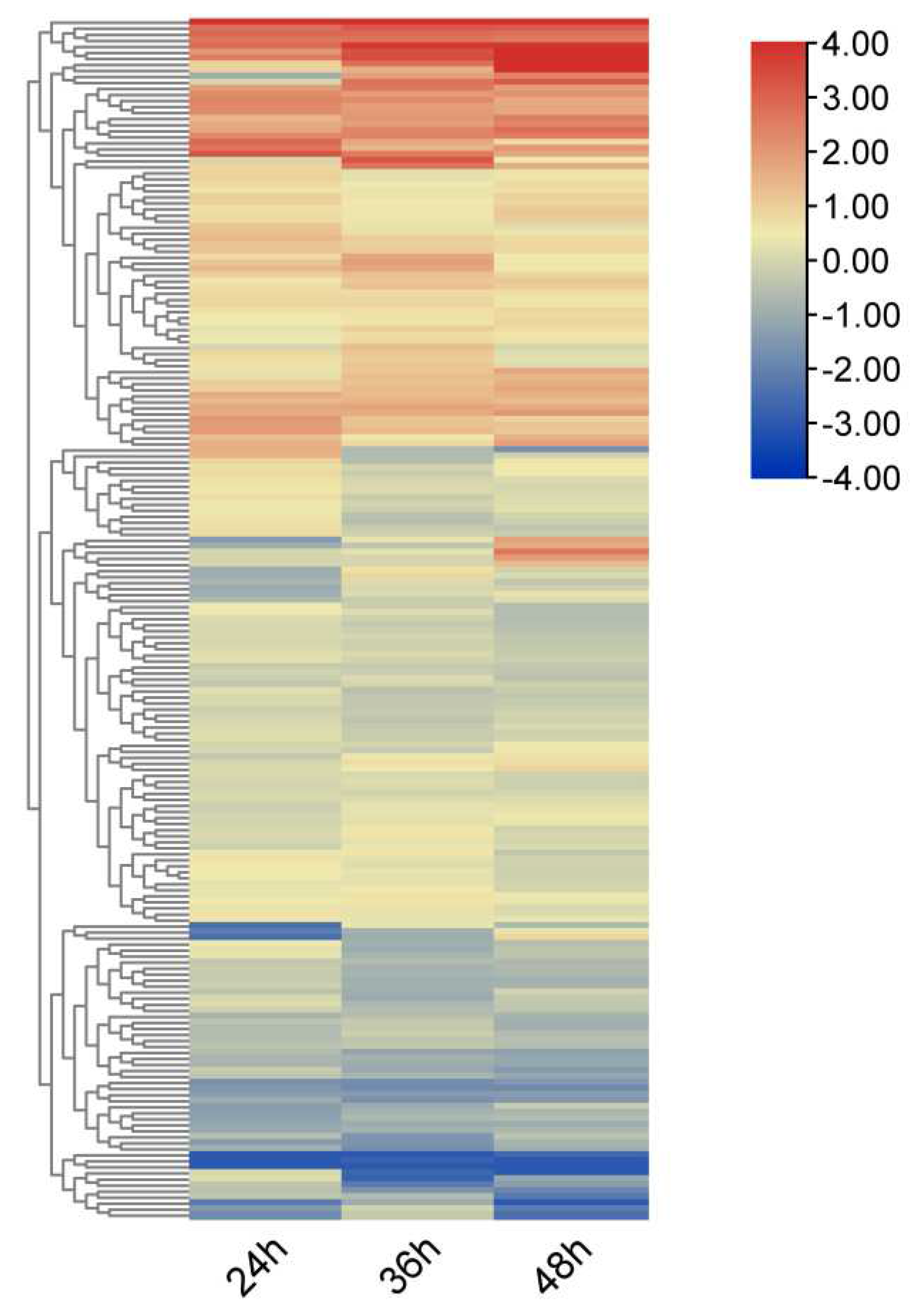
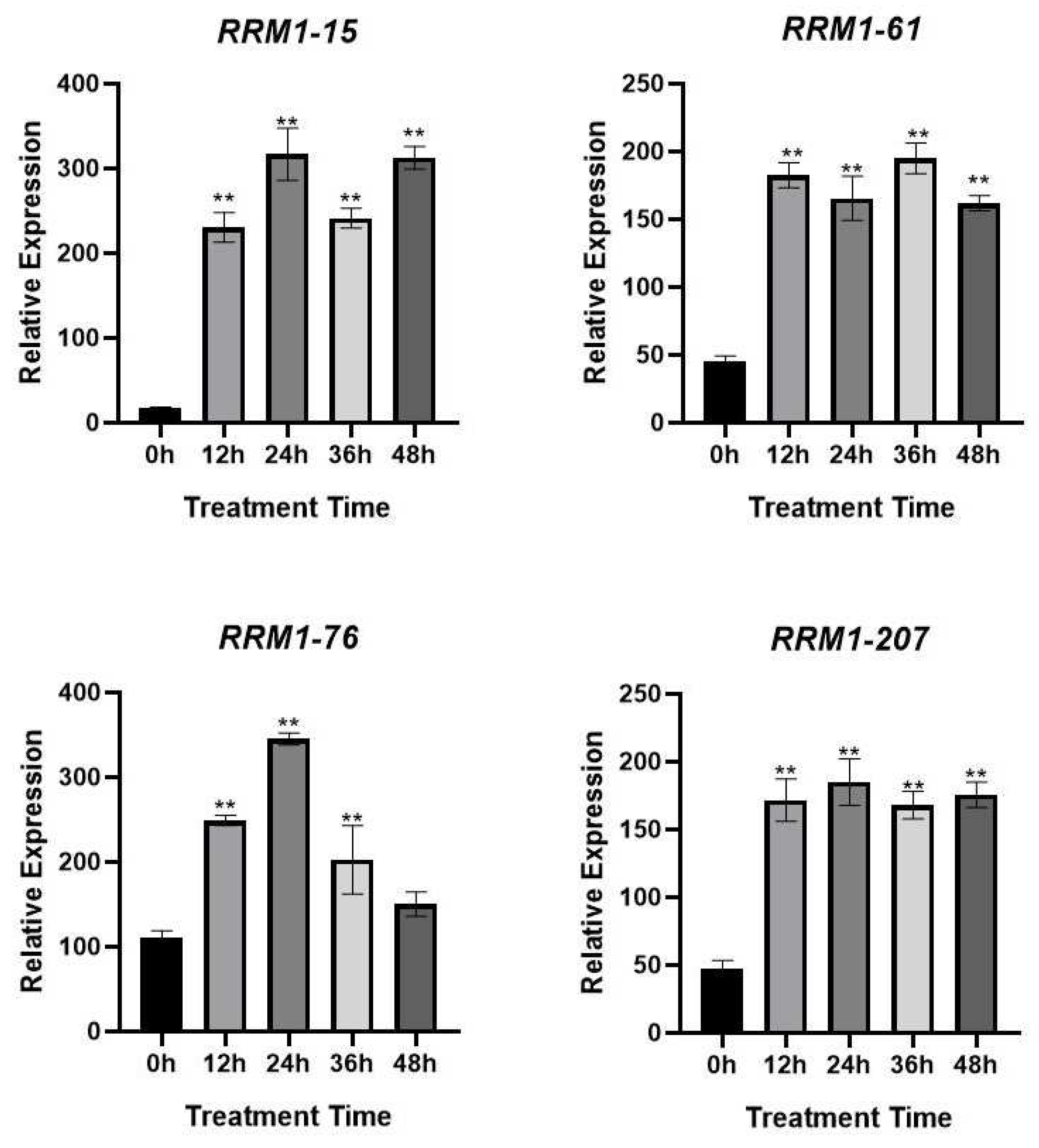
3.7. Expression pattern of RRM1 gene family in rice after treatment with blast fungus
3.8. Expression analysis of OsRRM1 gene in response to biological stress
4. Discussion
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
Appendix A
References
- Latchman, D.S. Transcriptional Gene Regulation in Eukaryotes. In Els, 2011.
- Jeune, E.L.; Ladurner, A.G. Book Review. Protein Science 2004, 13, 1950–1952. [Google Scholar] [CrossRef]
- Jackson, D.A.; Pombo, A.; Iborra, F. The Balance Sheet for Transcription: An Analysis of Nuclear Rna Metabolism in Mammalian Cells. Faseb j 2000, 14, 242–254. [Google Scholar] [CrossRef] [PubMed]
- Lorković, Z.J.; Barta, A. Genome Analysis: Rna Recognition Motif (Rrm) and K Homology (Kh) Domain Rna-Binding Proteins from the Flowering Plant Arabidopsis Thaliana. Nucleic Acids Res 2002, 30, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Burd, C.G.; Dreyfuss, G. Conserved Structures and Diversity of Functions of Rna-Binding Proteins. Science 1994, 265, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Dreyfuss, G.; Swanson, M.S.; Piñol-Roma, S. Heterogeneous Nuclear Ribonucleoprotein Particles and the Pathway of Mrna Formation. Trends in Biochemical Sciences 1988, 13, 86–91. [Google Scholar] [CrossRef]
- Adam, S.A.; Nakagawa, T.; Swanson, M.S.; Woodruff, T.K.; Dreyfuss, G. Mrna Polyadenylate-Binding Protein: Gene Isolation and Sequencing and Identification of a Ribonucleoprotein Consensus Sequence. Mol Cell Biol 1986, 6, 2932–2943. [Google Scholar] [PubMed]
- Bandziulis, R.J.; Swanson, M.S.; Dreyfuss, G. Rna-Binding Proteins as Developmental Regulators. Genes Dev 1989, 3, 431–437. [Google Scholar] [CrossRef]
- Dreyfuss, G.; Kim, V.N.; Kataoka, N. Messenger-Rna-Binding Proteins and the Messages They Carry. Nat Rev Mol Cell Biol 2002, 3, 195–205. [Google Scholar] [CrossRef]
- Gomes, J.-E.; Encalada, S.E.; Swan, K.A.; Shelton, C.A.; Carter, J.C.; Bowerman, B. The Maternal Gene Spn-4 Encodes a Predicted Rrm Protein Required for Mitotic Spindle Orientation and Cell Fate Patterning in Early C. Elegans Embryos. Development 2021, 21, 4301–4314. [Google Scholar]
- Zhan, X.; Qian, B.; Cao, F.; Wu, W.; Yang, L.; Guan, Q.; Gu, X.; Wang, P.; Okusolubo, T.A.; Dunn, S.L.; Zhu, J.K.; Zhu, J. An Arabidopsis Pwi and Rrm Motif-Containing Protein Is Critical for Pre-Mrna Splicing and Aba Responses. Nat Commun 2015, 6, 8139. [Google Scholar] [CrossRef]
- Paukku, K.; Backlund, M.; De Boer, R.A.; Kalkkinen, N.; Kontula, K.K.; Lehtonen, J.Y. Regulation of At1r Expression through Hur by Insulin. Nucleic Acids Res 2012, 40, 5250–5261. [Google Scholar] [CrossRef] [PubMed]
- O’Bryan, M.K.; Clark, B.J.; McLaughlin, E.A.; D’Sylva, R.J.; O’Donnell, L.; Wilce, J.A.; Sutherland, J.; O’Connor, A.E.; Whittle, B.; Goodnow, C.C.; Ormandy, C.J.; Jamsai, D. Rbm5 Is a Male Germ Cell Splicing Factor and Is Required for Spermatid Differentiation and Male Fertility. PLoS Genet 2013, 9, e1003628. [Google Scholar] [CrossRef] [PubMed]
- Maris, C.; Dominguez, C.; Allain, F.H. The Rna Recognition Motif, a Plastic Rna-Binding Platform to Regulate Post-Transcriptional Gene Expression. Febs j 2005, 272, 2118–2131. [Google Scholar] [CrossRef] [PubMed]
- Nagai, K.; Oubridge, C.; Jessen, T.H.; Li, J.; Evans, P.R. Crystal Structure of the Rna-Binding Domain of the U1 Small Nuclear Ribonucleoprotein A. Nature 1990, 348, 515–520. [Google Scholar] [CrossRef]
- Cléry, A.; Blatter, M.; Allain, F.H. Rna Recognition Motifs: Boring? Not Quite. Curr Opin Struct Biol 2008, 18, 290–298. [Google Scholar] [CrossRef]
- Birney, E.; Kumar, S.; Krainer, A.R. Analysis of the Rna-Recognition Motif and Rs and Rgg Domains: Conservation in Metazoan Pre-Mrna Splicing Factors. Nucleic Acids Research 1993, 21, 5803–5816. [Google Scholar] [CrossRef]
- Query, C.C.; Bentley, R.C.; Keene, J.D. A Common Rna Recognition Motif Identified within a Defined U1 Rna Binding Domain of the 70k U1 Snrnp Protein. Cell 1989, 57, 89–101. [Google Scholar] [CrossRef]
- Chambers, J.C.; Kenan, D.; Martin, B.J.; Keene, J.D. Genomic Structure and Amino Acid Sequence Domains of the Human La Autoantigen. J Biol Chem 1988, 263, 18043–18051. [Google Scholar] [CrossRef]
- 20. Sachs, Davis and Kornberg. A Single Domain of Yeast Poly(a)-Binding Protein Is Necessary and Sufficient for Rna Binding and Cell Viability. Mol.cell.biol.
- Kielkopf, C.L.; Lücke, S.; Green, M.R. U2af Homology Motifs: Protein Recognition in the Rrm World. Genes Dev 2004, 18, 1513–1526. [Google Scholar] [CrossRef]
- Zhai, K.; Deng, Y.; Liang, D.; Tang, J.; Liu, J.; Yan, B.; Yin, X.; Lin, H.; Chen, F.; Yang, D. Rrm Transcription Factors Interact with Nlrs and Regulate Broad-Spectrum Blast Resistance in Rice. Mol Cell 2019. [CrossRef]
- Jeon, J.; Lee, G.W.; Kim, K.T.; Park, S.Y.; Kim, S.; Kwon, S.; Huh, A.; Chung, H.; Lee, D.Y.; Kim, C.Y.; Lee, Y.H. Transcriptome Profiling of the Rice Blast Fungus Magnaporthe Oryzae and Its Host Oryza Sativa During Infection. Mol Plant Microbe Interact 2020, 33, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Manandhar, H.K.; Jørgensen, H.J.L.; Mathur, S.B.; Smedegaard-Petersen, V. Suppression of Rice Blast by Preinoculation with Avirulent Pyricularia Oryzae and the Nonrice Pathogen Bipolaris Sorokiniana. Phytopathology 1998, 7, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Nasir, F.; Tian, L.; Chang, C.; Li, X.; Gao, Y.; Tran, L.P.; Tian, C. Current Understanding of Pattern-Triggered Immunity and Hormone-Mediated Defense in Rice (Oryza Sativa) in Response to Magnaporthe Oryzae Infection. Semin Cell Dev Biol 2018, 83, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, H.; Urayama, S.I.; Higashiura, T.; Le, T.M.; Komatsu, K. Chrysoviruses in Magnaporthe Oryzae. Viruses 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Kim, D.S.; Hwang, B.K. The Pepper Rna-Binding Protein Carbp1 Functions in Hypersensitive Cell Death and Defense Signaling in the Cytoplasm. Plant J 2012, 72, 235–248. [Google Scholar] [CrossRef]
- Nina and F. J. C. O. i., P. Biology. Rna-Binding Proteins in Plants: The Tip of an Iceberg? 2002.
- Lorković, Z.J. Role of Plant Rna-Binding Proteins in Development, Stress Response and Genome Organization. 2009, 14, 229–236.
- Woloshen, V.; Huang, S.; Li, X. Review Article Rna-Binding Proteins in Plant Immunity. 2011.
- Zhai, K.; Deng, Y.; Liang, D.; Tang, J.; Liu, J.; Yan, B.; Yin, X.; Lin, H.; Chen, F.; Yang, D.; Xie, Z.; Liu, J.-Y.; Li, Q.; Zhang, L.; He, Z. Rrm Transcription Factors Interact with Nlrs and Regulate Broad-Spectrum Blast Resistance in Rice. Mol Cell 2019, 74, 996–1009. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. Tbtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Molecular Plant 2020, 13. [Google Scholar] [CrossRef]
- Hu, Y.; Zhu, N.; Wang, X.; Yi, Q.; Zhu, D.; Lai, Y.; Zhao, Y. Analysis of Rice Snf2 Family Proteins and Their Potential Roles in Epigenetic Regulation. Plant Physiol Biochem 2013, 70, 33–42. [Google Scholar] [CrossRef]
- Mimura, M.; Itoh, J. Genetic Interaction between Rice Plastochron Genes and the Gibberellin Pathway in Leaf Development. Rice (N Y) 2014, 7, 25. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Itoh, J.-I.; Miyoshi, K.; Kurata, N.; Alvarez, N.; Veit, B.; Nagato, Y. Plastochron2 Regulates Leaf Initiation and Maturation in Rice. The Plant Cell 2006, 18, 612–625. [Google Scholar] [CrossRef]
- Xiong, G.S.; Hu, X.M.; Jiao, Y.Q.; Yu, Y.C.; Chu, C.C.; Li, J.Y.; Qian, Q.; Wang, Y.H. Leafy Head2, Which Encodes a Putative Rna-Binding Protein, Regulates Shoot Development of Rice. Cell Res 2006, 16, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Lyu, J.; Wang, D.; Duan, P.; Liu, Y.; Huang, K.; Zeng, D.; Zhang, L.; Dong, G.; Li, Y.; Xu, R.; Zhang, B.; Huang, X.; Li, N.; Wang, Y.; Qian, Q.; Li, Y. . Control of Grain Size and Weight by the Gsk2-Large1/Oml4 Pathway in Rice. Plant Cell 2020, 32, 1905–1918. [Google Scholar] [CrossRef]
- Isshiki, M.; Matsuda, Y.; Takasaki, A.; Wong, H.L.; Satoh, H.; Shimamoto, K.J.P.B. Du3, a Mrna Cap-Binding Protein Gene, Regulates Amylose Content in Japonica Rice Seeds. Plant Biotechnology 2008. [CrossRef]
- Yano, M.; Okuno, K.; Satoh, H.; Omura, T. Chromosomal Location of Genes Conditioning Low Amylose Content of Endosperm Starches in Rice, Oryza Sativa L. Theor Appl Genet 1988, 76, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Zhai, K.; Deng, Y.; Liang, D.; Tang, J.; Liu, J.; Yan, B.; Yin, X.; Lin, H.; Chen, F.; Yang, D.; Xie, Z.; Liu, J.Y.; Li, Q.; Zhang, L.; He, Z. Rrm Transcription Factors Interact with Nlrs and Regulate Broad-Spectrum Blast Resistance in Rice. Mol Cell 2019, 74, 996–1009. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, X.; Zhang, H.; Cheng, Z.; Liu, J.; Zhou, C.; Luo, S.; Luo, W.; Li, S.; Xing, X.; Chang, Y.; Shi, C.; Ren, Y.; Zhu, S.; Lei, C.; Guo, X.; Wang, J.; Zhao, Z.; Wang, H.; Zhai, H.; Lin, Q.; Wan, J. Dwarf and High Tillering1 Represses Rice Tillering through Mediating the Splicing of D14 Pre-Mrna. Plant Cell 2022, 34, 3301–3318. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lan, J.; Huang, Y.; Cao, P.; Zhou, C.; Ren, Y.; He, N.; Liu, S.; Tian, Y.; Nguyen, T.; Jiang, L.; Wan, J. Wsl5, a Pentatricopeptide Repeat Protein, Is Essential for Chloroplast Biogenesis in Rice under Cold Stress. J Exp Bot 2018, 69, 3949–3961. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Ding, S.; Reiland, S.; Rödiger, A.; Roschitzki, B.; Xue, P.; Gruissem, W.; Lu, C.; Baginsky, S. Identification and Characterization of Chloroplast Casein Kinase Ii from Oryza Sativa (Rice). J Exp Bot 2015, 66, 175–187. [Google Scholar] [CrossRef]
- Conservation and Divergence of Fca Function between Arabidopsis and Rice. Plant Molecular Biology 2005, 58, 823–838. [CrossRef]
- Chen, S.Y.; Wang, Z.Y.; Cai, X.L. Osrrm, a Spen-Like Rice Gene Expressed Specifically in the Endosperm. Cell Res 2007, 17, 713–721. [Google Scholar] [CrossRef]
- Chen, S.-Y.; Wang, Z.-Y.; Cai, X.-L. Osrrm, a Spen-Like Rice Gene Expressed Specifically in the Endosperm. Cell Res 2007, 17, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Kwak, K.J.; Jung, H.J.; Lee, K.H.; Kim, Y.S.; Kim, W.Y.; Ahn, S.J.; Kang, H. The Minor Spliceosomal Protein U11/U12-31k Is an Rna Chaperone Crucial for U12 Intron Splicing and the Development of Dicot and Monocot Plants. PLoS One 2012, 7, e43707. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Vega-Sánchez, M.E.; Park, C.H.; Bellizzi, M.; Guo, Z.; Wang, G.L. Rbs1, an Rna Binding Protein, Interacts with Spin1 and Is Involved in Flowering Time Control in Rice. PLoS One 2014, 9, e87258. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kaur, C.; Singla-Pareek, S.L.; Sopory, S.K. Ossro1a Interacts with Rna Binding Domain-Containing Protein (Osrbd1) and Functions in Abiotic Stress Tolerance in Yeast. Front Plant Sci 2016, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Ren, L.; Zhao, Y.; You, H.; Zhou, Y.; Tang, D.; Du, G.; Shen, Y.; Li, Y.; Cheng, Z. Reproductive Cells and Peripheral Parietal Cells Collaboratively Participate in Meiotic Fate Acquisition in Rice Anthers. Plant J 2021, 108, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, S.; Sato, Y.; Asano, T.; Nagamura, Y.; Nonomura, K. Rice Mel2, the Rna Recognition Motif (Rrm) Protein, Binds in Vitro to Meiosis-Expressed Genes Containing U-Rich Rna Consensus Sequences in the 3’-Utr. Plant Mol Biol 2015, 89, 293–307. [Google Scholar] [CrossRef]
- Nonomura, K.; Eiguchi, M.; Nakano, M.; Takashima, K.; Komeda, N.; Fukuchi, S.; Miyazaki, S.; Miyao, A.; Hirochika, H.; Kurata, N. A Novel Rna-Recognition-Motif Protein Is Required for Premeiotic G1/S-Phase Transition in Rice (Oryza Sativa L.). PLoS Genet 2011, 7, e1001265. [Google Scholar] [CrossRef]
- Sahi, C.; Agarwal, M.; Singh, A.; Grover, A. Molecular Characterization of a Novel Isoform of Rice (Oryza Sativa L.) Glycine Rich-Rna Binding Protein and Evidence for Its Involvement in High Temperature Stress Response. Plant Science 2007, 173, 144–155. [Google Scholar] [CrossRef]
- Cléry, A.; Blatter, M.; Allain, F.H.T. Rna Recognition Motifs: Boring? Not Quite. Curr Opin Struct Biol 2008, 18, 290–298. [Google Scholar] [CrossRef]
- Lorković, Z.J.; Barta, A. Genome Analysis: Rna Recognition Motif (Rrm) and K Homology (Kh) Domain Rna-Binding Proteins from the Flowering Plant Arabidopsis Thaliana. Nucleic Acids Research 2002, 30, 623–635. [Google Scholar] [CrossRef]
- Shi, X.; Hanson, M.R.; Bentolila, S. Functional Diversity of Arabidopsis Organelle-Localized Rna-Recognition Motif-Containing Proteins. Wiley Interdiscip Rev Rna 2017, e1420. [Google Scholar] [CrossRef]
- Shi, X.; Hanson, M.R.; Bentolila, S. Functional Diversity of Arabidopsis Organelle-Localized Rna-Recognition Motif-Containing Proteins. Wiley Interdiscip Rev Rna 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Qian, B.; Cao, F.; Wu, W.; Yang, L.; Guan, Q.; Gu, X.; Wang, P.; Okusolubo, T.A.; Dunn, S.L.; Zhu, J.-K.; Zhu, J. An Arabidopsis Pwi and Rrm Motif-Containing Protein Is Critical for Pre-Mrna Splicing and Aba Responses. Nat Commun 2015, 6, 8139. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, S.J.; Jang, B.; Jung, C.H.; Ahn, S.-J.; Goh, C.H.; Cho, K.; Han, O.; Kang, H. Functional Characterization of a Glycine-Rich Rna-Binding Protein 2 in Arabidopsis Thaliana under Abiotic Stress Conditions. The Plant journal 2007, 3, 439–451. [Google Scholar] [CrossRef]
- Xu, T.; Lee, K.; Gu, L.; Kim, J.-I.; Kang, H. Functional Characterization of a Plastid-Specific Ribosomal Protein Psrp2 in Arabidopsis Thaliana under Abiotic Stress Conditions. Plant Physiology and Biochemistry 2013, 73, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Cold Shock Domain Proteins and Glycine-Rich Rna-Binding Proteins from Arabidopsis Thaliana Can Promote the Cold Adaptation Process in Escherichia Coli. Nucleic Acids Research 2007.
- Wang, S.; Bai, G.; Wang, S.; Yang, L.; Yang, F.; Wang, Y.; Zhu, J.K.; Hua, J. Chloroplast Rna-Binding Protein Rbd1 Promotes Chilling Tolerance through 23s Rrna Processing in Arabidopsis. PLoS Genet 2016, 12, e1006027. [Google Scholar] [CrossRef]
- Shi, X.; Germain, A.; Hanson, M.R.; Bentolila, S. Rna Recognition Motif-Containing Protein Orrm4 Broadly Affects Mitochondrial Rna Editing and Impacts Plant Development and Flowering. Plant Physiol 2016, 170, 294–309. [Google Scholar] [CrossRef]
- Vermel, M.; Guermann, B.; Delage, L.; Grienenberger, J.M.; Maréchal-Drouard, L.; Gualberto, J.M. A Family of Rrm-Type Rna-Binding Proteins Specific to Plant Mitochondria. Proc Natl Acad Sci U S A 2002, 99, 5866–5871. [Google Scholar] [CrossRef]
- Kwak, K.J.; Kim, Y.O.; Kang, H. Characterization of Transgenic Arabidopsis Plants Overexpressing Gr-Rbp4 under High Salinity, Dehydration, or Cold Stress. J Exp Bot 2005, 56, 3007–3016. [Google Scholar] [CrossRef]
- Lorković, Z.J. Role of Plant Rna-Binding Proteins in Development, Stress Response and Genome Organization. Trends Plant Sci 2009, 14, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Shuai, *!!! REPLACE !!!*; Wan, *!!! REPLACE !!!*; Shu, *!!! REPLACE !!!*; Wan, *!!! REPLACE !!!*; Leiyun, *!!! REPLACE !!!*; Yang, *!!! REPLACE !!!*; Fen, *!!! REPLACE !!!*; Genetics, Y.J.P. Shuai; Wan; Shu; Wan; Leiyun; Yang; Fen; Genetics, Y.J.P. Chloroplast Rna-Binding Protein Rbd1 Promotes Chilling Tolerance through 23s Rrna Processing in Arabidopsis. 2016.

| Gene | F-primer | R-primer |
|---|---|---|
| RRM1-15 | GGATGTGACTGAAGCTCGGGTGATC | GGATGTGACTGAAGCTCGGGTGATC |
| RRM1-61 | GGAGGTCTTGGAAGCCAAGGTCATC | CCATCCATGTCAGCGCCATCAAG |
| RRM1-76 | CACTGAAGCAAAGGTGGTTTTTGAC | GAGCTTTATCGACAGTGATCGCC |
| RRM1-207 | CTTGGATGGAAAGGATCTCGATGG | CATAGCCACCGCCTCCATAG |
| Actin | CCAATCGTGAGAAGATGACCCA | CCATCAGGAAGCTCGTAGCTCT |
| Gene | RAP | Number of Amino Acid | Molecular Weight | pI | subcellular localization |
|---|---|---|---|---|---|
| RRM1-1 | Os01g0101600 | 978 | 106323.33 | 9.56 | nucleus |
| RRM1-2 | Os01g0155600 | 324 | 36893.16 | 11.27 | nucleus |
| RRM1-3 | Os01g0209400 | 308 | 33656.38 | 8.94 | nucleus |
| RRM1-4 | Os01g0265800 | 490 | 49279.96 | 5.09 | nucleus |
| RRM1-5 | Os01g0316600 | 124 | 13965.3 | 9.91 | chloroplast |
| RRM1-6 | Os01g0367300 | 698 | 79763.67 | 10.57 | nucleus |
| RRM1-7 | Os01g0502800 | 53 | 5837.75 | 10.27 | chloroplast |
| RRM1-8 | Os01g0614500 | 447 | 44269.69 | 8.43 | nucleus |
| RRM1-9 | Os01g0619000 | 163 | 18016.54 | 5.76 | extracellular space |
| RRM1-10[33] | Os01g0636700 | 469 | 52108.23 | 8.63 | nucleus |
| RRM1-11 | Os01g0867800 | 439 | 49316.8 | 6.46 | nucleus |
| RRM1-12 | Os01g0876500 | 100 | 11385 | 7.72 | chloroplast |
| RRM1-13 | Os01g0876800 | 300 | 31951.79 | 8.91 | extracellular space |
| RRM1-14[34,35,36] | Os01g0907900 | 683 | 71779.19 | 6.37 | nucleus |
| RRM1-15 | Os01g0916600 | 150 | 15546.9 | 8.01 | chloroplast thylakoid lumen |
| RRM1-16 | Os01g0938200 | 460 | 48888.22 | 8.72 | nucleus |
| RRM1-17 | Os01g0945800 | 363 | 40073.63 | 6.61 | nucleus |
| RRM1-18 | Os01g0956600 | 608 | 68184.84 | 7.8 | nucleus |
| RRM1-19 | Os01g0958500 | 310 | 31802.78 | 8.34 | nucleus |
| RRM1-20 | Os01g0959000 | 432 | 48111.49 | 12.37 | chloroplast thylakoid lumen |
| RRM1-21 | Os01g0974701 | 116 | 12459.19 | 9.74 | mitochondrion |
| RRM1-22 | Os02g0122800 | 249 | 28953.55 | 10 | nucleus |
| RRM1-23 | Os02g0131700 | 448 | 49261.45 | 5.02 | nucleus |
| RRM1-24 | Os02g0167500 | 957 | 105767.07 | 7.88 | extracellular space |
| RRM1-25 | Os02g0179900 | 240 | 28105.95 | 8.85 | nucleus |
| RRM1-26 | Os02g0221500 | 397 | 40265.08 | 5.63 | nucleus |
| RRM1-27 | Os02g0244600 | 359 | 38737.53 | 5.62 | nucleus |
| RRM1-28 | Os02g0252100 | 265 | 30466.57 | 11.09 | nucleus |
| RRM1-29 | Os02g0319100 | 811 | 90295.23 | 6.27 | nucleus |
| RRM1-30 | Os02g0497700 | 480 | 50879.51 | 5.01 | nucleus |
| RRM1-31[37] | Os02g0517531 | 1001 | 110368.83 | 6.39 | nucleus |
| RRM1-32 | Os02g0536400 | 656 | 74812.42 | 9.43 | nucleus |
| RRM1-33 | Os02g0567900 | 259 | 28284.5 | 9.18 | nucleus |
| RRM1-34 | Os02g0602600 | 386 | 41584.82 | 7.67 | nucleus |
| RRM1-35 | Os02g0610400 | 467 | 51689.83 | 5.56 | nucleus |
| RRM1-36 | Os02g0610600 | 200 | 22797.31 | 11.33 | nucleus |
| RRM1-37[38,39] | Os02g0612300 | 243 | 28573.22 | 5.44 | chloroplast |
| RRM1-38 | Os02g0714000 | 287 | 30609.94 | 9.32 | nucleus |
| RRM1-39 | Os02g0719800 | 428 | 47331.12 | 5.57 | nucleus |
| RRM1-40 | Os02g0730800 | 399 | 43547.8 | 6.15 | extracellular space |
| RRM1-41 | Os02g0755400 | 176 | 18512.61 | 9.99 | mitochondrion |
| RRM1-42 | Os02g0757900 | 212 | 24083.82 | 5.07 | nucleus |
| RRM1-43 | Os02g0788300 | 295 | 32235.18 | 7.72 | nucleus |
| RRM1-44 | Os02g0788400 | 289 | 32009.09 | 8.66 | nucleus |
| RRM1-45 | Os02g0789400 | 185 | 21023.33 | 11.24 | nucleus |
| RRM1-46 | Os02g0815200 | 316 | 34612.01 | 5.17 | chloroplast thylakoid lumen |
| RRM1-47 | Os03g0123200 | 252 | 28108.69 | 7.64 | nucleus |
| RRM1-48 | Os03g0136800 | 296 | 32305.94 | 9.02 | nucleus |
| RRM1-49 | Os03g0174100 | 416 | 46056.33 | 5.35 | nucleus |
| RRM1-50 | Os03g0265600 | 125 | 13993.55 | 7.86 | chloroplast |
| RRM1-51 | Os03g0278300 | 238 | 24720.42 | 9.83 | chloroplast |
| RRM1-52 | Os03g0278500 | 647 | 72627.76 | 8.43 | nucleus |
| RRM1-53 | Os03g0278800 | 173 | 18433.86 | 9.3 | chloroplast outer membrane |
| RRM1-54 | Os03g0285900 | 330 | 37042.2 | 11 | nucleus |
| RRM1-55 | Os03g0286500 | 310 | 32704.09 | 9 | extracellular space |
| RRM1-56 | Os03g0298800 | 232 | 26100.86 | 9.44 | chloroplast |
| RRM1-57 | Os03g0326600 | 467 | 51073.78 | 9.06 | nucleus |
| RRM1-58 | Os03g0344100 | 264 | 29782.1 | 10.08 | nucleus |
| RRM1-59 | Os03g0363800 | 243 | 27781.69 | 10.83 | nucleus |
| RRM1-60 | Os03g0374575 | 217 | 25589.48 | 11.17 | nucleus |
| RRM1-61 | Os03g0376600 | 265 | 28556.57 | 4.5 | chloroplast outer membrane |
| RRM1-62 | Os03g0376900 | 464 | 49564.37 | 6.39 | nucleus |
| RRM1-63 | Os03g0388000 | 205 | 24739.51 | 10.27 | nucleus |
| RRM1-64 | Os03g0418800 | 523 | 56761.18 | 8.75 | chloroplast |
| RRM1-65 | Os03g0566500 | 429 | 46194.37 | 9.62 | chloroplast |
| RRM1-66 | Os03g0569900 | 402 | 43945.82 | 5.34 | extracellular space |
| RRM1-67 | Os03g0670700 | 196 | 20375.4 | 6.73 | nucleus |
| RRM1-68 | Os03g0681900 | 308 | 34036.6 | 9.05 | nucleus |
| RRM1-69[40] | Os03g0713600 | 284 | 30904.71 | 5.06 | nucleus |
| RRM1-70 | Os03g0748900 | 278 | 29986.94 | 9.23 | nucleus |
| RRM1-71 | Os03g0801800 | 959 | 105396.52 | 9.48 | nucleus |
| RRM1-72 | Os03g0809900 | 197 | 21969.34 | 5.2 | nucleus |
| RRM1-73 | Os03g0811700 | 130 | 14710.82 | 9.49 | chloroplast |
| RRM1-74 | Os03g0824300 | 523 | 58186.08 | 7.22 | nucleus |
| RRM1-75 | Os03g0826400 | 312 | 36258.57 | 9.25 | nucleus |
| RRM1-76 | Os03g0836200 | 205 | 21823.38 | 8.29 | nucleus |
| RRM1-77 | Os03g0854300 | 441 | 48288.94 | 10.11 | nucleus |
| RRM1-78 | Os04g0118900 | 245 | 28783.89 | 9.94 | nucleus |
| RRM1-79 | Os04g0306800 | 649 | 72026.14 | 9.09 | nucleus |
| RRM1-80 | Os04g0372800 | 486 | 51446 | 5.1 | nucleus |
| RRM1-81 | Os04g0394300 | 903 | 97243.83 | 8.7 | nucleus |
| RRM1-82 | Os04g0414300 | 137 | 15074.25 | 9.93 | chloroplast |
| RRM1-83 | Os04g0449900 | 387 | 41807.64 | 8.68 | extracellular space |
| RRM1-84 | Os04g0467300 | 484 | 51314.72 | 7.33 | nucleus |
| RRM1-85 | Os04g0496400 | 476 | 53576.63 | 4.69 | nucleus |
| RRM1-86 | Os04g0497600 | 435 | 48295.81 | 5.49 | nucleus |
| RRM1-87 | Os04g0504800 | 659 | 71231.24 | 8.95 | extracellular space |
| RRM1-88 | Os04g0510500 | 462 | 51785.72 | 5.01 | nucleus |
| RRM1-89 | Os04g0543200 | 774 | 86649.43 | 5.64 | nucleus |
| RRM1-90 | Os04g0591000 | 291 | 31672.86 | 6.05 | mitochondrion |
| RRM1-91 | Os04g0611500 | 536 | 60240.64 | 9.16 | nucleus |
| RRM1-92 | Os04g0620700 | 707 | 75253.44 | 4.85 | nucleus |
| RRM1-93 | Os04g0624800 | 376 | 40858.93 | 5.59 | nucleus |
| RRM1-94 | Os04g0625800 | 425 | 46195.8 | 5.99 | extracellular space |
| RRM1-95[41] | Os04g0636900 | 515 | 52204.84 | 5.79 | nucleus |
| RRM1-96 | Os04g0641400 | 144 | 16026.58 | 4.61 | nucleus |
| RRM1-97 | Os04g0682400 | 1008 | 110200.99 | 6.17 | nucleus |
| RRM1-98[42] | Os04g0684500 | 901 | 101135.53 | 6.65 | chloroplast inner membrane |
| RRM1-99 | Os05g0102800 | 955 | 104522 | 6.01 | nucleus |
| RRM1-100 | Os05g0105900 | 380 | 42434.11 | 12.18 | nucleus |
| RRM1-101 | Os05g0114500 | 290 | 32890.31 | 6.85 | nucleus |
| RRM1-102 | Os05g0120100 | 323 | 36222.41 | 10.83 | nucleus |
| RRM1-103 | Os05g0140500 | 204 | 22104.33 | 5.18 | nucleus |
| RRM1-104 | Os05g0154800 | 253 | 28203.66 | 9.2 | cytoplasm |
| RRM1-105 | Os05g0162600 | 338 | 39019.1 | 9.83 | nucleus |
| RRM1-106 | Os05g0223200 | 104 | 11486.44 | 8.03 | nucleus |
| RRM1-107 | Os05g0223300 | 102 | 11702.99 | 5.06 | nucleus |
| RRM1-108 | Os05g0303700 | 254 | 29800.11 | 8.77 | nucleus |
| RRM1-109 | Os05g0364600 | 319 | 36105.16 | 11.2 | nucleus |
| RRM1-110 | Os05g0373400 | 466 | 50213.29 | 8.1 | nucleus |
| RRM1-111 | Os05g0376000 | 209 | 23394.61 | 9.14 | nucleus |
| RRM1-112 | Os05g0437300 | 444 | 49754.23 | 6.41 | nucleus |
| RRM1-113[40] | Os06g0112400 | 261 | 27763.35 | 6.23 | nucleus |
| RRM1-114 | Os06g0127500 | 265 | 28209.55 | 7.14 | nucleus |
| RRM1-115 | Os06g0151200 | 300 | 32650.85 | 5 | nucleus |
| RRM1-116 | Os06g0170500 | 482 | 54009.89 | 8.12 | nucleus |
| RRM1-117 | Os06g0187900 | 185 | 21183.36 | 11.29 | nucleus |
| RRM1-118 | Os06g0219600 | 204 | 23178.94 | 5.19 | nucleus |
| RRM1-119 | Os06g0220600 | 343 | 36170.91 | 9.63 | chloroplast outer membrane |
| RRM1-120 | Os06g0248200 | 164 | 17952.57 | 5.98 | nucleus |
| RRM1-121 | Os06g0256200 | 294 | 31817.7 | 10.97 | nucleus |
| RRM1-122 | Os06g0566100 | 292 | 29810.49 | 9.33 | nucleus |
| RRM1-123 | Os06g0589700 | 399 | 43823.12 | 9.17 | nucleus |
| RRM1-124 | Os06g0622900 | 275 | 29594.2 | 8.39 | nucleus |
| RRM1-125 | Os06g0670400 | 469 | 53864.27 | 5.38 | nucleus |
| RRM1-126 | Os06g0670500 | 564 | 64975.32 | 5.63 | nucleus |
| RRM1-127 | Os06g0687500 | 219 | 23922.07 | 9.52 | endomembrane system |
| RRM1-128 | Os06g0698400 | 123 | 13222.7 | 5 | nucleus |
| RRM1-129 | Os06g0724600 | 164 | 18503.88 | 10.31 | nucleus |
| RRM1-130 | Os07g0102500 | 438 | 47703.93 | 9.53 | nucleus |
| RRM1-131 | Os07g0124600 | 377 | 41006.31 | 6.68 | nucleus |
| RRM1-132 | Os07g0158300 | 364 | 39084.91 | 4.61 | mitochondrion |
| RRM1-133 | Os07g0180800 | 411 | 46253.74 | 9.65 | nucleus |
| RRM1-134 | Os07g0237100 | 340 | 36144.67 | 10.27 | chloroplast |
| RRM1-135 | Os07g0281000 | 486 | 54334.99 | 6.72 | nucleus |
| RRM1-136 | Os07g0296200 | 394 | 43291.14 | 8.3 | nucleus |
| RRM1-137 | Os07g0516900 | 251 | 27613.79 | 6.3 | extracellular space |
| RRM1-138 | Os07g0549800 | 133 | 14421.25 | 9.41 | chloroplast outer membrane |
| RRM1-139 | Os07g0583500 | 474 | 54197.46 | 6.55 | extracellular space |
| RRM1-140 | Os07g0584500 | 472 | 50477.44 | 5.94 | nucleus |
| RRM1-141 | Os07g0602600 | 238 | 23564.23 | 8.54 | mitochondrion |
| RRM1-142 | Os07g0603100 | 569 | 62175.74 | 6.15 | nucleus |
| RRM1-143 | Os07g0615400 | 427 | 46723.56 | 7.19 | nucleus |
| RRM1-144 | Os07g0623300 | 275 | 32242.91 | 11.35 | nucleus |
| RRM1-145[43] | Os07g0631900 | 264 | 28099.31 | 4.75 | chloroplast thylakoid lumen |
| RRM1-146 | Os07g0633200 | 213 | 24820.57 | 10.68 | nucleus |
| RRM1-147 | Os07g0663300 | 427 | 46493.89 | 9.17 | nucleus |
| RRM1-148 | Os07g0673500 | 296 | 33141.48 | 10.64 | nucleus |
| RRM1-149 | Os08g0113200 | 838 | 95016.62 | 5.47 | endomembrane system |
| RRM1-150 | Os08g0116400 | 302 | 32739.26 | 6.4 | nucleus |
| RRM1-151 | Os08g0117100 | 319 | 35941.88 | 6.02 | chloroplast outer membrane |
| RRM1-152 | Os08g0139000 | 111 | 11938.8 | 9.55 | chloroplast outer membrane |
| RRM1-153 | Os08g0190200 | 442 | 47809.63 | 5.86 | extracellular space |
| RRM1-154 | Os08g0192900 | 572 | 60393.68 | 4.98 | nucleus |
| RRM1-155 | Os08g0314800 | 660 | 71558.46 | 7.55 | nucleus |
| RRM1-156 | Os08g0320100 | 350 | 36738.65 | 9.22 | nucleus |
| RRM1-157 | Os08g0385900 | 279 | 32947.48 | 11.88 | nucleus |
| RRM1-158 | Os08g0412200 | 214 | 25104.07 | 10.05 | chloroplast |
| RRM1-159 | Os08g0416400 | 503 | 54742.2 | 7.66 | nucleus |
| RRM1-160 | Os08g0427900 | 286 | 30491.17 | 11.05 | nucleus |
| RRM1-161 | Os08g0436000 | 461 | 49888.14 | 6.46 | nucleus |
| RRM1-162 | Os08g0483200 | 269 | 29132.14 | 9.39 | mitochondrion |
| RRM1-163 | Os08g0486200 | 289 | 33541.09 | 11.8 | nucleus |
| RRM1-164 | Os08g0490300 | 603 | 64733.85 | 6.09 | nucleus |
| RRM1-165 | Os08g0492100 | 362 | 38125.83 | 9.22 | nucleus |
| RRM1-166 | Os08g0504600 | 684 | 75299.38 | 6.19 | nucleus |
| RRM1-167 | Os08g0520300 | 447 | 48765.28 | 6.86 | nucleus |
| RRM1-168 | Os08g0547000 | 294 | 31708.05 | 7.08 | nucleus |
| RRM1-169 | Os08g0557100 | 194 | 21388.83 | 4.95 | chloroplast |
| RRM1-170 | Os08g0567200 | 235 | 26254.56 | 9.77 | nucleus |
| RRM1-171 | Os09g0115400 | 662 | 71630.27 | 6.45 | mitochondrion |
| RRM1-172[44] | Os09g0123200 | 738 | 79658.29 | 9.09 | nucleus |
| RRM1-173 | Os09g0279500 | 245 | 26681.14 | 8.53 | chloroplast thylakoid lumen |
| RRM1-174[45] | Os09g0298700 | 1005 | 110844.59 | 6.79 | nucleus |
| RRM1-175 | Os09g0299500 | 160 | 17315.17 | 5.76 | extracellular space |
| RRM1-176 | Os09g0314500 | 353 | 38868.39 | 5.96 | nucleus |
| RRM1-177 | Os09g0462700 | 441 | 46949.78 | 8.52 | chloroplast |
| RRM1-178 | Os09g0476100 | 604 | 64263.07 | 6.3 | nucleus |
| RRM1-179 | Os09g0491756 | 290 | 34087.52 | 8.92 | nucleus |
| RRM1-180 | Os09g0513700 | 375 | 43193.42 | 9.74 | nucleus |
| RRM1-181[46] | Os09g0516300 | 900 | 97198.57 | 6.85 | nucleus |
| RRM1-182 | Os09g0527100 | 149 | 16616.66 | 8.8 | nucleus |
| RRM1-183 | Os09g0527500 | 235 | 25960.25 | 8.81 | nucleus |
| RRM1-184[47] | Os09g0549500 | 276 | 29500.33 | 9.18 | nucleus |
| RRM1-185 | Os09g0565200 | 322 | 35425.05 | 4.41 | mitochondrion |
| RRM1-186 | Os10g0115600 | 463 | 55113.96 | 9.1 | nucleus |
| RRM1-187 | Os10g0151800 | 438 | 47821.62 | 4.98 | nucleus |
| RRM1-188 | Os10g0167500 | 374 | 40267.56 | 3.97 | nucleus |
| RRM1-189 | Os10g0321700 | 317 | 32244.11 | 4.59 | chloroplast thylakoid lumen |
| RRM1-190 | Os10g0439600 | 330 | 34829.59 | 4.96 | nucleus |
| RRM1-191 | Os10g0457000 | 355 | 38849.39 | 8.55 | nucleus |
| RRM1-192 | Os10g0470900 | 464 | 45620.47 | 6.24 | nucleus |
| RRM1-193 | Os10g0569200 | 719 | 83181.8 | 4.98 | nucleus |
| RRM1-194 | Os11g0100200 | 219 | 24033.05 | 9.87 | nucleus |
| RRM1-195 | Os11g0133600 | 298 | 32998.39 | 7.65 | nucleus |
| RRM1-196 | Os11g0139500 | 189 | 21471.25 | 4.13 | extracellular space |
| RRM1-197 | Os11g0176100 | 495 | 52955.01 | 6.43 | extracellular space |
| RRM1-198[48] | Os11g0250000 | 441 | 48446.94 | 5.68 | nucleus |
| RRM1-199 | Os11g0549537 | 242 | 26479.77 | 6.08 | chloroplast |
| RRM1-200 | Os11g0620100 | 441 | 47561.26 | 6.86 | nucleus |
| RRM1-201 | Os11g0636900 | 550 | 61141.76 | 7.78 | nucleus |
| RRM1-202 | Os11g0637700 | 467 | 49048.64 | 8.44 | nucleus |
| RRM1-203 | Os11g0704700 | 511 | 57960.23 | 10.14 | chloroplast |
| RRM1-204[49] | Os12g0100100 | 228 | 24809.9 | 9.87 | nucleus |
| RRM1-205 | Os12g0131000 | 300 | 33277.87 | 8.81 | chloroplast |
| RRM1-206 | Os12g0136200 | 502 | 55072.87 | 5.03 | nucleus |
| RRM1-207 | Os12g0502200 | 258 | 25044.52 | 4.74 | mitochondrion |
| RRM1-208 | Os12g0572400 | 263 | 30186.19 | 10.9 | nucleus |
| RRM1-209[50,51,52] | Os12g0572800 | 1160 | 127816.97 | 8.61 | plasma membrane |
| RRM1-210 | Os12g0577100 | 414 | 47380.57 | 9.1 | nucleus |
| RRM1-211 | Os12g0587100 | 947 | 106893.09 | 9.14 | nucleus |
| RRM1-212[53] | Os12g0632000 | 162 | 16083.1 | 6.31 | nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





