Submitted:
05 June 2023
Posted:
06 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacteria Growth
2.2. Plant Growth
2.3. Acetylene Reduction Assay (ARA)
2.4. Microbial 59Fe Assimilation –
2.5. Luciferase Chemiluminescence ATP Assay
2.6. Spectrophotometric Auxin Assay
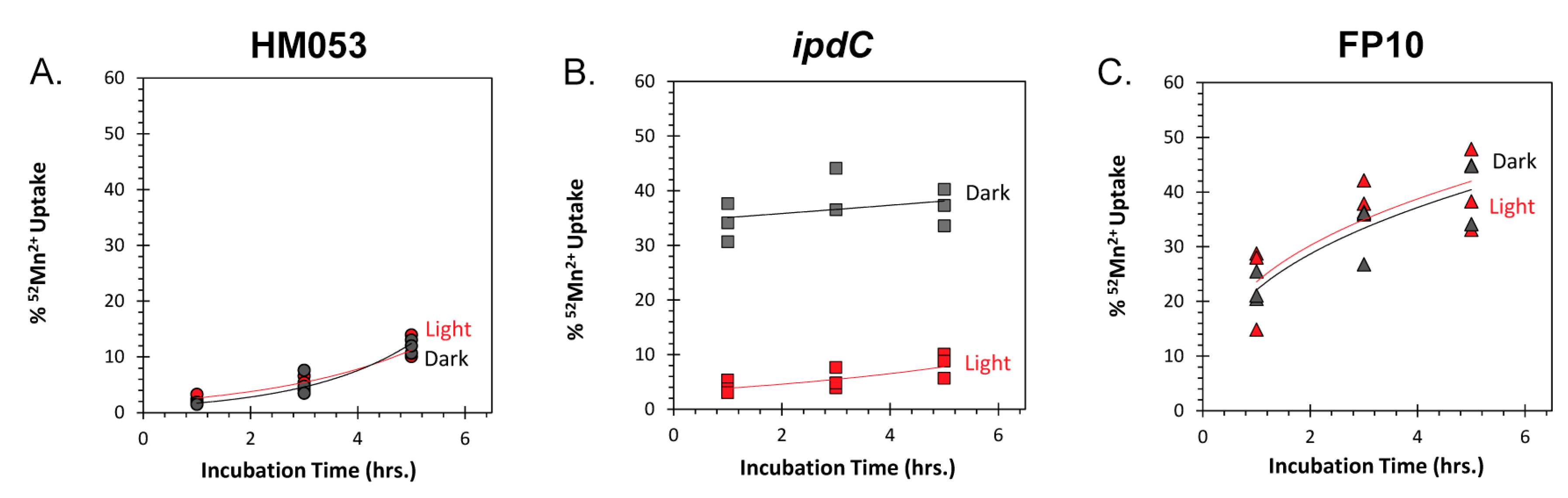
2.7. Bacteria 52Mn Assimilation
2.8. Statistical Analysis
3. Results and Discussion
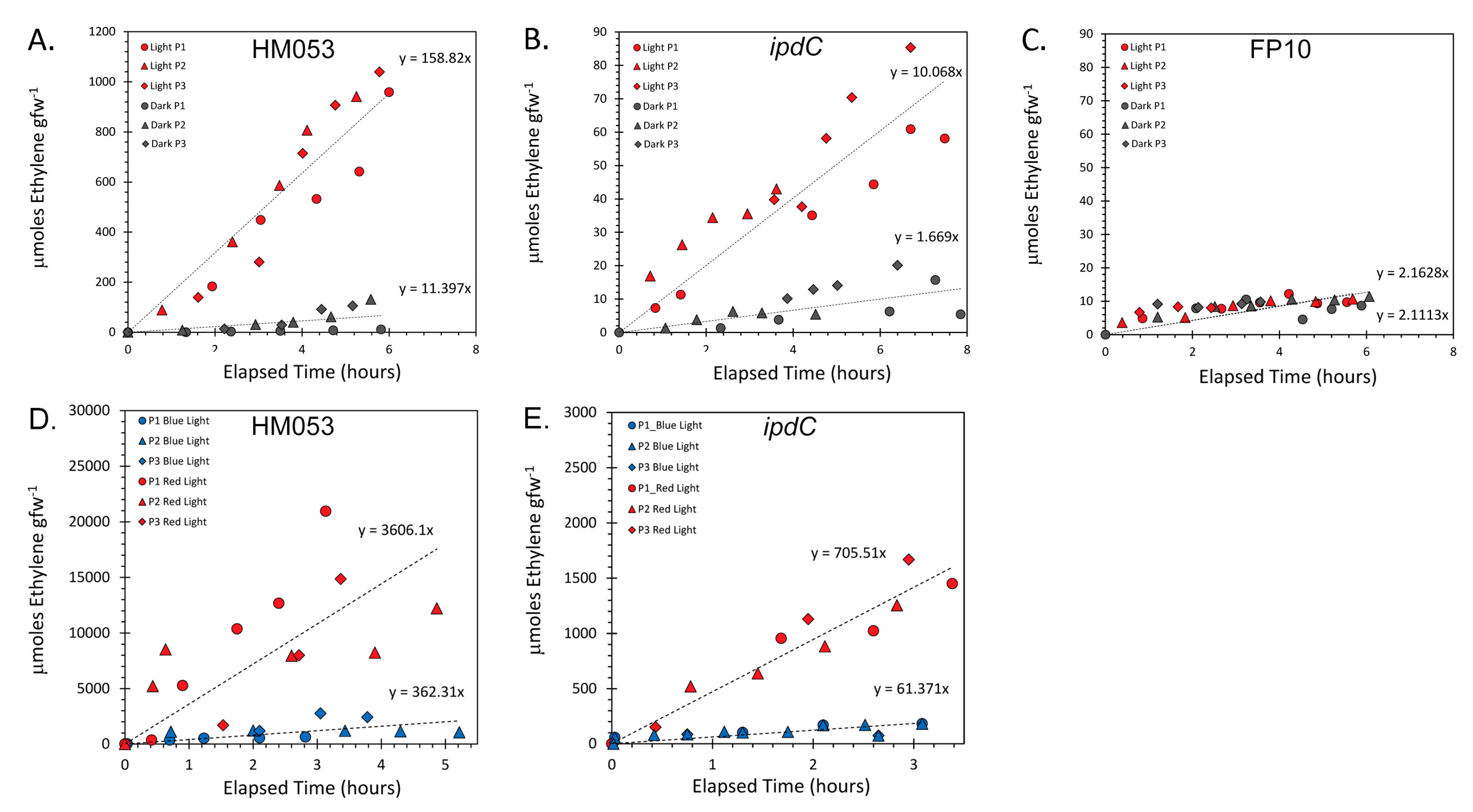
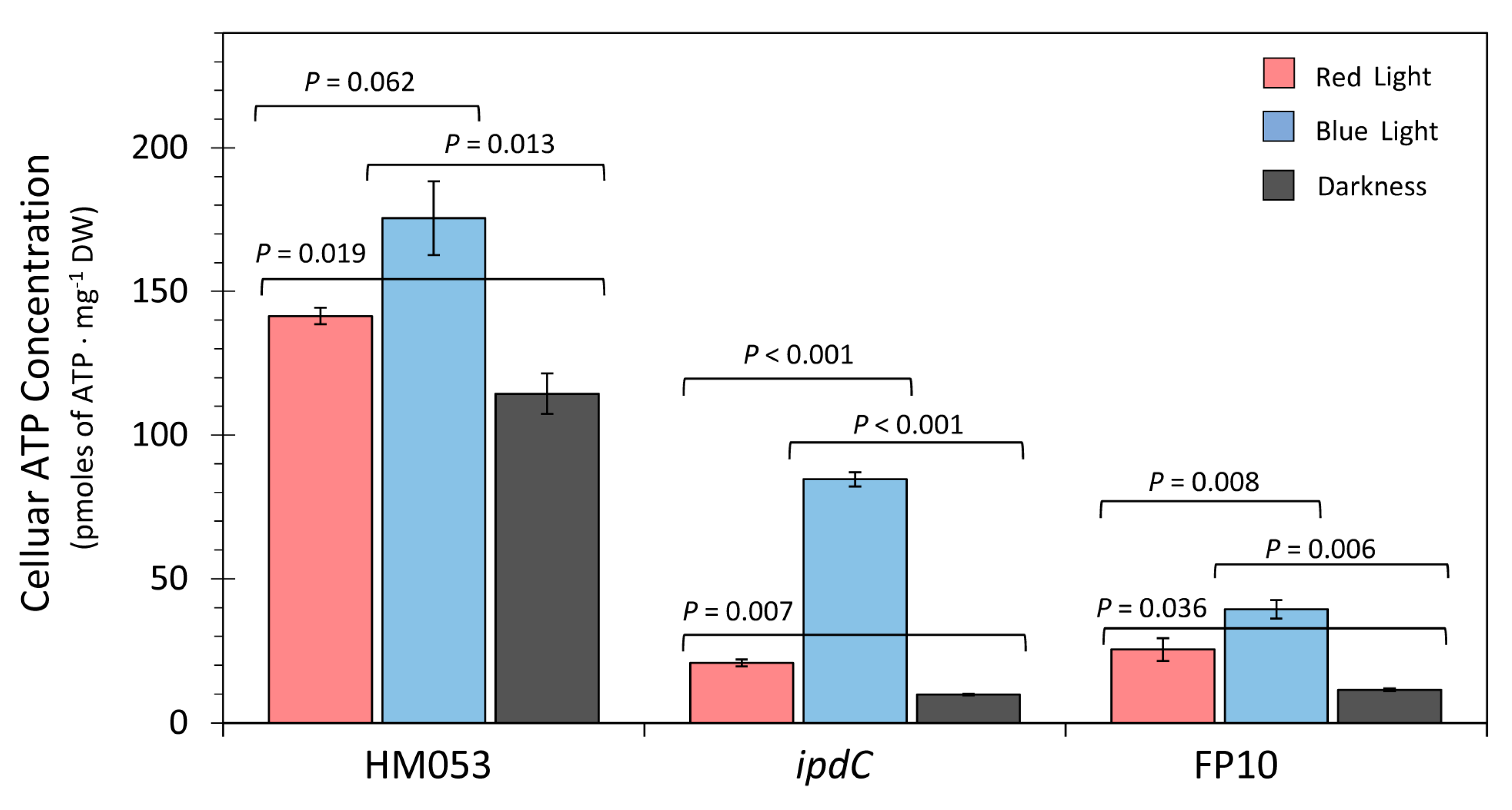
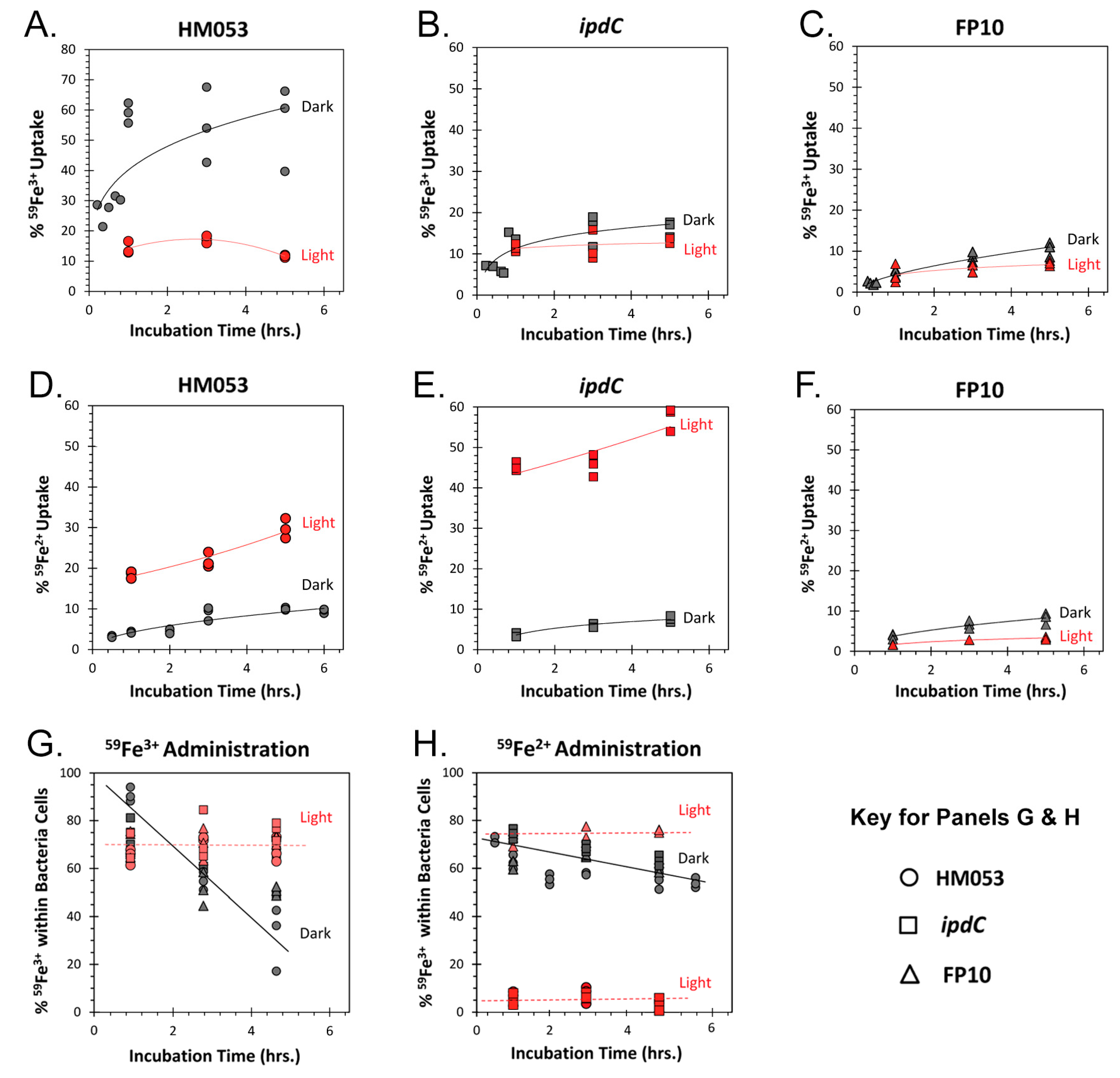
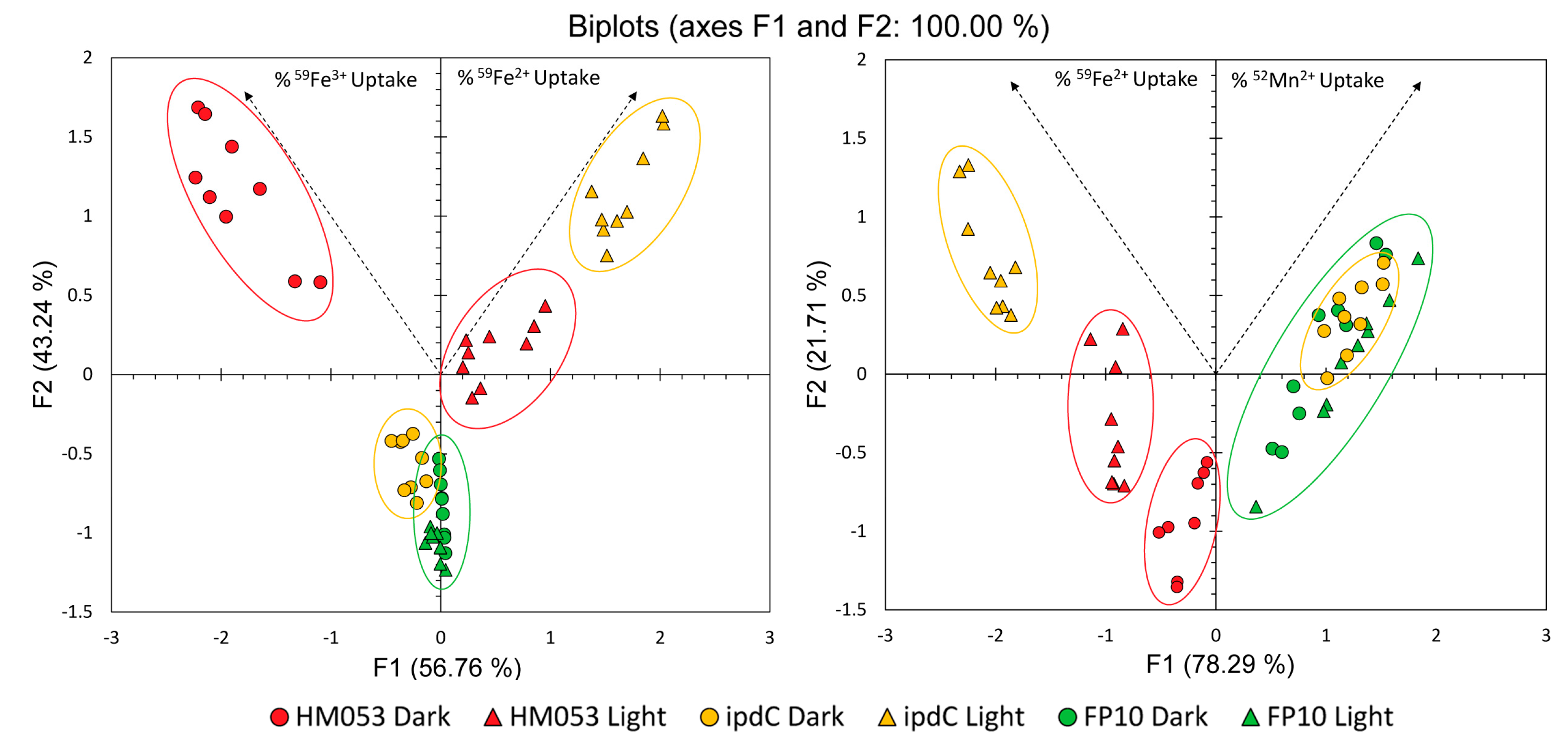
3.1. Light Dependencies of BNF and its Supporting Processes in A. brasilense
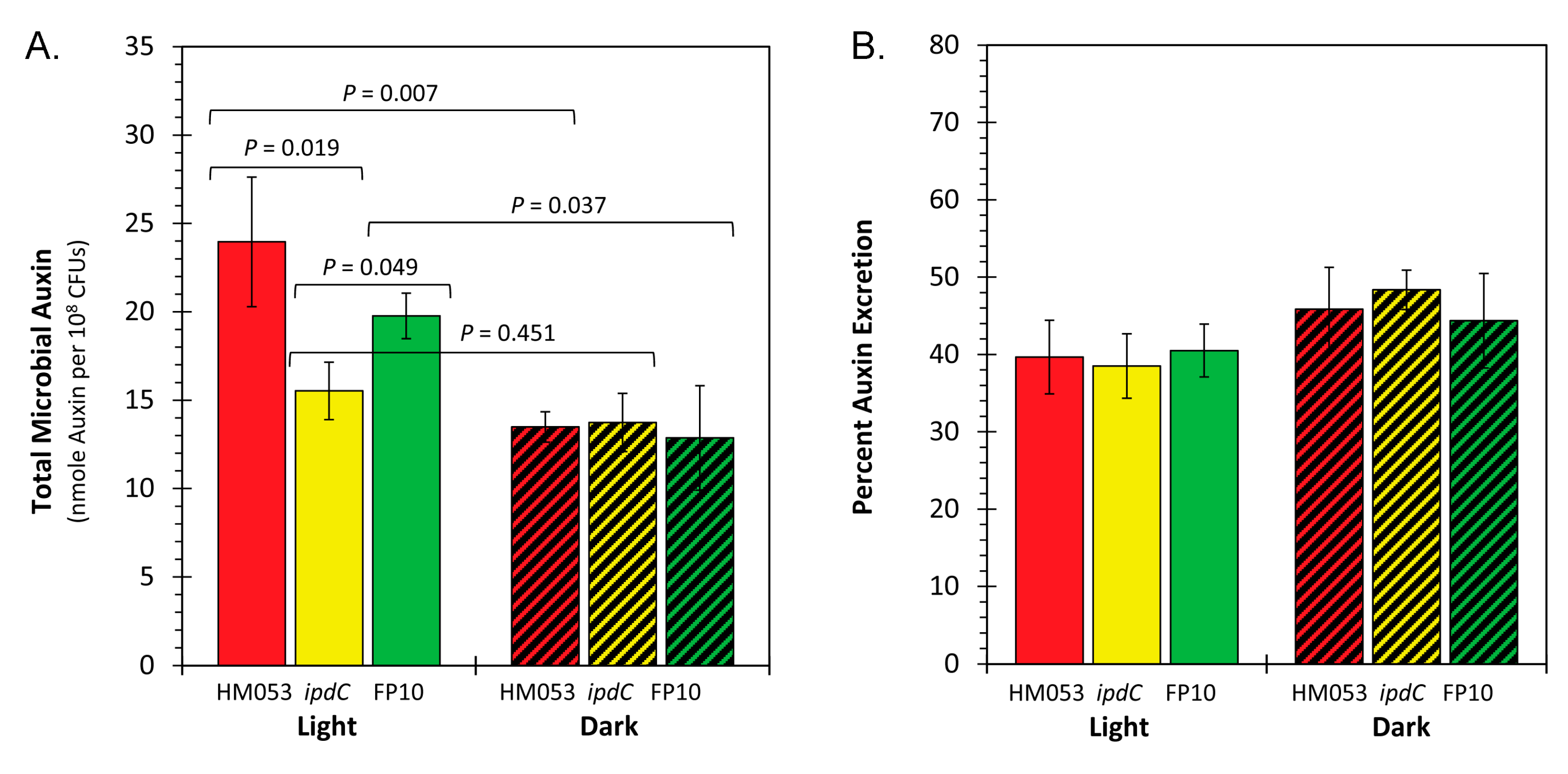
3.2. Light Dependencies of Auxin Biosynthesis and its Supporting Processes in A. brasilense
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, S.; Kateriya, S.; Singh, V.S.; Tanwar, M.; Agarwal, S.; Singh, H.; Khurana, J.P.; Amla, D.V.; Tripathi, A.K. Bacteriophytochrome controls carotenoid-independent response to photodynamic stress in a non-photosynthetic rhizobacterium, Azospirillum brasilense Sp7. Scientific Reports 2012, 2, 872. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Ha, J.H.; Kim, S.G.; Choi, H.K.; Kim, Z.H.; Han, Y.-J.; Kim, J.-I.; Oh, Y.; Fragoso, V.; Shin, K.; Hyeon, T.; Choi, H.-G.; Oh, K.-H.; Baldwin, I.T.; Park, C.-M. Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots. Sci Signal. 2016, 9, ra106. [Google Scholar] [CrossRef] [PubMed]
- van Gelderen, K.; Kang, C.; Pierik, R. Light Signaling, Root Development, and Plasticity. Plant Physiol. 2018, 176, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
- Kakuszi, A.; Sárvári, É.; Solti, Á.; Czégény, G.; Hideg, É.; Hunyadi-Gulyás, É.; Bóka, K.; Böddi, B. Light piping driven photosynthesis in the soil: Low-light adapted active photosynthetic apparatus in the under-soil hypocotyl segments of bean (Phaseolus vulgaris). Journal of Photochemistry and Photobiology B: Biology. 2016, 161, 422–429. [Google Scholar] [CrossRef]
- Waller, S.; Wilder, S.L.; Schueller, M.J.; Ferrieri, R.A. Plants use the suberin biopolymer to conduct light. Polymers 2022, 14, 5387. [Google Scholar] [CrossRef]
- Sun, Q.; Yoda, K.; Suzuki, H. Internal axial light conduction in the stems and roots of herbaceous plants. Journal of Experimental Botany 2005, 56, 191–203. [Google Scholar] [CrossRef]
- Okon, Y. Azospirillum/Plant Associations; CRC Press: Boca Raton, FL, USA, 1993; 192p. [Google Scholar]
- Okon, Y.; Vanderleyden, J. Root-associated Azospirillum species can stimulate plants. ASM News 1997, 63, 366–370. [Google Scholar]
- Steenhoudt, O.; Vanderleyden, J. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical, and ecological aspects. FEMS Microbiol Rev. 2000, 24, 487–506. [Google Scholar] [CrossRef]
- James, E.; Baldani, J. The role of biological nitrogen fixation by non-legumes in the sustainable production of food and biofuels. Plant and Soil 2012, 356, 1–3. [Google Scholar] [CrossRef]
- Richardson, A.; Barea, J.; Mcneill, A.; Prigent-Combaret, C.; Richardson, A.E.; Barea, J.M.; McNeill, A.M.; Prigent-Combaret, C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant and Soil 2009, 321, 305–339. [Google Scholar] [CrossRef]
- Pankievicz, V.C.S.; do Amaral, F.P.; Santos, K.F.D.N.; Agtuca, B.; Xu, Y.; Schueller, M.J.; Arisi, A.C.M.; Steffens, M.B.R.; de Souza, E.M.; Pedrosa, F.O.; Ferrieri, R.A.; Stacey, G. Robust biological nitrogen fixation in a model grass–bacterial association. The Plant Journal 2015, 81, 907–919. [Google Scholar] [CrossRef] [PubMed]
- Okon, Y.; Labandera-Gonzalez, C.A. Agronomic applications of Azospirillum: an evaluation of 20 years worldwide field inoculation. Soil Biol Biochem. 1994, 26, 1591–1601. [Google Scholar] [CrossRef]
- Pedraza, R.O.; Bellone, C.H.; Carrizo de Bellone, S.; Boa Sorte, P.M.F.; Teixeira, K. Azospirillum inoculation and nitrogen fertilization effect on grain yield and on the diversity of endophytic bacteria in the phyllosphere of rice rainfed crop. European Journal of Soil Biology 2009, 45, 36–43. [Google Scholar] [CrossRef]
- Housh, A.B.; Powell, G.; Scott, S.; Anstaett, A.; Gerheart, A.; Benoit, M.; Waller, S.; Powell, A.; Guthrie, J.M.; Higgins, B.; Wilder, S.L.; Schueller, M.J.; Ferrieri, R.A. Functional mutants of Azospirillum brasilense elicit beneficial physiological and metabolic responses in Zea mays contributing to increased host iron assimilation. The ISME Journal 2021, 15, 1505–1522. [Google Scholar] [CrossRef] [PubMed]
- Dobbelaere, S.; Croonenborghs, A.; Thys, A.; Ptacek, D.; Vanderleyden, J.; Dutto, P.; Labandera-Gonzalez, C.; Caballero-Mellado, J.; Aguirre, J.F.; Kapulnik, Y.; Brener, S.; Burdman, S.; Kadouri, D.; Sarig, S.; Okon, Y. Responses of agronomically important crops to inoculation with Azospirillum. Functional Plant Biol. 2001, 28, 871–879. [Google Scholar] [CrossRef]
- Suhameena, B.; Devi, S.; Gowri, R.; Kumar, S.D. Utilization of Azospirillum as a biofertilizer – An overview. International Journal of Pharmaceutical Sciences Review and Research 2020, 62, 141–145. [Google Scholar]
- Giraud, E.; Lavergne, J.; Verméglio, A. Chapter 9 - Characterization of Bacteriophytochromes from Photosynthetic Bacteria: Histidine Kinase Signaling Triggered by Light and Redox Sensing. Methods in Enzymology 2010, 471, 135–159. [Google Scholar]
- Carithers, R.P.; Yoch, D.C.; Arnon, D.I. Two Forms of Nitrogenase from the Photosynthetic Bacterium Rhodospirillum rubrum. J Bacteriol. 1979, 137, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Romina, M.; Gastón, L.; Belén, R.; Susana, R.; Verónica, M.; Fabricio, C. Evaluation of growth and motility in non-photosynthetic Azospirillum brasilense exposed to red, blue, and white light. Archives of Microbiology 2020, 202, 1193–1201. [Google Scholar] [CrossRef]
- Machado, H.B.; Funayama, S.; Rigo, L.U.; Pedrosa, F.O. Excretion of ammonium by Azospirillum brasilense mutants resistant to ethylenediamine. Can J Microbiol. 1991, 37, 549–553. [Google Scholar] [CrossRef]
- Santos, A.R.S.; Etto, R.M.; Furmam, R.W.; Freitas, D.L.; de Santos, K.F.; d’Eça, N.; de Souza, E.M.; Pedrosa, F.O.; Ayub, R.A.; Steffens, M.B.R.; Galvão, C.W. Labeled Azospirillum brasilense wild type and excretion-ammonium strains in association with barley roots. Plant Physiology and Biochemistry 2017, 118, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Näsvall, J.; Knöppel, A.; Andersson, D.I. Duplication-Insertion Recombineering: a fast and scar-free method for efficient transfer of multiple mutations in bacteria. Nucleic Acids Res. 2017, 45, e3. [Google Scholar] [CrossRef]
- Pedrosa, F.O.; Yates, M.G. Regulation of nitrogen fixation (nif) genes of Azospirillum brasilense by nifA and ntr (gln) type gene products. FEMS Microbiology Letters 1984, 23, 95–101. [Google Scholar] [CrossRef]
- Mempin, R.; Tran, H.; Chen, C.; Gong, H.; Ho, K.K.; Lu, S. Release of extracellular ATP by bacteria during growth. BMC Microbiology 2013, 13, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.-C.; Ho, W.-M.; Chen, Y.-H.; Hu, M.-L. A convenient one-step extraction of cellular ATP using boiling water for the Luciferin-Luciferase assay of ATP. Anal. Biochem. 2003, 306, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Ehmann, A. The van URK-Salkowski reagent — a sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification of indole derivatives. J Chromatography 1977, A132, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, A.-L.; Hong, Y.-K.; Shin, J.-H.; Joo, S.-H. A highly efficient auxin-producing bacterial strain and its effect on plant growth. J Genet Eng Biotechnol. 2021, 19, 179–188. [Google Scholar] [CrossRef]
- Graves, S.A.; Hernandez, R.; Fonslet, J.; England, C.G.; Valdovinos, H.F.; Ellison, P.A.; Barnhart, T.E.; Elema, D.R.; Theuer, C.P.; Cai, W.; Nickles, R.J.; Severin, G.W. Novel preparation methods of Mn-52 for immunoPET imaging. Bioconjugate Chemistry 2015, 26, 2118–2124. [Google Scholar] [CrossRef]
- Inomura, K.; Deutsch, C.; Wilson, S.T.; Masuda, T.; Lawrenz, E.; Bučinská, L.; Sobotka, R.; Gauglitz, J.M.; Saito, M.A.; Prášil, O.; Follows, M.J. Quantifying oxygen management and temperature and light dependencies of nitrogen fixation by Crocosphaera watsonii. mSphere 2019, 4, e00531–19. [Google Scholar] [CrossRef]
- Fixen, K.R.; Zheng, Y.; Harris, D.F.; Shaw, S.; Yang, Z.Y.; Dean, D.R.; Seefeldt, L.C.; Harwood, C.S. Light-driven carbon dioxide reduction to methane by nitrogenase in a photosynthetic bacterium. Proc Nat’l Acad Sci. (USA) 2016, 113, 10163–10167. [Google Scholar] [CrossRef]
- Zhang, Y.; Burris, R.H.; Ludden, P.W.; Roberts, G.P. Regulation of nitrogen fixation in Azospirillum brasilense. FEMS Microbiol Lett. 1997, 152, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Vekshin, N.L. Light-dependent ATP synthesis in mitochondria. Biochem Int. 1991, 25, 603–611. [Google Scholar] [PubMed]
- Fu, H.A.; Hartmann, A.; Lowery, R.G.; Fitzmaurice, W.P.; Roberts, G.P.; Burris, R.H. Posttranslational regulatory system for nitrogenase activity in Azospirillum spp. J Bacteriol. 1989, 171, 4679–4685. [Google Scholar] [CrossRef]
- Klassen, G.; de Souza, E.M.; Yates, M.G.; Rigo, L.U.; Inaba, J.; Pedrosa, F.O. Control of nitrogenase reactivation by the GlnZ protein in Azospirillum brasilense. J. Bacteriol. 2001, 183, 6710–6713. [Google Scholar] [CrossRef] [PubMed]
- Huergo, L.F.; Chubatsu, L.S.; de Souza, E.M.; Pedrosa, F.O.; Steffens, M.B.R.; Merrick, M. Interactions between PII proteins and the nitrogenase regulatory enzymes DraT and DraG in Azospirillum brasilense. FEBS Letters 2006, 580, 5232–5236. [Google Scholar] [CrossRef]
- Wenke, B.B.; Spatzal, T.; Rees, D.C. Site-specific oxidation state assignments of the iron atoms in the [4Fe:4S]2+/1+/0 states of the nitrogenase Fe-protein. Angewandte Chemie. International Edition 2019, 58, 3894–3897. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.W.; Bartel, B. Auxin: regulation, action, and interaction. Ann Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef]
- McSteen, P. Auxin and monocot development. Cold Spring Harbor Perspective Biol. 2010, 2, a001479. [Google Scholar] [CrossRef]
- Agtuca, B.; Rieger, E.; Hilger, K.; Song, L.; Robert, C.A.M.; Erb, M.; Karve, A.; Ferrieri, R.A. Carbon-11 reveals opposing roles of auxin and salicylate in regulating leaf physiology, leaf metabolism and resource allocation patterns that impact root growth in Zea mays. J Plant Growth Regulation 2014, 33, 328–339. [Google Scholar] [CrossRef]
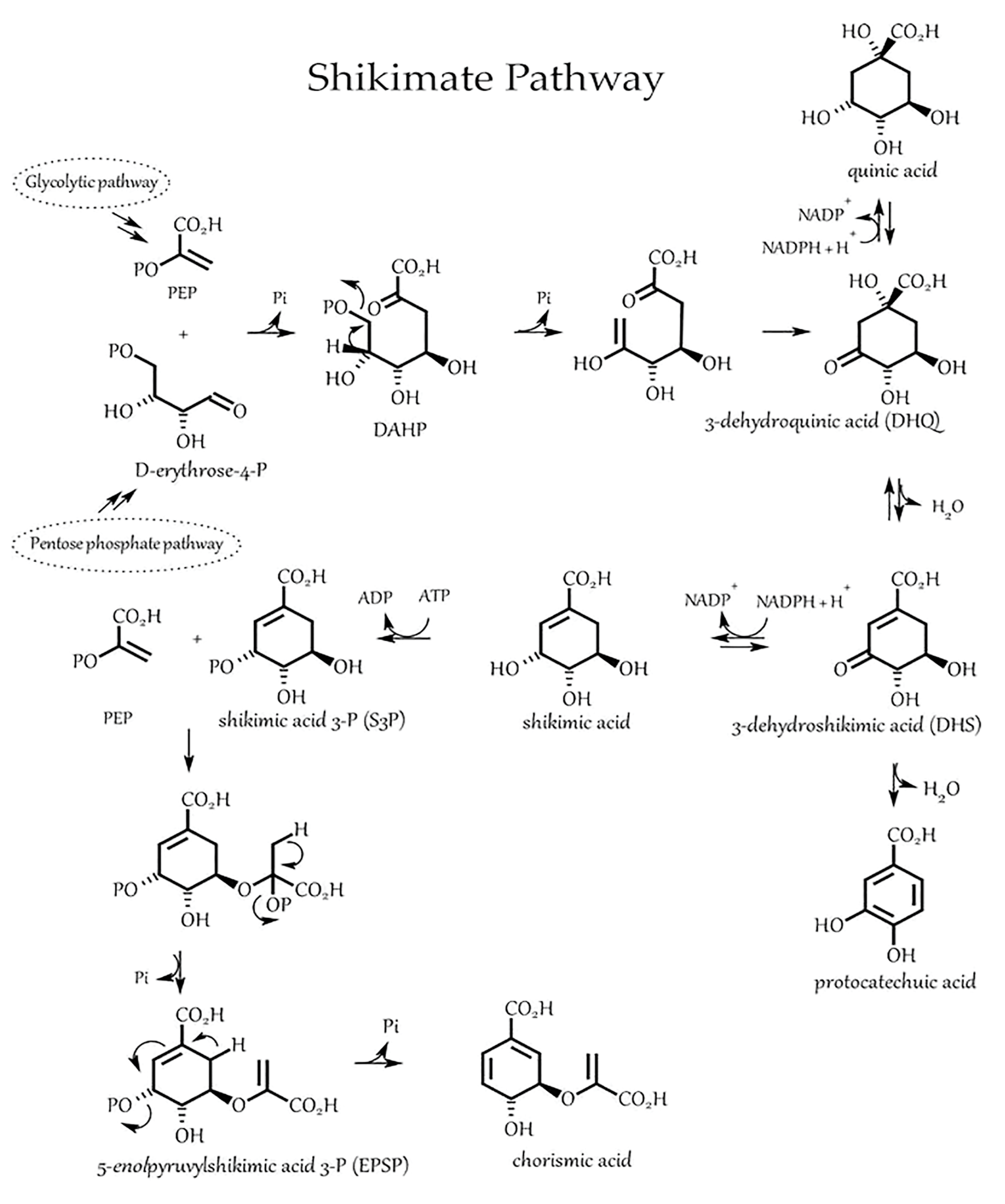
- Schmid, J.; Amrhein, N. Molecular organization of the shikimate pathway in higher plants. Phytochemistry 1995, 39, 737–749. [Google Scholar] [CrossRef]
- Bender, S.L.; Mehdi, S.; Knowles, J.R. Dehydroquinate synthase: The role of divalent metal cations and of nicotinamide adenine dinucleotide in catalysis. Biochemistry 1989, 28, 7555–7560. [Google Scholar] [CrossRef] [PubMed]
- Schneider, D.A.; Gourse, R.L. Relationship between growth rate and ATP concentration in Eschericha coli. J Biological Chemistry 2004, 279, 8262–8268. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




