Submitted:
03 June 2023
Posted:
05 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. BGL Engineering Strategies
2.1. Directed Evolution
2.1.1. Generation of Diverse Mutants
2.1.2. Mutants Screening
2.1.3. Machine Learning-Assisted Directed Evolution
2.2. Rational Design
2.2.1. Structural Analysis
2.2.2. Multiple Sequence Alignment (MSA)
2.2.3. Computational Approaches
2.2.4. Site-Directed Mutagenesis (SDM)
2.3. Semi-Rational Design
3. Engineering of BGL Functionalities
3.1. Enhancing Activity
3.2. Improving Product Tolerance
3.3. Improving Transglycosylation
3.4. Improving Thermostability
3.5. Improving Catalytic Performance in Unconventional Phase
3.6. Improving pH Stability
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, J.; Kumar, V.; Prasad, R.; Gaur, N.A. Engineering of Saccharomyces cerevisiae as a consolidated bioprocessing host to produce cellulosic ethanol: Recent advancements and current challenges. Biotechnol. Adv. 2022, 56, 107925. [Google Scholar] [CrossRef]
- Gutierrez-Gutierrez, D.A.; Fuentes-Garibay, J.A.; Viader-Salvado, J.M.; Guerrero-Olazaran, M. Biochemical characterization of the beta-glucosidase Glu1B from Coptotermes formosanus produced in Pichia pastoris. Enzym. Microb. Technol. 2023, 163, 110155. [Google Scholar] [CrossRef]
- Wang, H.; Lin, X.; Li, S.; Lin, J.; Xie, C.; Liu, D.; Yao, D. Rational molecular design for improving digestive enzyme resistance of beta-glucosidase from Trichoderma viride based on inhibition of bound state formation. Enzym. Microb. Technol. 2020, 133, 109465. [Google Scholar] [CrossRef]
- Amer Ahmed, F. Microbial β-Glucosidase: Sources, Production and Applications. Sci. Educ. Publ. Co. Ltd. 2017. [Google Scholar]
- Zhang, R.; Cao, C.; Bi, J.; Li, Y. Fungal cellulases: Protein engineering and post-translational modifications. Appl. Microbiol. Biotechnol. 2022, 106, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Huang, Q.; Li, H.; Zhao, X. Co-evolution of beta-glucosidase activity and product tolerance for increasing cellulosic ethanol yield. Biotechnol. Lett. 2020, 42, 2239–2250. [Google Scholar] [CrossRef] [PubMed]
- Godse, R.; Bawane, H.; Tripathi, J.; Kulkarni, R. Unconventional beta-Glucosidases: A Promising Biocatalyst for Industrial Biotechnology. Appl. Biochem. Biotechnol. 2021, 193, 2993–3016. [Google Scholar] [CrossRef]
- Chen, H.; Jin, X.; Zhu, L.; Lu, Y.; Ma, Z.; Liu, S.; Chen, X. Glycosyl hydrolase catalyzed glycosylation in unconventional media. Appl. Microbiol. Biotechnol. 2020, 104, 9523–9534. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, Q.; Zhou, Y.; Li, J.; Gao, R.; Guo, Z. Engineering T. naphthophila β-glucosidase for enhanced synthesis of galactooligosaccharides by site-directed mutagenesis. Biochem. Eng. J. 2017, 127, 1–8. [Google Scholar] [CrossRef]
- Boudabbous, M.; Ben Hmad, I.; Saibi, W.; Mssawra, M.; Belghith, H.; Gargouri, A. Trans-glycosylation capacity of a highly glycosylated multi-specific beta-glucosidase from Fusarium solani. Bioprocess. Biosyst. Eng. 2017, 40, 559–571. [Google Scholar] [CrossRef]
- Rocha, R.E.O.; Mariano, D.C.B.; Almeida, T.S.; CorreaCosta, L.S.; Fischer, P.H.C.; Santos, L.H.; Caffarena, E.R.; da Silveira, C.H.; Lamp, L.M.; Fernandez-Quintero, M.L.; et al. Thermostabilizing mechanisms of canonical single amino acid substitutions at a GH1 beta-glucosidase probed by multiple MD and computational approaches. Proteins 2023, 91, 218–236. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Blasco, G.; Sanz-Aparicio, J.; Gonzalez, B.; Hermoso, J.A.; Polaina, J. Directed evolution of beta -glucosidase A from Paenibacillus polymyxa to thermal resistance. J. Biol. Chem. 2000, 275, 13708–13712. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, W.; Ying, Y.; Hao, J. A Novel Glucose-Tolerant GH1 beta-Glucosidase and Improvement of Its Glucose Tolerance Using Site-Directed Mutation. Appl. Biochem. Biotechnol. 2020, 192, 999–1015. [Google Scholar] [CrossRef] [PubMed]
- Li, S.F.; Cheng, F.; Wang, Y.J.; Zheng, Y.G. Strategies for tailoring pH performances of glycoside hydrolases. Crit. Rev. Biotechnol. 2023, 43, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Ju, X.; Fu, J.; Meng, F.; Yan, L.; Li, L. Surface charge engineering of β-glucosidase using rational design improves catalytic capacity and ionic liquid tolerance. J. Mol. Liq. 2022, 367, 120577. [Google Scholar] [CrossRef]
- Arnold, F.H. Directed Evolution: Bringing New Chemistry to Life. Angew. Chem. Int. Ed. Engl. 2018, 57, 4143–4148. [Google Scholar] [CrossRef]
- Romero, P.A.; Arnold, F.H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 2009, 10, 866–876. [Google Scholar] [CrossRef]
- Yoav, S.; Stern, J.; Salama-Alber, O.; Frolow, F.; Anbar, M.; Karpol, A.; Hadar, Y.; Morag, E.; Bayer, E.A. Directed Evolution of Clostridium thermocellum beta-Glucosidase A Towards Enhanced Thermostability. Int. J. Mol. Sci. 2019, 20, 4701. [Google Scholar] [CrossRef]
- Liu, X.; Cao, L.; Zeng, J.; Liu, Y.; Xie, W. Improving the cellobiose-hydrolysis activity and glucose-tolerance of a thermostable beta-glucosidase through rational design. Int. J. Biol. Macromol. 2019, 136, 1052–1059. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, K.; Li, H.; Yi, S.; Zhao, X. Enhancing cellulosic ethanol production through coevolution of multiple enzymatic characteristics of beta-glucosidase from Penicillium oxalicum 16. Appl. Microbiol. Biotechnol. 2020, 104, 8299–8308. [Google Scholar] [CrossRef]
- Cao, L.; Chen, R.; Huang, X.; Li, S.; Zhang, S.; Yang, X.; Qin, Z.; Kong, W.; Xie, W.; Liu, Y. Engineering of beta-Glucosidase Bgl15 with Simultaneously Enhanced Glucose Tolerance and Thermostability To Improve Its Performance in High-Solid Cellulose Hydrolysis. J. Agric. Food Chem. 2020, 68, 5391–5401. [Google Scholar] [CrossRef]
- Santos, C.A.; Morais, M.A.B.; Terrett, O.M.; Lyczakowski, J.J.; Zanphorlin, L.M.; Ferreira-Filho, J.A.; Tonoli, C.C.C.; Murakami, M.T.; Dupree, P.; Souza, A.P. An engineered GH1 beta-glucosidase displays enhanced glucose tolerance and increased sugar release from lignocellulosic materials. Sci. Rep. 2019, 9, 4903. [Google Scholar] [CrossRef] [PubMed]
- Lenz, F.; Zurek, P.; Umlauf, M.; Tozakidis, I.E.P.; Jose, J. Tailor-made β-glucosidase with increased activity at lower temperature without loss of stability and glucose tolerance. Green. Chem. 2020, 22, 2234–2243. [Google Scholar] [CrossRef]
- Yin, B.; Hui, Q.; Kashif, M.; Yu, R.; Chen, S.; Ou, Q.; Wu, B.; Jiang, C. Simultaneous Enhancement of Thermostability and Catalytic Activity of a Metagenome-Derived beta-Glucosidase Using Directed Evolution for the Biosynthesis of Butyl Glucoside. Int. J. Mol. Sci. 2019, 20, 6224. [Google Scholar] [CrossRef] [PubMed]
- Sneha Sawant, N.A. Annamma Anil Odaneth,Sanjeev Kumar Chandrayan, Syed Shams Yazdani,Arvind Lali, Role of N166 residue in β–glucosidase catalysis and glucose tolerance. Appl. Biotechnol. Bioeng. 2019. [Google Scholar]
- Mendez-Liter, J.A.; Nieto-Dominguez, M.; Fernandez de Toro, B.; Gonzalez Santana, A.; Prieto, A.; Asensio, J.L.; Canada, F.J.; de Eugenio, L.I.; Martinez, M.J. A glucotolerant beta-glucosidase from the fungus Talaromyces amestolkiae and its conversion into a glycosynthase for glycosylation of phenolic compounds. Microb. Cell Fact. 2020, 19, 127. [Google Scholar] [CrossRef]
- Noguchi, T.; Saito, H.; Nishiyama, R.; Yoshida, N.; Matsubayashi, T.; Teshima, Y.; Yamada, C.; Hiramatsu, S.; Yamada, K.; Kagawa, Y. Isolation of a cellulase hyperproducing mutant strain of Trichoderma reesei. Bioresour. Technol. Rep. 2021, 15, 100733. [Google Scholar] [CrossRef]
- Niu, K.; Liu, Z.; Feng, Y.; Gao, T.; Wang, Z.; Zhang, P.; Du, Z.; Gao, D.; Fang, X. A novel strategy for efficient disaccharides synthesis from glucose by β-glucosidase. Bioresour. Bioprocess. 2020, 7, 45. [Google Scholar] [CrossRef]
- Deng, P.; Meng, C.; Wu, Y.; Xu, J.; Tang, X.; Zhang, X.; Xiao, Y.; Wang, X.; Fang, Z.; Fang, W. An unusual GH1 beta-glucosidase from marine sediment with beta-galactosidase and transglycosidation activities for superior galacto-oligosaccharide synthesis. Appl. Microbiol. Biotechnol. 2020, 104, 4927–4943. [Google Scholar] [CrossRef]
- Fang, W.; Yang, Y.; Zhang, X.; Yin, Q.; Zhang, X.; Wang, X.; Fang, Z.; Yazhong, X. Improve ethanol tolerance of beta-glucosidase Bgl1A by semi-rational engineering for the hydrolysis of soybean isoflavone glycosides. J. Biotechnol. 2016, 227, 64–71. [Google Scholar] [CrossRef]
- Chen, J.J.; Liang, X.; Chen, T.J.; Yang, J.L.; Zhu, P. Site-Directed Mutagenesis of a beta-Glycoside Hydrolase from Lentinula Edodes. Molecules 2018, 24, 59. [Google Scholar] [CrossRef]
- Zong, Z.; Gao, L.; Cai, W.; Yu, L.; Cui, C.; Chen, S.; Zhang, D. Computer-Assisted Rational Modifications to Improve the Thermostability of β-Glucosidase from Penicillium piceum H16. BioEnergy Res. 2015, 8, 1384–1390. [Google Scholar] [CrossRef]
- Lundemo, P.; Adlercreutz, P.; Karlsson, E.N. Improved transferase/hydrolase ratio through rational design of a family 1 beta-glucosidase from Thermotoga neapolitana. Appl. Env. Microbiol. 2013, 79, 3400–3405. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, R.; Kim, I.W.; Tiwari, M.K.; Gao, H.; Mardina, P.; Das, D.; Kumar, A.; Jeya, M.; Kim, S.Y.; Kim, Y.S.; et al. Tyr320 is a molecular determinant of the catalytic activity of beta-glucosidase from Neosartorya fischeri. Int. J. Biol. Macromol. 2020, 151, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Ehrenreich, C.L. Site-directed mutagenesis studies on a novel dual domain β-galactosidase/ β-glucosidase open reading frame identified from a dairy run-off metagenome. Stellenbosch Univ. 2021. [Google Scholar]
- Gao, X.; Wu, J.; Wu, D. Rational design of the beta-galactosidase from Aspergillus oryzae to improve galactooligosaccharide production. Food Chem. 2019, 286, 362–367. [Google Scholar] [CrossRef]
- Li, S.; Yang, X.; Wu, X.; Xu, S.; Liu, Y. Simultaneously optimizing multiple properties of beta-glucosidase Bgl6 using combined (semi-)rational design strategies and investigation of the underlying mechanisms. Bioresour. Technol. 2023, 374, 128792. [Google Scholar] [CrossRef]
- Bansal, S.; Kundu, B. Protein engineering: Methods and applications. In Advances in Protein Molecular and Structural Biology Methods, 2022; pp 641–668.
- McCullum, E.O.; Williams, B.A.; Zhang, J.; Chaput, J.C. Random mutagenesis by error-prone PCR. Methods Mol. Biol. 2010, 634, 103–109. [Google Scholar] [PubMed]
- James, A.; Wells, M.V.; David, B. Powers, Cassette mutagenesis: An effkient method for generation of multiple mutations at defined sites. Gene 1985. [Google Scholar]
- Joyce, H.G.; Lebbink, T.K. Improving Low-Temperature Catalysis in the Hyperthermostable Pyrococcus furiosus β-Glucosidase CelB by Directed Evolution. BIOCHEMISTRY 2000. [Google Scholar]
- Zhao, H.; Giver, L.; Shao, Z.; et al. , Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat. Biotechnol. 1998, 16, 258–261. [Google Scholar] [CrossRef]
- Zhixin Shao, H.Z. Random-priming in vitro recombination: An effective tool for directed evolution. Nucleic Acids Res. 1998, 26, 2. [Google Scholar]
- Packer, M.S.; Rees, H.A.; Liu, D.R. Phage-assisted continuous evolution of proteases with altered substrate specificity. Nat. Commun. 2017, 8, 956. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, A.; Sindhu, R.; Binod, P.; Sukumaran, R.K.; Pandey, A. Strategies for design of improved biocatalysts for industrial applications. Bioresour. Technol. 2017, 245 Pt B, 1304–1313. [Google Scholar] [CrossRef]
- Yi, D.; Bayer, T.; Badenhorst, C.P.S.; Wu, S.; Doerr, M.; Hohne, M.; Bornscheuer, U.T. Recent trends in biocatalysis. Chem. Soc. Rev. 2021, 50, 8003–8049. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, A.; Arun, K.B.; Binod, P.; Sirohi, R.; Tarafdar, A.; Reshmy, R.; Kumar Awasthi, M.; Sindhu, R. Design of novel enzyme biocatalysts for industrial bioprocess: Harnessing the power of protein engineering, high throughput screening and synthetic biology. Bioresour. Technol. 2021, 325, 124617. [Google Scholar] [CrossRef]
- Hardiman, E.; Gibbs, M.; Reeves, R.; Bergquist, P. Directed evolution of a thermophilic beta-glucosidase for cellulosic bioethanol production. Appl. Biochem. Biotechnol. 2010, 161, 301–312. [Google Scholar] [CrossRef]
- Zeng, W.; Guo, L.; Xu, S.; Chen, J.; Zhou, J. High-Throughput Screening Technology in Industrial Biotechnology. Trends Biotechnol. 2020, 38, 888–906. [Google Scholar] [CrossRef]
- Yao, B.; Zhao, J.; Ding, S.; Giel, M.C.; Zhang, G.; Ding, D.; Tang, Y.; Weng, Z.H.; Hong, Y. A novel red-emitting aggregation-induced emission probe for determination of beta-glucosidase activity. Biomaterials 2023, 295, 122046. [Google Scholar] [CrossRef]
- Yang, K.K.; Wu, Z.; Arnold, F.H. Machine-learning-guided directed evolution for protein engineering. Nat. Methods 2019, 16, 687–694. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef]
- Das, R.; Baker, D. Macromolecular modeling with rosetta. Annu. Rev. Biochem. 2008, 77, 363–382. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Richard, A. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar]
- Marcel, L.; Verdonk, J.C.C.; Michael, J. Improved Protein–Ligand Docking Using GOLD. Proteins-Struct. Funct. Bioinform. 2003, 52, 609–623. [Google Scholar]
- Amorim, A.R.; Zafalon, G.F.D.; Contessoto, A.G.; Valencio, C.R.; Sato, L.M. Metaheuristics for multiple sequence alignment: A systematic review. Comput. Biol. Chem. 2021, 94, 107563. [Google Scholar] [CrossRef] [PubMed]
- Daugelaite, J.; O’ Driscoll, A.; Sleator, R.D. An Overview of Multiple Sequence Alignments and Cloud Computing in Bioinformatics. ISRN Biomath. 2013, 2013, 1–14. [Google Scholar] [CrossRef]
- Hospital, A.; Goni, J.R.; Orozco, M.; Gelpi, J.L. Molecular dynamics simulations: Advances and applications. Adv. Appl. Bioinform. Chem. 2015, 8, 37–47. [Google Scholar]
- Lidan, Y.; Hongwei, Y. From molecular engineering to process engineering: Development of high-throughput screening methods in enzyme directed evolution. Appl. Microbiol. Biotechnol. 2017. [Google Scholar]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Leman, J.K.; Weitzner, B.D.; Lewis, S.M.; Adolf-Bryfogle, J.; Alam, N.; Alford, R.F.; Aprahamian, M.; Baker, D. Macromolecular modeling and design in Rosetta: Recent methods and frameworks. Nat. Methods 2020, 17, 665–680. [Google Scholar] [CrossRef]
- Chowdhury, R.; Maranas, C.D. From directed evolution to computational enzyme engineering—A review. AIChE J. 2019, 66. [Google Scholar] [CrossRef]
- Jonathan Greenhalgh1, A.S. Data-driven Protein Engineering. John Wiley Sons Ltd. 2021. [Google Scholar]
- Song, H.; Bremer, B.J.; Hinds, E.C.; Raskutti, G.; Romero, P.A. Inferring Protein Sequence-Function Relationships with Large-Scale Positive-Unlabeled Learning. Cell Syst. 2021, 12, 92–101. [Google Scholar] [CrossRef]
- Robert, M.; Horton, H.D.H.; Steffan, N. Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene 1989. [Google Scholar]
- Guo, W.; Xie, B.; Jiang, M.; Zhu, X.J.; Qiu, M.; Dai, Z.M. An improved overlap extension PCR for simultaneous multiple sites large fragments insertion, deletion and substitution. Sci. Rep. 2019, 9, 15637. [Google Scholar] [CrossRef]
- Dadwal, A.; Sharma, S.; Satyanarayana, T. Progress in Ameliorating Beneficial Characteristics of Microbial Cellulases by Genetic Engineering Approaches for Cellulose Saccharification. Front. Microbiol. 2020, 11, 1387. [Google Scholar] [CrossRef]
- Ali, M.; Ishqi, H.M.; Husain, Q. Enzyme engineering: Reshaping the biocatalytic functions. Biotechnol. Bioeng. 2020, 117, 1877–1894. [Google Scholar] [CrossRef]
- Zhou, H.Y.; Chen, Q.; Zhang, Y.F.; Chen, D.D.; Yi, X.N.; Chen, D.S.; Cheng, X.P.; Li, M.; Wang, H.Y.; Chen, K.Q.; et al. Improving the catalytic activity of beta-glucosidase from Coniophora puteana via semi-rational design for efficient biomass cellulose degradation. Enzym. Microb. Technol. 2023, 164, 110188. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.J.; Kan, C.N.E.; Luo, C.; Kazlauskas, R.J. Consensus Finder web tool to predict stabilizing substitutions in proteins. Methods Enzym. 2020, 643, 129–148. [Google Scholar]
- Manfred, T.; Reetz, M.B. Expanding the Range of Substrate Acceptance of Enzymes: Combinatorial Active-Site Saturation Test. Angew. Chem. Int. Ed. 2005. [Google Scholar]
- Li, D.; Wu, Q.; Reetz, M.T. Focused rational iterative site-specific mutagenesis (FRISM). Methods Enzym. 2020, 643, 225–242. [Google Scholar]
- Contreras, F.; Pramanik, S.; Rozhkova, A.M.; Zorov, I.N.; Korotkova, O.; Sinitsyn, A.P.; Schwaneberg, U.; Davari, M.D. Engineering Robust Cellulases for Tailored Lignocellulosic Degradation Cocktails. Int. J. Mol. Sci. 2020, 21, 1589. [Google Scholar] [CrossRef] [PubMed]
- Kao, M.R.; Kuo, H.W.; Lee, C.C.; Huang, K.Y.; Huang, T.Y.; Li, C.W.; Chen, C.W.; Wang, A.H.; Yu, S.M.; Ho, T.D. Chaetomella raphigera beta-glucosidase D2-BGL has intriguing structural features and a high substrate affinity that renders it an efficient cellulase supplement for lignocellulosic biomass hydrolysis. Biotechnol. Biofuels 2019, 12, 258. [Google Scholar] [CrossRef]
- Sousa, A.S.; Melo, R.R.; Miyamoto, R.Y.; Morais, M.A.B.; Andrade, L.P.; Milan, N.; Avila, M.C.; Souza, C.M.; Adão, R.C.; Scarpassa, J.A.; et al. A rationally identified marine GH1 β-glucosidase has distinguishing functional features for simultaneous saccharification and fermentation. Biofuels Bioprod. Biorefining 2020, 14, 1163–1179. [Google Scholar] [CrossRef]
- Sinha, S.K.; Goswami, S.; Das, S.; Datta, S. Exploiting non-conserved residues to improve activity and stability of Halothermothrix orenii beta-glucosidase. Appl. Microbiol. Biotechnol. 2017, 101, 1455–1463. [Google Scholar] [CrossRef]
- Kao, M.R.; Yu, S.M.; Ho, T.U.D. Improvements of the productivity and saccharification efficiency of the cellulolytic beta-glucosidase D2-BGL in Pichia pastoris via directed evolution. Biotechnol. Biofuels 2021, 14, 126. [Google Scholar] [CrossRef]
- Xia, W.; Bai, Y.; Shi, P. Improving the Substrate Affinity and Catalytic Efficiency of beta-Glucosidase Bgl3A from Talaromyces leycettanus JCM12802 by Rational Design. Biomolecules 2021, 11, 1882. [Google Scholar] [CrossRef]
- Larue, K.; Melgar, M.; Martin, V.J. Directed evolution of a fungal beta-glucosidase in Saccharomyces cerevisiae. Biotechnol. Biofuels 2016, 9, 52. [Google Scholar] [CrossRef]
- Sinha, S.K.; Das, S.; Konar, S.; Ghorai, P.K.; Das, R.; Datta, S. Elucidating the regulation of glucose tolerance in a beta-glucosidase from Halothermothrix orenii by active site pocket engineering and computational analysis. Int. J. Biol. Macromol. 2020, 156, 621–632. [Google Scholar] [CrossRef]
- Pang, P.; Cao, L.C.; Liu, Y.H.; Xie, W.; Wang, Z. Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism. J. Struct. Biol. 2017, 198, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Long, L.; Liang, M.; Li, H.; Chen, Y.; Zheng, M.; Ni, H.; Li, Q.; Zhu, Y. Characterization of a glucose-stimulated beta-glucosidase from Microbulbifer sp. ALW1. Microbiol. Res. 2021, 251, 126840. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.; Das, S.; Datta, S. Understanding the role of residues around the active site tunnel towards generating a glucose-tolerant beta-glucosidase from Agrobacterium tumefaciens 5A. Protein Eng. Des. Sel. 2017, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Amano, Y.; Nozaki, K. Improvements in Glucose Sensitivity and Stability of Trichoderma reesei beta-Glucosidase Using Site-Directed Mutagenesis. PLoS ONE 2016, 11, e0147301. [Google Scholar]
- Meleiro, L.P.; Salgado, J.C.S.; Maldonado, R.F.; Carli, S.; Moraes, L.A.B.; Ward, R.J.; Jorge, J.A.; Furriel, R.P.M. Engineering the GH1 beta-glucosidase from Humicola insolens: Insights on the stimulation of activity by glucose and xylose. PLoS ONE 2017, 12, e0188254. [Google Scholar] [CrossRef]
- Goswami, S.; Manna, B.; Chattopadhyay, K.; Ghosh, A.; Datta, S. Role of Conformational Change and Glucose Binding Sites in the Enhanced Glucose Tolerance of Agrobacterium tumefaciens 5A GH1 beta-Glucosidase Mutants. J. Phys. Chem. B 2021, 125, 9402–9416. [Google Scholar] [CrossRef] [PubMed]
- Mariano, D.; Pantuza, N.; Santos, L.H.; Rocha, R.E.O.; de Lima, L.H.F.; Bleicher, L.; de Melo-Minardi, R.C. Glutantbetaase: A database for improving the rational design of glucose-tolerant beta-glucosidases. BMC Mol. Cell Biol. 2020, 21, 50. [Google Scholar] [CrossRef]
- Kaushal, G.; Rai, A.K.; Singh, S.P. A novel beta-glucosidase from a hot-spring metagenome shows elevated thermal stability and tolerance to glucose and ethanol. Enzym. Microb. Technol. 2021, 145, 109764. [Google Scholar] [CrossRef]
- Correa, T.L.R.; Franco Cairo, J.P.L.; Cota, J.; Damasio, A.; Oliveira, L.C.; Squina, F.M. A novel mechanism of beta-glucosidase stimulation through a monosaccharide binding-induced conformational change. Int. J. Biol. Macromol. 2021, 166, 1188–1196. [Google Scholar] [CrossRef]
- Kuusk, S.; Valjamae, P. When substrate inhibits and inhibitor activates: Implications of beta-glucosidases. Biotechnol. Biofuels 2017, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Lundemo, P.; Karlsson, E.N.; Adlercreutz, P. Eliminating hydrolytic activity without affecting the transglycosylation of a GH1 beta-glucosidase. Appl. Microbiol. Biotechnol. 2017, 101, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Xue, M.; Xie, F.; Zhang, M.; Zhao, H.; Zhou, T. Engineering Thermotoga maritima beta-glucosidase for improved alkyl glycosides synthesis by site-directed mutagenesis. J. Ind. Microbiol. Biotechnol. 2021, 48, 5–6. [Google Scholar] [CrossRef]
- Lyu, J.; Zhang, J.; Zhu, J.; Chen, S.; Han, T.; Zhang, Y.; Gao, R.; Xie, G.; Guo, Z. Molecular dynamics simulation guided distal mutation of Thermotoga naphthophila beta-glucosidase for significantly enhanced synthesis of galactooligosaccharides and expanded product scope. Int. J. Biol. Macromol. 2022, 210, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Anica Dadwal, S.S. Tulasi Satyanarayana Thermostable cellulose saccharifying microbial enzymes: Characteristics, recent advances and biotechnological applications. Int. J. Biol. Macromol. 2021, 188, 226–244. [Google Scholar] [CrossRef] [PubMed]
- Rahban, M.; Zolghadri, S.; Salehi, N.; Ahmad, F.; Haertle, T.; Rezaei-Ghaleh, N.; Sawyer, L.; Saboury, A.A. Thermal stability enhancement: Fundamental concepts of protein engineering strategies to manipulate the flexible structure. Int. J. Biol. Macromol. 2022, 214, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Matsuzawa, T.; Yaoi, K. Rational protein design for thermostabilization of glycoside hydrolases based on structural analysis. Appl. Microbiol. Biotechnol. 2018, 102, 8677–8684. [Google Scholar] [CrossRef]
- Sharma, S.; Vaid, S.; Bhat, B.; Singh, S.; Bajaj, B.K. Thermostable Enzymes for Industrial Biotechnology. In Advances in Enzyme Technology, 2019; pp. 469–495.
- Ahmed, A.; Sumreen, A.; Bibi, A.; Nasim, F. u. H.; Batool, K. In silico Approach to Elucidate Factors Associated with GH1 β-Glucosidase Thermostability. J. Pure Appl. Microbiol. 2019, 13, 1953–1968. [Google Scholar] [CrossRef]
- Hu, Y.; Kang, G.; Wang, L.; Gao, M.; Wang, P.; Yang, D.; Huang, H. Current Status of Mining, Modification, and Application of Cellulases in Bioactive Substance Extraction. Curr. Issues Mol. Biol. 2021, 43, 687–703. [Google Scholar] [CrossRef]
- Prakash Chandra Rathi, D.M. A GUI for interactive Constraint Network Analysis and protein engineering for improving thermostability. Bioinformatics 2015, 31, 2394–2396. [Google Scholar] [CrossRef]
- Mamonova, T.B.; Glyakina, A.V.; Kurnikova, M.G.; Galzitskaya, O.V. Flexibility and mobility in mesophilic and thermophilic homologous proteins from molecular dynamics and FoldUnfold method. J. Bioinform. Comput. Biol. 2010, 8, 377–394. [Google Scholar] [CrossRef]
- Modarres, H.P.; Mofrad, M.R.; Sanati-Nezhad, A. Protein thermostability engineering. RSC Adv. 2016, 6, 115252–115270. [Google Scholar] [CrossRef]
- Wijma, H.J.; Floor, R.J.; Jekel, P.A.; Baker, D.; Marrink, S.J.; Janssen, D.B. Computationally designed libraries for rapid enzyme stabilization. Protein Eng. Des. Sel. 2014, 27, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Goldenzweig, A.; Goldsmith, M.; Hill, S.E.; Gertman, O.; Laurino, P.; Ashani, Y.; Dym, O.; Unger, T.; Albeck, S.; Prilusky, J.; et al. Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability. Mol. Cell 2016, 63, 337–346. [Google Scholar] [CrossRef]
- Mazurenko, S. Predicting protein stability and solubility changes upon mutations: Data perspective. ChemCatChem 2020, 12, 5590–5598. [Google Scholar] [CrossRef]
- Mendez-Liter, J.A.; Gil-Munoz, J.; Nieto-Dominguez, M.; Barriuso, J.; de Eugenio, L.I.; Martinez, M.J. A novel, highly efficient beta-glucosidase with a cellulose-binding domain: Characterization and properties of native and recombinant proteins. Biotechnol. Biofuels 2017, 10, 256. [Google Scholar] [CrossRef] [PubMed]
- Peishan Huang, S.K.S.C. Evaluating Protein Engineering Thermostability Prediction Tools Using an Independently Generated Dataset. ACS Omega 2020, 5, 6487–6493. [Google Scholar] [CrossRef]
- Okereke, O.E.; Gupta, M.; Ogunyewo, O.A.; Sharma, K.; Yazdani, S.S. Profiling of the β-glucosidases identified in the genome of Penicillium funiculosum: Insights from genomics, transcriptomics, proteomics and homology modelling studies. bioRxiv 2022. [Google Scholar] [CrossRef]
- Sinha, S.K.; Datta, M.; Datta, S. A glucose tolerant β-glucosidase from Thermomicrobium roseum that can hydrolyze biomass in seawater. Green. Chem. 2021, 23, 7299–7311. [Google Scholar] [CrossRef]
- Cao, L.; Li, S.; Huang, X.; Qin, Z.; Kong, W.; Xie, W.; Liu, Y. Enhancing the Thermostability of Highly Active and Glucose-Tolerant beta-Glucosidase Ks5A7 by Directed Evolution for Good Performance of Three Properties. J. Agric. Food Chem. 2018, 66, 13228–13235. [Google Scholar] [CrossRef]
- Matsuzawa, T.; Watanabe, M.; Yaoi, K. Improved thermostability of a metagenomic glucose-tolerant beta-glycosidase based on its X-ray crystal structure. Appl. Microbiol. Biotechnol. 2017, 101, 8353–8363. [Google Scholar] [CrossRef]
- Amaike Campen, S.; Lynn, J.; Sibert, S.J.; Srikrishnan, S.; Phatale, P.; Feldman, T.; Guenther, J.M.; Hiras, J.; Tran, Y.T.A.; Singer, S.W.; et al. Expression of naturally ionic liquid-tolerant thermophilic cellulases in Aspergillus niger. PLoS ONE 2017, 12, e0189604. [Google Scholar] [CrossRef]
- Batra, J.; Mishra, S. Organic solvent tolerance and thermostability of a β-glucosidase co-engineered by random mutagenesis. J. Mol. Catal. B: Enzym. 2013, 96, 61–66. [Google Scholar] [CrossRef]
- Li, Y.; Hu, X.; Sang, J.; Zhang, Y.; Zhang, H.; Lu, F.; Liu, F. An acid-stable beta-glucosidase from Aspergillus aculeatus: Gene expression, biochemical characterization and molecular dynamics simulation. Int. J. Biol. Macromol. 2018, 119, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Michał Rostkowski, M.H.O. Graphical analysis of pH-dependent properties of proteins predicted using PROPKA. BMC Struct. Biol. 2011, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Solhtalab, M.; Flannelly, D.F.; Aristilde, L. Substrate binding versus escape dynamics in a pH-affected fungal beta-glucosidase revealed by molecular dynamics simulations. Carbohydr. Res. 2019, 472, 127–131. [Google Scholar] [CrossRef]
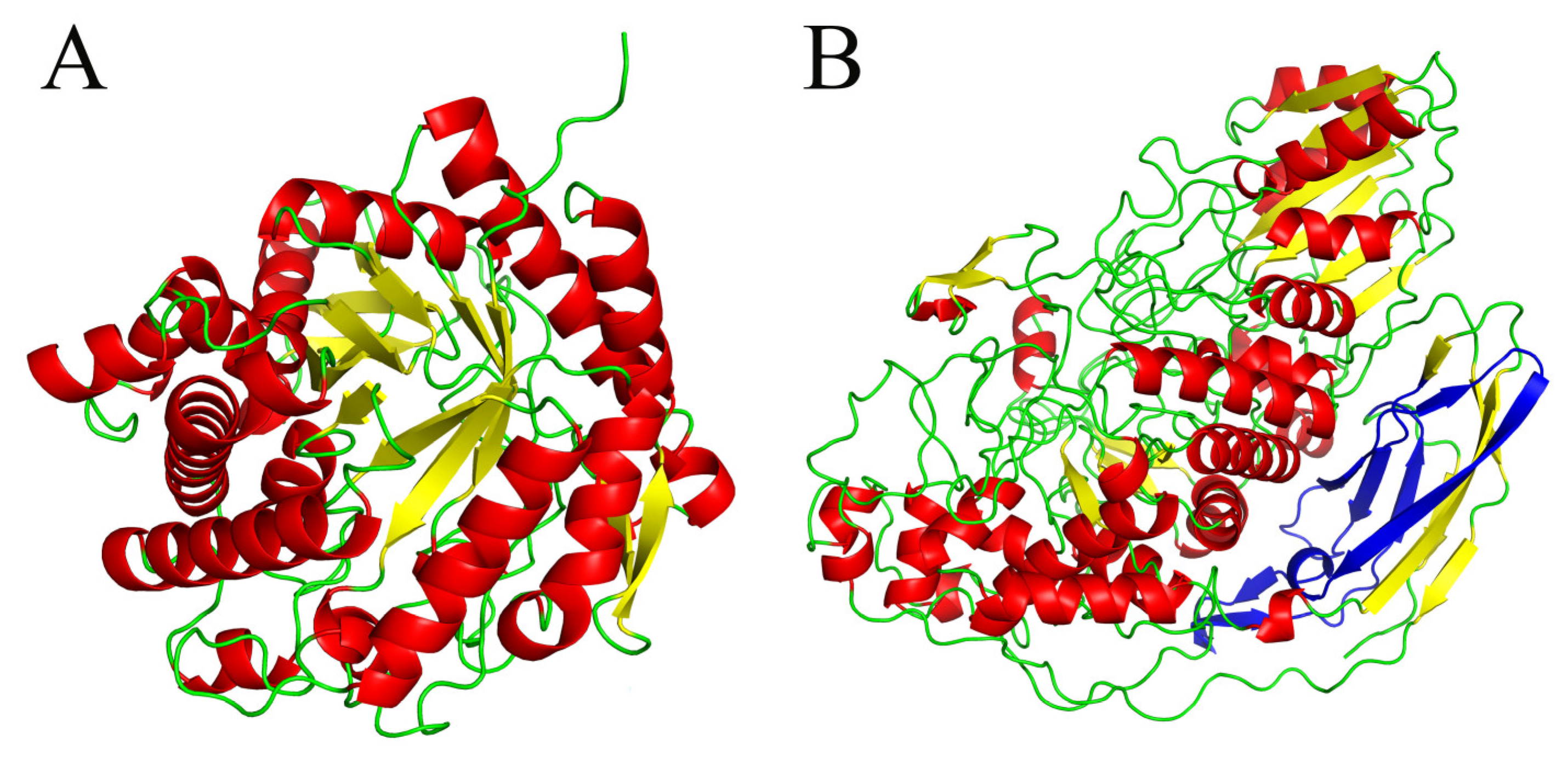
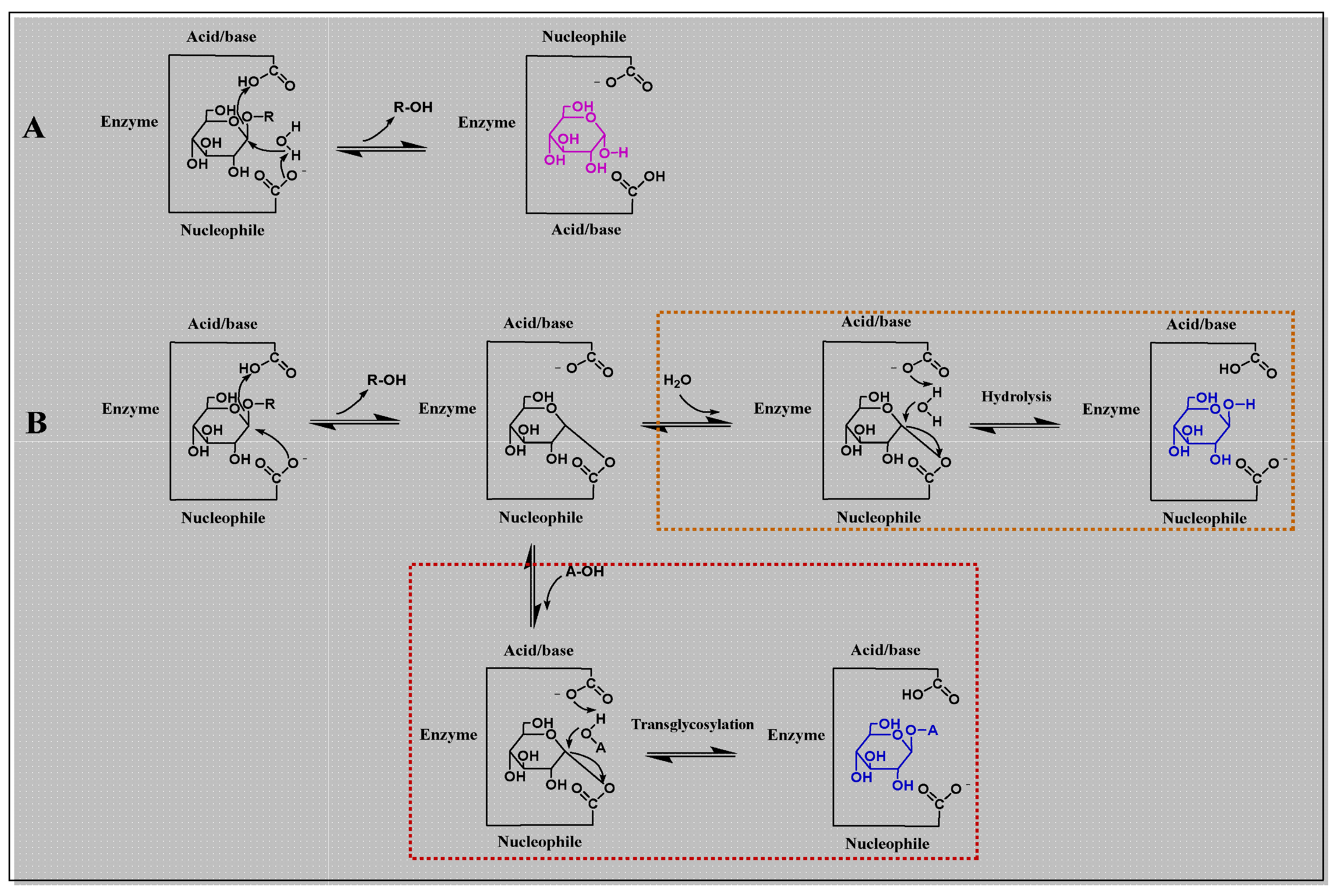
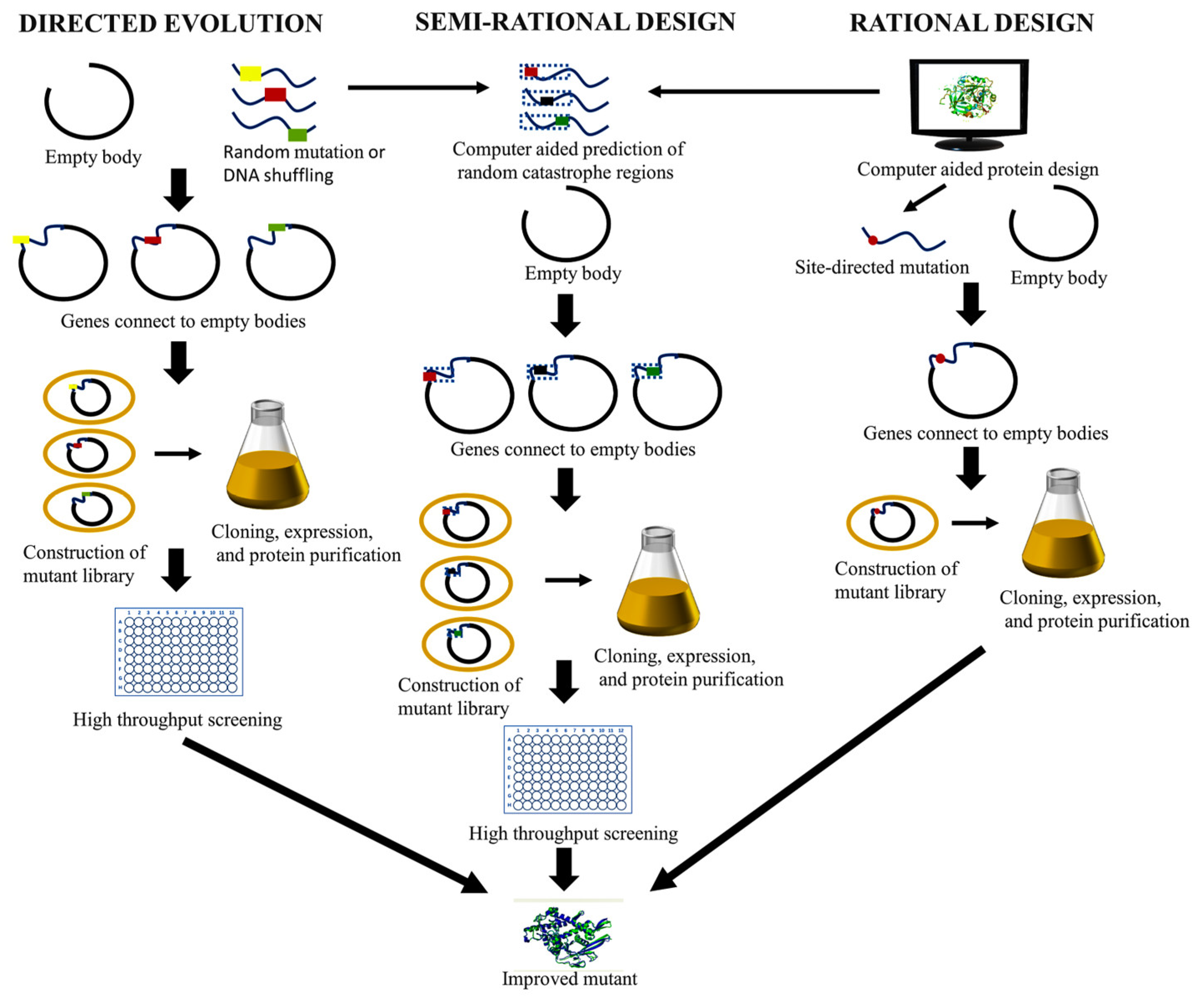
| Organism source | Engineering methods | High throughput screening method | Reference |
|---|---|---|---|
| Clostridium thermocellum | Directed evolution Error-prone PCR |
Assay medium screening method(0.02% Magenta GlcA) | [18] |
| Alteromonas sp. L82 | Rational design Site-Directed Mutation |
--- | [13] |
| Metagenomic library of Turpan Depression | Rational design Site-Directed Mutation |
--- | [19] |
| P. oxalicum 16 | Directed evolution Error-prone PCR |
Assay medium screening method 6-(β-D-glucopyranosyloxy)-7-hydroxy-2H -1-benzopyran-2-one | [20] |
| Thermotoga naphthophila RKU-10 | Rational design Site-Directed Mutation |
--- | [9] |
| Soil Macrogenome Library | Directed evolution Error-prone PCR |
Double assay medium screening method (0.1% hesperidin) | [21] |
| Trichoderma harzianum | Rational design Site-Directed Mutation |
--- | [22] |
| Caldicellulosirutor saccharolyticus | Semi-rational design Site-Directed Mutation |
Cell Surface Display Fluorescence detection medium screening method (pNPG) |
[23] |
| GenBank FJ686869 | Directed evolution Error-prone PCR |
Assay medium screening method (0.1% hesperidin) | [24] |
|
Paenibacillus polymyxa |
Rational design Site-Directed Mutation |
--- | [25] |
|
Talaromyces amestolkiae |
Rational design Site-Directed Mutation |
--- | [26] |
| Trichoderma reesei | Directed evolution (UV light, N-methyl-N′-nitro-N-nitrosoguanidine) |
Detection medium screening method (phosphoric acid-swollen cellulose) | [27] |
| T. reesei | Rational design (Hydropathy index for enzyme activity) Site-Directed Mutation |
--- | [28] |
| Bacillus sp. D1 | Semi-rational design Site-Directed mutagenesis |
--- | [29] |
| Marine microbial metagenomic library | Semi-rational design Site-Directed mutagenesis |
--- | [30] |
| Lentinula edodes | Rational design Site-Directed mutagenesis |
--- | [31] |
| Penicillium piceum H16 | Rational design Site-Directed Mutation |
--- | [32] |
|
Thermotoga neapolitana |
Rational design Site-Directed Mutation |
--- | [33] |
|
Neosartorya fischeri |
Rational design Site-Directed Mutation |
--- | [34] |
| A dairy run-off metagenome | Rational design Site-Directed Mutation |
--- | [35] |
| Aspergillus oryzae | Rational design Site-Directed Mutation |
--- | [36] |
| Metagenomic library of Turpan Depression |
Semi-rational design Site-Directed Mutation |
--- | [37] |
| Organism | Strategy | Mutations | Molecular effects | References |
|---|---|---|---|---|
| Halothermothrix orenii | Rational design OEP |
V169C, I246A | Lack of stable polar contacts; Reduction of side chain volume |
[79] |
| Coniophora puteana | Semi-rational design (HotSpot, Alanine scanning technique) SDM |
Q20C, A240S | A combination of structural changes in the active pocket and protein-ligand interactions | [72] |
|
Chaetomella raphigera (D2-BGL) |
Directed evolution Error-prone PCR |
F256M/Y260D /D224G |
F256 and Y260 on a short loop related to the high substrate affinity of the enzyme | [80] |
| Metagenomic library of Turpan soil (Bgl1317) |
Rational design SDM |
A397R, L188A, A262S | Increase in the polarity of residues and hydrogen bonding contacts | [19] |
| Talaromyces leycettanus JCM12802 | Rational design OEP |
M36E, M36N, F66Y, E168Q | Increase in hydrophobic stacking interactions and hydrogen bonding networks of active centers | [81] |
| Marine bacteria (bgla) |
Rational design OEP |
F171W | Increase in volume of side chains near the active site | [13] |
| P. oxalicum 16 | Directed evolution Error-prone PCR |
M280T/V484L /D589E |
Increase in the number of hydrogen bonds formed by the substrate to increase the binding free energy | [6] |
| C. saccharolyticus | Directed evolution Error-prone PCR, Random drift mutagenesis |
--- | Smaller residues near catalytic residues allow more flexibility in the active site or more access to the substrate | [48] |
| Aspergillus niger (BGL1) | Directed evolution Error-prone PCR |
Q140L A480V, K494Q N557D |
Multiple hydrogen bond numbers to improve substrate affinity in the substrate binding pocket | [82] |
| P. oxalicum 16 | Directed evolution Error-prone PCR |
G414S/D421V/ T441S |
Tighter active site pockets | [20] |
| Pyrococcus furiosus (CelB) | Directed evolution DNA shuffling |
N415S | --- | [41] |
| C. saccharolyticus (CsBglA) | Semi-rational design (SDM combined with Random mutagenesis) |
L64R/Y73F/ T221N/H324L |
--- | [23] |
| Organism source | Improved engineering methods | Mutation sites | Molecular effects | References |
|---|---|---|---|---|
| Metagenomic library of Turpan soil |
Rational design SDM |
L188A,A262S | Active site metastable interactions | [19] |
| H. orenii | SDM MD simulation |
--- | Conservative + 2 subsite hydrophobic residues | [83] |
|
Agrobacterium tumefaciens 5A |
Rational design SDM OEP |
W127F, C174V, V176A, L178A, L178E, H229S |
Increase in the hydrophobicity of the aglycone-binding sites and gatekeeper regions | [86] |
|
Trichoderma Harzianum |
Rational design SDM |
L167W/P172L | Replacement of gatekeeper residues to alter active site accessibility | [22] |
| T. Cel1A (Bgl II) | Rational design SDM OEP |
L167W/P172L | Replacement of gatekeeper residues to narrow the entrance to the active pocket | [87] |
| Humicola insolens (Bglhi) | Directed evolution Error-prone PCR |
H307Y, D237V, N235S |
Increasing trans-glycosylation Unbinding of unproductive substrates |
[88] |
| A. tumefaciens 5A | --- | --- | Presence of separate glucose binding sites | [89] |
| Marine microbial metagenome |
Rational design SDM |
H228T | Interaction leading to glucose excretion by slingshot mechanism | [90] |
| H. orenii | Rational design SDM |
V169C/E173L/ I246A |
Increasing backbone kinetics of active channel residues and flexibility of active site pockets | [83] |
| GenBank MK490918 (Bgl15) |
Directed evolution Error-prone PCR Petri-dish-based double-layer high-throughput screening |
S167V/W178L | Increasing transglycosylation activity | [21] |
| Hot-spring metagenome (BglM) | --- | --- | The narrow space between the remnants of the gatekeeper’s base | [91] |
| Organism source | Improved engineering methods | Mutation sites | Molecular effects |
References |
|---|---|---|---|---|
| T. amestolkiae | Rational design SDM |
E521G | Stimulating glycosyl donor departure Absence of side chains to reduce steric hindrance | [26] |
|
T. naphthophila RKU-10 (Tn0602) |
Rational design SDM |
F226G/F414S | Reducing steric hindrance and removing interactions at the aglycone-binding sites | [96] |
|
T. naphthophila RKU-10 (Tn0602) |
Rational design SDM |
F414S | Improving hydrophilicity of the lumen of the -1 subsite | [9] |
| Thermotoga maritima (TmBglA) | Rational design SDM |
N222F/Y295F / F414S |
Creating a more suitable environment for hexanol in the active center pocket to inhibit hydrolysis | [95] |
| A. niger (BGL1) | Directed evolution Error-prone PCR |
Y305C | Reducing hydrolytic activity | [82] |
| T. neapolitana | Rational design SDM |
N220F, N220R, N220Y | Inhibiting hydrolysis | [94] |
|
T. reesei (TrCel1b) |
Rational design SDM HIFEA Strategy |
I177S/I174S/ W173H |
Inhibition of hydrophilicity of key amino acid residues in the catalytic sites | [28] |
| Method | Access | Description | Reference |
|---|---|---|---|
| Constraint network analysis (CAN) |
--- | Local and global flexibility/stiffness properties of proteins calculated by the graph theory-based rigidity analysis of thermal unfolding simulation. | [103] |
| MD simulation | e.g., GROMACS | Analysis of protein unfolding pathways at higher temperatures. | [70] |
| B-Fitter | https://www.kofo.mpg.de/en/research/organic-synthesis | Calculate and average the B-factor values for all atoms in an amino acid. | [70] |
| FoldUnfold | http://bioinfo.protres.ru/ogu/ | Use the expected average number of contacts per residue calculated from the amino acid sequence as an indicator of whether a given region is folded or unfolded. | [104] |
| PredyFlexy | https://www.dsimb.inserm.fr/dsimb_tools/predyflexy/ | Combine the B-factor with the state of motion of amino acid residues during molecular dynamics simulations. | [102] |
| FIRST | -- | Representation of protein structure as a set of constraints on bond-angle interactions, identification of rigid and flexible regions of protein conformation by CAN. | [105] |
| FlexPred | https://kiharalab.org/flexPred/ | Flexibility in predicting elastic residues using SVM algorithm. | [102] |
| Rosetta Design | Rosetta software | Design of thermally stable proteins based on iterative sidechain optimization and backbone relaxation through optimizing packing and idealizing backbone conformation |
[98] |
| FRESCO | --- | Combined with MD simulations to predict flexible regions of proteins that can incorporate stable disulfide bonds. | [106] |
| HINGEprot | http://bioinfo3d.cs.tau.ac.il/HingeProt/ | Predict the hinge region of a protein. | [102] |
| PROSS | http://pross.weizmann.ac.il | Calculation of ΔΔG and thus analysis of potential stable mutation locations using Rosetta combination sequences. | [107] |
| FireProtDB | https://loschmidt.chemi.muni.cz/fireprotdb/ | Numerical data, structural information for mutation experiments with a variety of proteins. | [108] |
| Organism source | Improved engineering methods | Mutation sites | Molecular effects | References |
|---|---|---|---|---|
| Penicillium funiculosum (PfBgl3A) | Rational design SDM |
--- | --- | [111] |
| A. tumefaciens 5A | Rational design SDM OEP |
W127F, V176A, L178A, L178E | Enhancement of hydrophobic interactions | [86] |
| Metagenomic library of Turpan Depression |
Directed evolution Quikchange |
V174C/A404V/ L441F |
Enhancement of hydrophobic interactions within the enzyme | [84] |
| Thermomicrobium roseum (B9L147) | Rational design SDM OEP |
V169C | -- | [112] |
| H. orenii | Rational design SDM |
V169C/E173L/ I246A |
Increase in hydrophobic interactions | [83] |
| GenBank MK490918 (Bgl15) |
Directed evolution Error-prone PCR Petri-dish-based double-layer high-throughput screening |
S39T/L42N/ V167C/W178L/ A251L/E319A/ E326P/A396V/ L433F |
Increasing hydrophobic interactions and formation of more additional hydrogen bonds | [21] |
| P. piceum H16 | Rational design Proline theory Computer-assisted virtual saturation mutation |
S507F/Q512W/ S514W |
Mutation of glycine by proline reducing conformational entropy Increased hydrophobic interactions |
[32] |
| C. thermocellum | Directed evolution Error-prone PCR |
A17S/K268N | Increasing hydrophobic interactions | [18] |
| GenBank FJ686869 (Bgl1D) |
Directed evolution DNA shuffling |
S28T/Y37H/ D44E/R91G/ L115N |
Enhancing interaction with protein structure around water molecules and introduction of more hydrogen bonds | [24] |
| GenBank HV348683 (Ks5A7) |
Directed evolution Error-prone PCR |
T167I/V181F/ K186T/A187E/ A298G |
Increasing hydrophobic interactions with the protein core | [113] |
| MeBglD2 | Rational design Directed evolution |
His8/Asn59/ Gly295 |
Increasing hydrophobic interactions with the protein core | [114] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





