Submitted:
20 April 2023
Posted:
21 April 2023
You are already at the latest version
Abstract
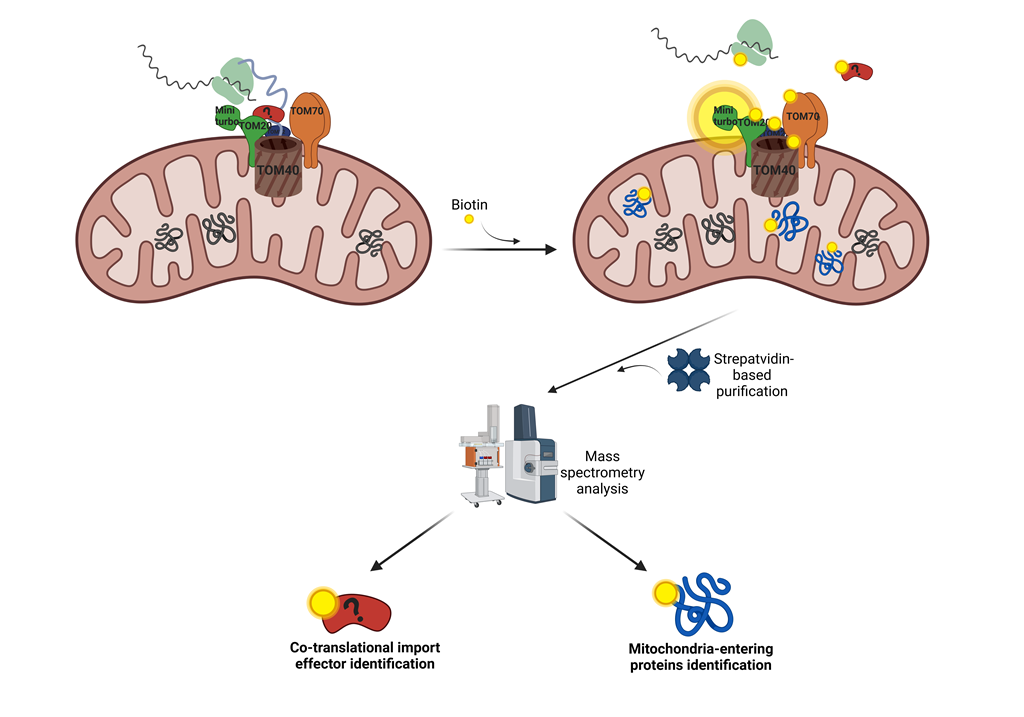
Keywords:
1. INTRODUCTION
2. RESULTS
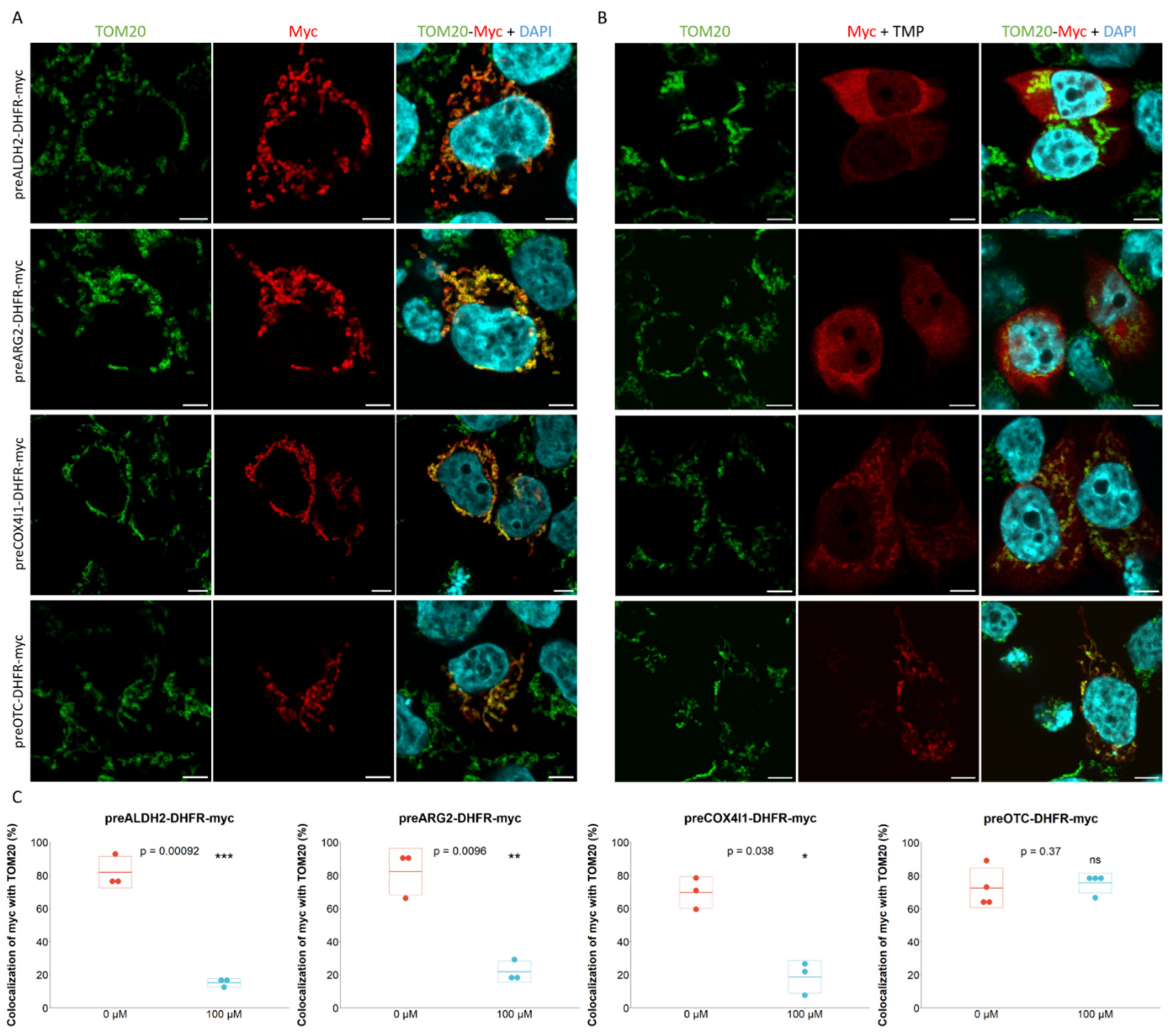
2.1. Characterization of Mitochondrial Post- and Co-Translational Import Reporters
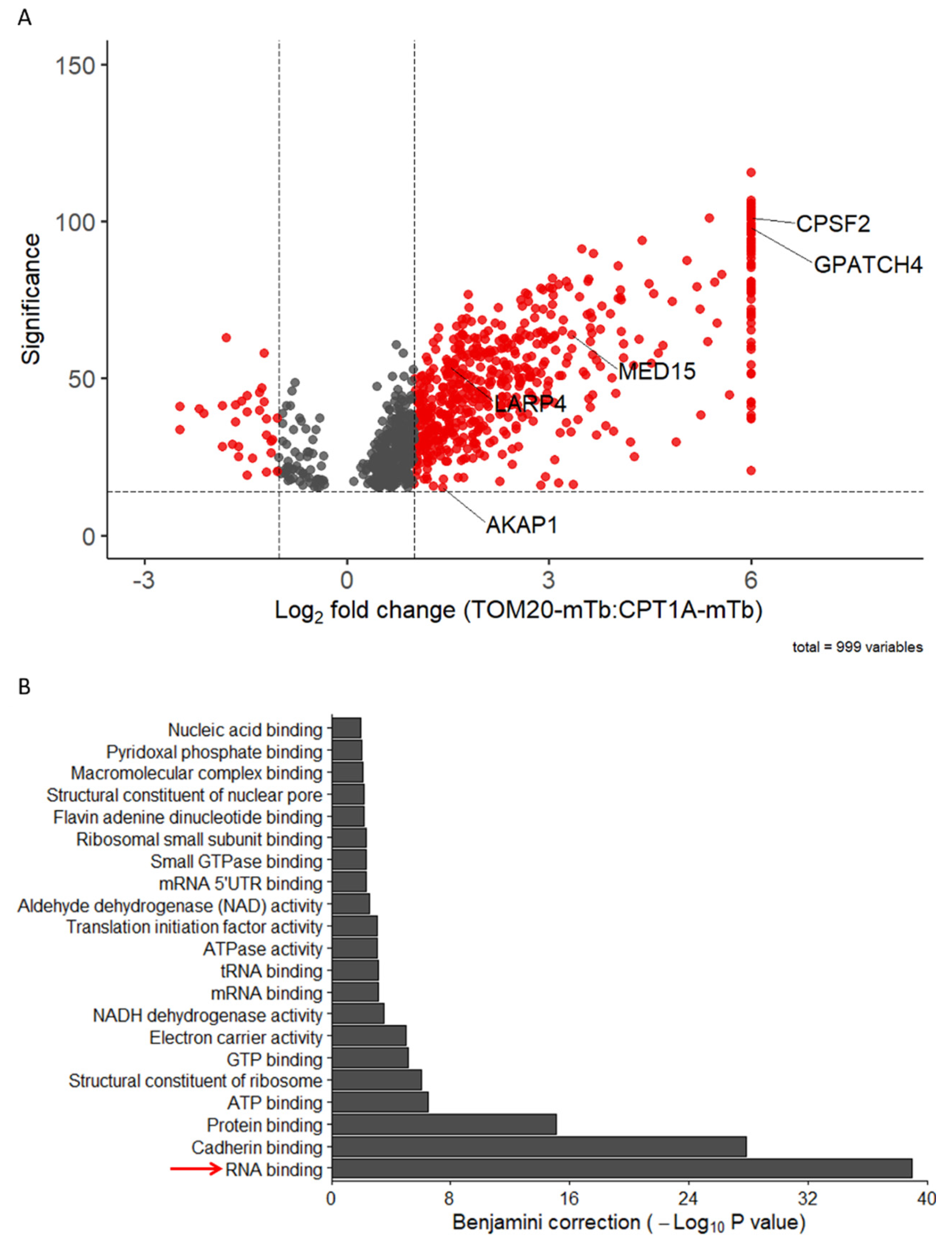
2.2. TOM20 Proxisome Characterization as a Strategy to Identify Effectors of the Mitochondrial Co-Translational Import
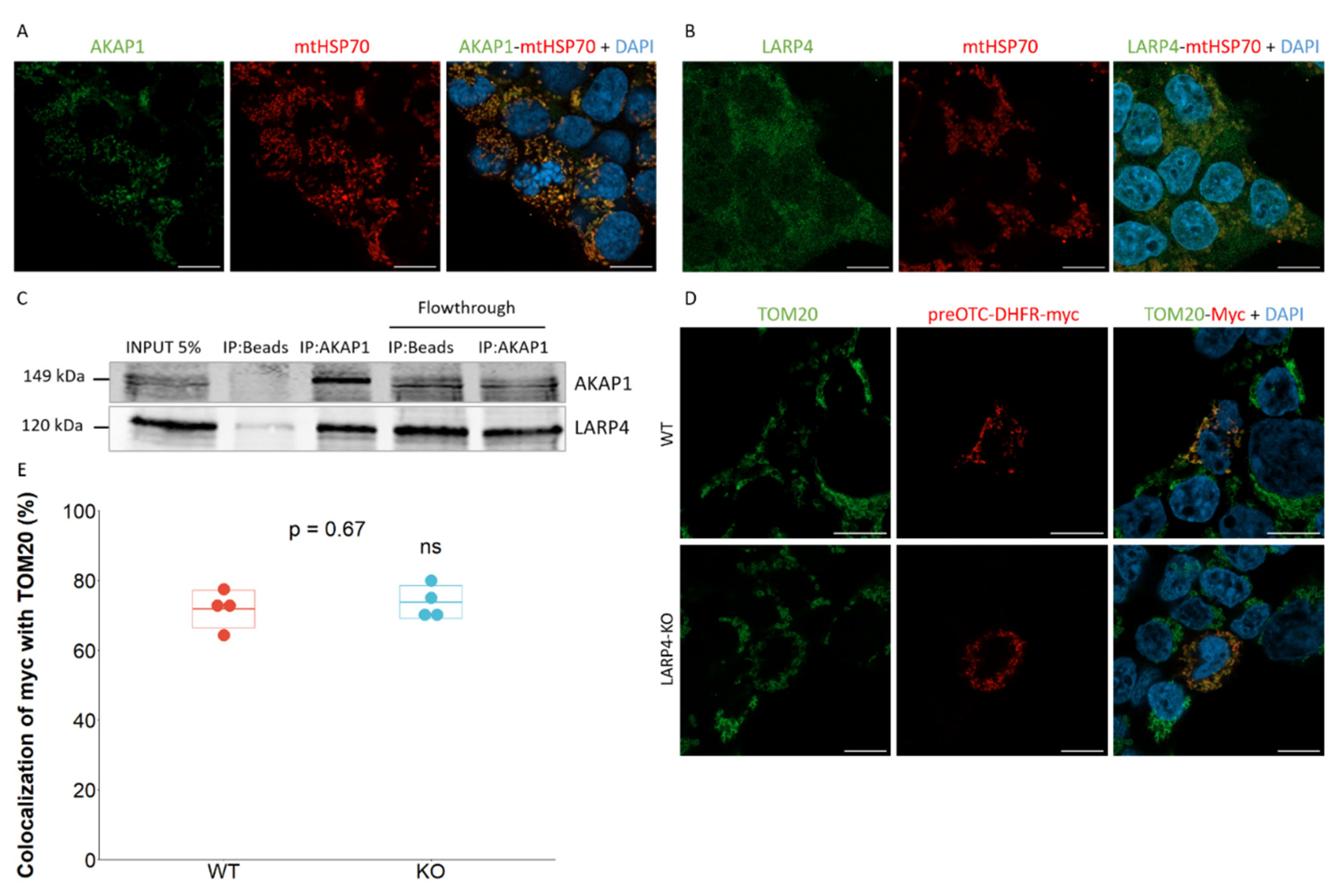
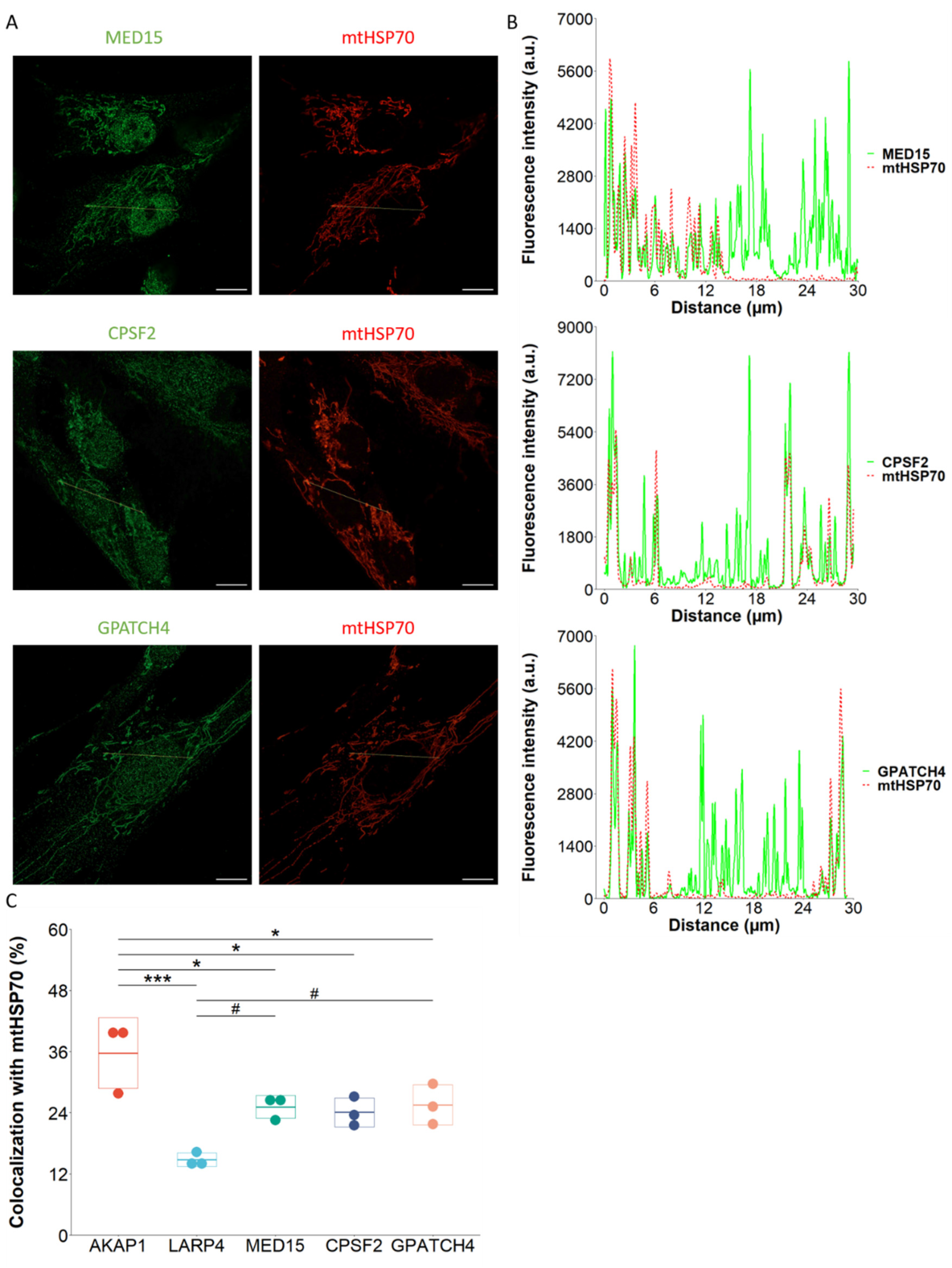
2.3. AKAP1-LARP4 Couple Does not Mediate Mitochondrial Co-Translational Import in Human Cells
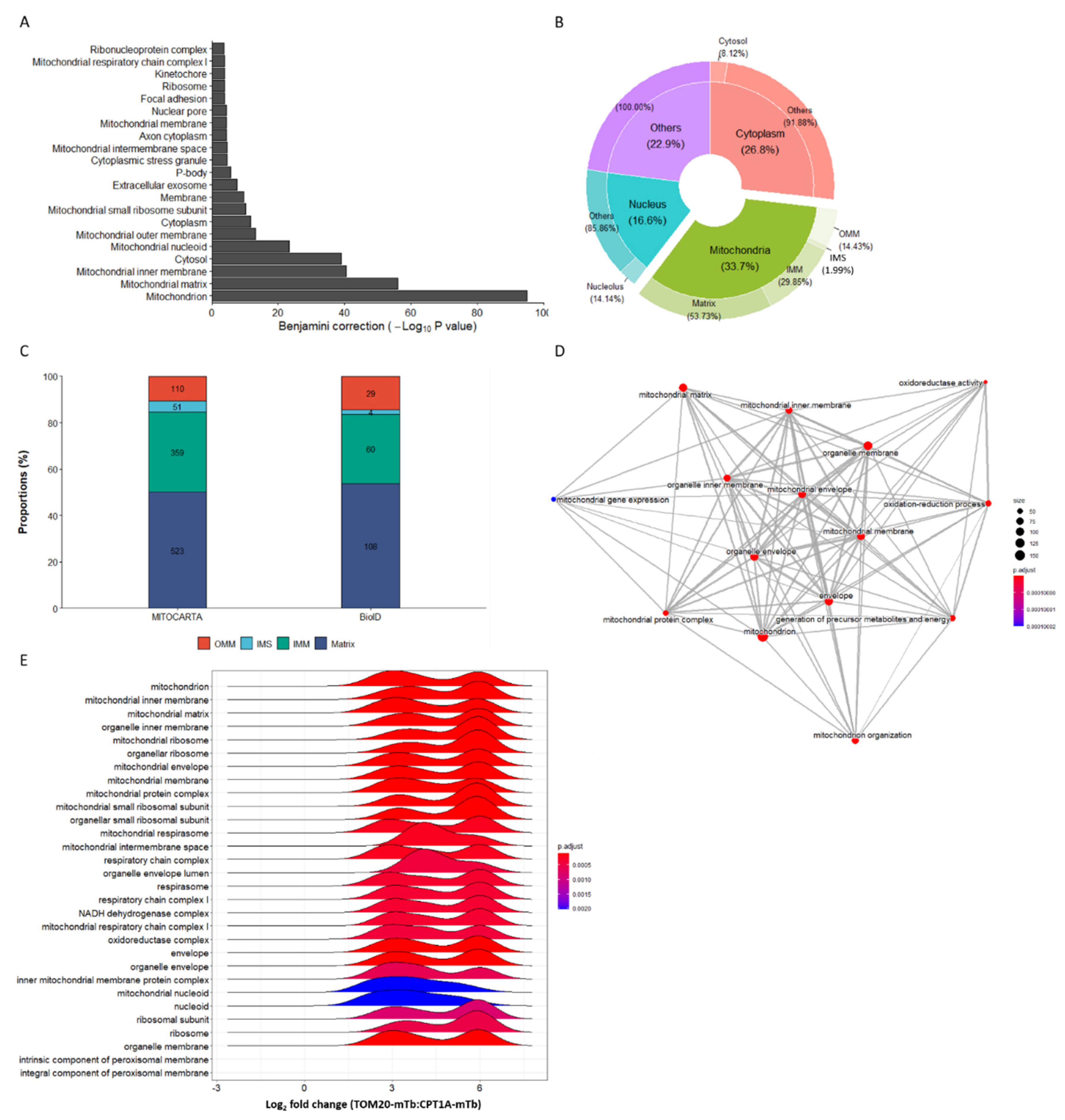
2.4. TOM20-mTb-Based BioID Unravels New Proteins Localized in the Mitochondria
2.5. TOM20-mTb as a Tool to Detect Protein Entry Inside Mitochondria
3. DISCUSSION
4. MATERIALS AND METHODS
4.1. Isolation of Primary Fibroblasts
4.2. Cell Culture
4.3. Generation of KO Cell Lines
4.4. Generation of Endogenous BioID Cell Lines
4.5. Repair Template Plasmids and Reporters Cloning
4.6. Proximity Labeling
4.7. Mass Spectrometry Analyses
4.8. Cell Transfection with Reporter Constructs
4.9. Immunofluorescence Analysis and Confocal Microscopy Observation
4.10. Image Analysis
4.11. Western Blotting
4.12. Immunoprecipitation
4.13. Data Analyses
Supplementary Materials
Author Contributions
Funding
Ethical Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vandemoortele, G.; De Sutter, D.; Moliere, A.; Pauwels, J.; Gevaert, K.; Eyckerman, S. A Well-Controlled BioID Design for Endogenous Bait Proteins. J. Proteome Res. 2018, acs.jproteome.8b00367. [CrossRef]
- Sen, A.; Kallabis, S.; Gaedke, F.; Jüngst, C.; Boix, J.; Nüchel, J.; Maliphol, K.; Hofmann, J.; Schauss, A. C.; Krüger, M.; Wiesner, R. J.; Pla-Martín, D. Mitochondrial Membrane Proteins and VPS35 Orchestrate Selective Removal of MtDNA. Nat. Commun. 2022 131 2022, 13 (1), 1–20. [CrossRef]
- Yoshinaka, T.; Kosako, H.; Yoshizumi, T.; Furukawa, R.; Hirano, Y.; Kuge, O.; Tamada, T.; Koshiba, T. Structural Basis of Mitochondrial Scaffolds by Prohibitin Complexes: Insight into a Role of the Coiled-Coil Region. iScience 2019, 19, 1065–1078. [CrossRef]
- Jiang, S.; Koolmeister, C.; Misic, J.; Siira, S.; Kühl, I.; Ramos, E. S.; Miranda, M.; Jiang, M.; Posse, V.; Lytovchenko, O.; Atanassov, I.; Schober, F. A.; Wibom, R.; Hultenby, K.; Milenkovic, D.; Gustafsson, C. M.; Filipovska, A.; Larsson, N. TEFM Regulates Both Transcription Elongation and RNA Processing in Mitochondria. EMBO Rep. 2019, e48101. [CrossRef]
- Rhee, H. W.; Zou, P.; Udeshi, N. D.; Martell, J. D.; Mootha, V. K.; Carr, S. A.; Ting, A. Y. Proteomic Mapping of Mitochondria in Living Cells via Spatially Restricted Enzymatic Tagging. Science (80-. ). 2013, 339 (6125), 1328–1331. [CrossRef]
- Kim, D. I.; Birendra, K. C.; Zhu, W.; Motamedchaboki, K.; Doye, V.; Roux, K. J. Probing Nuclear Pore Complex Architecture with Proximity-Dependent Biotinylation. Proc. Natl. Acad. Sci. U. S. A. 2014, 111 (24), E2453–E2461. [CrossRef]
- Kaewsapsak, P.; Shechner, D. M.; Mallard, W.; Rinn, J. L.; Ting, A. Y. Live-Cell Mapping of Organelle-Associated RNAs via Proximity Biotinylation Combined with Protein-RNA Crosslinking. Elife 2017, 6. [CrossRef]
- Kwak, C.; Shin, S.; Park, J. S.; Jung, M.; My Nhung, T. T.; Kang, M. G.; Lee, C.; Kwon, T. H.; Park, S. K.; Mun, J. Y.; Kim, J. S.; Rhee, H. W. Contact-ID, a Tool for Profiling Organelle Contact Sites, Reveals Regulatory Proteins of Mitochondrial-Associated Membrane Formation. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (22), 12109–12120. [CrossRef]
- Cho, K. F.; Branon, T. C.; Rajeev, S.; Svinkina, T.; Udeshi, N. D.; Thoudam, T.; Kwak, C.; Rhee, H. W.; Lee, I. K.; Carr, S. A.; Ting, A. Y. Split-TurboID Enables Contact-Dependent Proximity Labeling in Cells. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (22), 12143–12154. [CrossRef]
- Kim, J.; Cantor, A. B.; Orkin, S. H.; Wang, J. Use of in Vivo Biotinylation to Study Protein–Protein and Protein–DNA Interactions in Mouse Embryonic Stem Cells. Nat. Protoc. 2009, 4 (4), 506–517. [CrossRef]
- Titeca, K.; Lemmens, I.; Tavernier, J.; Eyckerman, S. Discovering Cellular Protein-protein Interactions: Technological Strategies and Opportunities. Mass Spectrom. Rev. 2019, 38 (1), 79–111. [CrossRef]
- Koshiba, T.; Kosako, H. Mass Spectrometry-Based Methods for Analysing the Mitochondrial Interactome in Mammalian Cells. Journal of Biochemistry. Oxford University Press March 1, 2020, pp 225–231. [CrossRef]
- Lönn, P.; Landegren, U. Close Encounters - Probing Proximal Proteins in Live or Fixed Cells. Trends Biochem. Sci. 2017, 42 (7), 504–515. [CrossRef]
- Hung, V.; Lam, S. S.; Udeshi, N. D.; Svinkina, T.; Guzman, G.; Mootha, V. K.; Carr, S. A.; Ting, A. Y. Proteomic Mapping of Cytosol-Facing Outer Mitochondrial and ER Membranes in Living Human Cells by Proximity Biotinylation. Elife 2017, 6. [CrossRef]
- Han, S.; Udeshi, N. D.; Deerinck, T. J.; Svinkina, T.; Ellisman, M. H.; Carr, S. A.; Ting, A. Y. Proximity Biotinylation as a Method for Mapping Proteins Associated with MtDNA in Living Cells. Cell Chem. Biol. 2017, 24 (3), 404–414. [CrossRef]
- Fazal, F. M.; Han, S.; Parker, K. R.; Kaewsapsak, P.; Xu, J.; Boettiger, A. N.; Chang, H. Y.; Ting, A. Y. Atlas of Subcellular RNA Localization Revealed by APEX-Seq. Cell 2019, 178 (2), 473-490.e26. [CrossRef]
- Williams, C. C.; Jan, C. H.; Weissman, J. S. Targeting and Plasticity of Mitochondrial Proteins Revealed by Proximity-Specific Ribosome Profiling. Science 2014, 346 (6210), 748–751. [CrossRef]
- Vardi-Oknin, D.; Arava, Y. Characterization of Factors Involved in Localized Translation Near Mitochondria by Ribosome-Proximity Labeling. Front. Cell Dev. Biol. 2019, 7, 305. [CrossRef]
- Yoo, C. M.; Rhee, H. W. APEX, a Master Key to Resolve Membrane Topology in Live Cells. Biochemistry 2020, 59 (3). [CrossRef]
- Lee, S. Y.; Kang, M. G.; Park, J. S.; Lee, G.; Ting, A. Y.; Rhee, H. W. APEX Fingerprinting Reveals the Subcellular Localization of Proteins of Interest. Cell Rep. 2016, 15 (8), 1837–1847. [CrossRef]
- Branon, T. C.; Bosch, J. A.; Sanchez, A. D.; Udeshi, N. D.; Svinkina, T.; Carr, S. A.; Feldman, J. L.; Perrimon, N.; Ting, A. Y. Efficient Proximity Labeling in Living Cells and Organisms with TurboID. Nature Biotechnology. Nature Publishing Group October 1, 2018, pp 880–898. [CrossRef]
- Zhao, X.; Bitsch, S.; Kubitz, L.; Schmitt, K.; Deweid, L.; Roehrig, A.; Barazzone, E. C.; Valerius, O.; Kolmar, H.; Béthune, J. UltraID: A Compact and Efficient Enzyme for Proximity-Dependent Biotinylation in Living Cells. bioRxiv 2021, 2021.06.16.448656. [CrossRef]
- Avendaño-Monsalve, M. C.; Ponce-Rojas, J. C.; Funes, S. From Cytosol to Mitochondria: The Beginning of a Protein Journey. Biological Chemistry. De Gruyter May 1, 2020, pp 645–661. [CrossRef]
- Pfanner, N.; Warscheid, B.; Wiedemann, N. Mitochondrial Proteins: From Biogenesis to Functional Networks. Nat. Rev. Mol. Cell Biol. 2019. [CrossRef]
- Becker, T.; Song, J.; Pfanner, N. Versatility of Preprotein Transfer from the Cytosol to Mitochondria. Trends in Cell Biology. Elsevier Ltd July 1, 2019, pp 534–548. [CrossRef]
- Pfanner, N.; Geissler, A. Versatility of the Mitochondrial Protein Import Machinery. Nat. Rev. Mol. Cell Biol. 2001, 2 (5), 339–349. [CrossRef]
- Dudek, J.; Rehling, P.; van der Laan, M. Mitochondrial Protein Import: Common Principles and Physiological Networks. Biochimica et Biophysica Acta - Molecular Cell Research. Elsevier B.V. 2013, pp 274–285. [CrossRef]
- Tucker, K.; Park, E. Cryo-EM Structure of the Mitochondrial Protein-Import Channel TOM Complex at near-Atomic Resolution. Nat. Struct. Mol. Biol. 2019, 26 (12), 1158–1166. [CrossRef]
- Araiso, Y.; Tsutsumi, A.; Qiu, J.; Imai, K.; Shiota, T.; Song, J.; Lindau, C.; Wenz, L.-S.; Sakaue, H.; Yunoki, K.; Kawano, S.; Suzuki, J.; Wischnewski, M.; Schütze, C.; Ariyama, H.; Ando, T.; Becker, T.; Lithgow, T.; Wiedemann, N.; Pfanner, N.; Kikkawa, M.; Endo, T. Structure of the Mitochondrial Import Gate Reveals Distinct Preprotein Paths. Nature 2019, 1–1. [CrossRef]
- Opaliński, Ł.; Song, J.; Priesnitz, C.; Wenz, L.-S.; Oeljeklaus, S.; Warscheid, B.; Pfanner, N.; Becker, T. Recruitment of Cytosolic J-Proteins by TOM Receptors Promotes Mitochondrial Protein Biogenesis. Cell Rep. 2018, 25 (8), 2036-2043.e5. [CrossRef]
- Hansen, K. G.; Herrmann, J. M. Transport of Proteins into Mitochondria. Protein J. 2019 383 2019, 38 (3), 330–342. [CrossRef]
- Zhang, Y.; Xu, H. Translational Regulation of Mitochondrial Biogenesis. Biochem. Soc. Trans. 2016, 44 (6), 1717–1724. [CrossRef]
- Lesnik, C.; Golani-Armon, A.; Arava, Y. Localized Translation near the Mitochondrial Outer Membrane: An Update. RNA Biol. 2015, 12 (8), 801–809. [CrossRef]
- Lesnik, C.; Cohen, Y.; Atir-Lande, A.; Schuldiner, M.; Arava, Y. OM14 Is a Mitochondrial Receptor for Cytosolic Ribosomes That Supports Co-Translational Import into Mitochondria. Nat. Commun. 2014, 5 (1), 5711. [CrossRef]
- Quenault, T.; Lithgow, T.; Traven, A. PUF Proteins: Repression, Activation and MRNA Localization. Trends Cell Biol. 2011, 21 (2), 104–112. [CrossRef]
- Saint-Georges, Y.; Garcia, M.; Delaveau, T.; Jourdren, L.; Le Crom, S.; Lemoine, S.; Tanty, V.; Devaux, F.; Jacq, C. Yeast Mitochondrial Biogenesis: A Role for the PUF RNA-Binding Protein Puf3p in MRNA Localization. PLoS One 2008, 3 (6), e2293. [CrossRef]
- Devaux, F.; Lelandais, G.; Garcia, M.; Goussard, S.; Jacq, C. Posttranscriptional Control of Mitochondrial Biogenesis: Spatio-Temporal Regulation of the Protein Import Process. FEBS Lett. 2010, 584 (20), 4273–4279. [CrossRef]
- Lapointe, C. P.; Stefely, J. A.; Jochem, A.; Hutchins, P. D.; Wilson, G. M.; Kwiecien, N. W.; Coon, J. J.; Wickens, M.; Pagliarini, D. J. Multi-Omics Reveal Specific Targets of the RNA-Binding Protein Puf3p and Its Orchestration of Mitochondrial Biogenesis. Cell Syst. 2018, 6 (1), 125-135.e6. [CrossRef]
- Eliyahu, E.; Pnueli, L.; Melamed, D.; Scherrer, T.; Gerber, A. P.; Pines, O.; Rapaport, D.; Arava, Y. Tom20 Mediates Localization of MRNAs to Mitochondria in a Translation-Dependent Manner. Mol. Cell. Biol. 2010, 30 (1), 284–294. [CrossRef]
- Gehrke, S.; Wu, Z.; Klinkenberg, M.; Sun, Y.; Auburger, G.; Guo, S.; Lu, B. PINK1 and Parkin Control Localized Translation of Respiratory Chain Component MRNAs on Mitochondria Outer Membrane. Cell Metab. 2015, 21 (1), 95–108. [CrossRef]
- Zhang, Y.; Wang, Z. H.; Liu, Y.; Chen, Y.; Sun, N.; Gucek, M.; Zhang, F.; Xu, H. PINK1 Inhibits Local Protein Synthesis to Limit Transmission of Deleterious Mitochondrial DNA Mutations. Mol. Cell 2019, 73 (6), 1127-1137.e5. [CrossRef]
- Zhang, Y.; Chen, Y.; Gucek, M.; Xu, H. The Mitochondrial Outer Membrane Protein MDI Promotes Local Protein Synthesis and MtDNA Replication. EMBO J. 2016, 35 (10), 1045–1057. [CrossRef]
- Sen, A.; Kalvakuri, S.; Bodmer, R.; Cox, R. T. Clueless, a Protein Required for Mitochondrial Function, Interacts with the PINK1-Parkin Complex in Drosophila. Dis. Model. Mech. 2015, 8 (6), 577–589. [CrossRef]
- Sen, A.; Cox, R. T. Clueless Is a Conserved Ribonucleoprotein That Binds the Ribosome at the Mitochondrial Outer Membrane. Biol. Open 2016, 5 (2), 195–203. [CrossRef]
- Mukhopadhyay, A.; Ni, L.; Weiner, H. A Co-Translational Model to Explain the in Vivo Import of Proteins into HeLa Cell Mitochondria. Biochem. J. 2004, 382 (1), 385–392. [CrossRef]
- Sylvestre, J.; Margeot, A.; Jacq, C.; Dujardin, G.; Corral-Debrinski, M. The Role of the 3′ Untranslated Region in MRNA Sorting to the Vicinity of Mitochondria Is Conserved from Yeast to Human Cells. Mol. Biol. Cell 2003, 14 (9), 3848–3856. [CrossRef]
- Matsumoto, S.; Uchiumi, T.; Saito, T.; Yagi, M.; Takazaki, S.; Kanki, T.; Kang, D. Localization of MRNAs Encoding Human Mitochondrial Oxidative Phosphorylation Proteins. Mitochondrion 2012, 12 (3), 391–398. [CrossRef]
- Jan, C. H.; Williams, C. C.; Weissman, J. S. Principles of ER Cotranslational Translocation Revealed by Proximity-Specific Ribosome Profiling. Science 2014, 346 (6210), 1257521. [CrossRef]
- Grevel, A.; Pfanner, N.; Becker, and T. Coupling of Import and Assembly Pathways in Mitochondrial Protein Biogenesis. Biol. Chem. 2019, 401 (1), 117–129.
- Priesnitz, C.; Becker, T. Pathways to Balance Mitochondrial Translation and Protein Import. Genes and Development. Cold Spring Harbor Laboratory Press October 1, 2018, pp 1285–1296. [CrossRef]
- Fujiki, M.; Verner, K. Coupling of Cytosolic Protein Synthesis and Mitochondrial Protein Import in Yeast. Evidence for Cotranslational Import in Vivo. J. Biol. Chem. 1993, 268 (3). [CrossRef]
- Baker, D. J.; Beddell, C. R.; Champness, J. N.; Goodford, P. J.; Norrington, F. E. A.; Smith, D. R.; Stammers, D. K. The Binding of Trimethoprim to Bacterial Dihydrofolate Reductase. FEBS Lett. 1981, 126 (1). [CrossRef]
- Donnelly, M. L. L.; Hughes, L. E.; Luke, G.; Mendoza, H.; Ten Dam, E.; Gani, D.; Ryan, M. D. The “cleavage” Activities of Foot-and-Mouth Disease Virus 2A Site-Directed Mutants and Naturally Occurring “2A-like” Sequences. J. Gen. Virol. 2001, 82 (5), 1027–1041. [CrossRef]
- Dennis, G.; Sherman, B. T.; Hosack, D. A.; Yang, J.; Gao, W.; Lane, H. C.; Lempicki, R. A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4 (5), 1–11. [CrossRef]
- Gabrovsek, L.; Collins, K. B.; Aggarwal, S.; Saunders, L. M.; Lau, H. T.; Suh, D.; Sancak, Y.; Trapnell, C.; Ong, S. E.; Smith, F. D.; Scott, J. D. A-Kinase-Anchoring Protein 1 (DAKAP1)-Based Signaling Complexes Coordinate Local Protein Synthesis at the Mitochondrial Surface. J. Biol. Chem. 2020, 295 (31), 10749–10765. [CrossRef]
- Calvo, S. E.; Clauser, K. R.; Mootha, V. K. MitoCarta2.0: An Updated Inventory of Mammalian Mitochondrial Proteins. Nucleic Acids Res. 2016, 44 (D1), D1251–D1257. [CrossRef]
- Rhee, H.-W.; Zou, P.; Udeshi, N. D.; Martell, J. D.; Mootha, V. K.; Carr, S. A.; Ting, A. Y. Proteomic Mapping of Mitochondria in Living Cells via Spatially Restricted Enzymatic Tagging. Science 2013, 339 (6125), 1328–1331. [CrossRef]
- Chan, X. C. Y.; Black, C. M.; Lin, A. J.; Ping, P.; Lau, E. Mitochondrial Protein Turnover: Methods to Measure Turnover Rates on a Large Scale. Journal of Molecular and Cellular Cardiology. 2015. [CrossRef]
- Li, J.; Cai, Z.; Vaites, L. P.; Shen, N.; Mitchell, D. C.; Huttlin, E. L.; Paulo, J. A.; Harry, B. L.; Gygi, S. P. Proteome-Wide Mapping of Short-Lived Proteins in Human Cells. Mol. Cell 2021, 81 (22), 4722-4735.e5. [CrossRef]
- Mathieson, T.; Franken, H.; Kosinski, J.; Kurzawa, N.; Zinn, N.; Sweetman, G.; Poeckel, D.; Ratnu, V. S.; Schramm, M.; Becher, I.; Steidel, M.; Noh, K. M.; Bergamini, G.; Beck, M.; Bantscheff, M.; Savitski, M. M. Systematic Analysis of Protein Turnover in Primary Cells. Nat. Commun. 2018 91 2018, 9 (1), 1–10. [CrossRef]
- Kleinjan, D. A.; Wardrope, C.; Nga Sou, S.; Rosser, S. J. Drug-Tunable Multidimensional Synthetic Gene Control Using Inducible Degron-Tagged DCas9 Effectors. Nat. Commun. 2017 81 2017, 8 (1), 1–9. [CrossRef]
- Richter-Dennerlein, R.; Oeljeklaus, S.; Lorenzi, I.; Ronsör, C.; Bareth, B.; Schendzielorz, A. B.; Wang, C.; Warscheid, B.; Rehling, P.; Dennerlein, S. Mitochondrial Protein Synthesis Adapts to Influx of Nuclear-Encoded Protein. Cell 2016, 167 (2), 471-483.e10. [CrossRef]
- Couvillion, M. T.; Soto, I. C.; Shipkovenska, G.; Churchman, L. S. Synchronized Mitochondrial and Cytosolic Translation Programs. Nature 2016, 533 (7604), 499–503. [CrossRef]
- Wanet, A.; Arnould, T.; Najimi, M.; Renard, P. Connecting Mitochondria, Metabolism, and Stem Cell Fate. Stem Cells Dev. 2015, 24 (17), 1957–1971. [CrossRef]
- Roberts, B.; Haupt, A.; Tucker, A.; Grancharova, T.; Arakaki, J.; Fuqua, M. A.; Nelson, A.; Hookway, C.; Ludmann, S. A.; Mueller, I. A.; Yang, R.; Horwitz, R.; Rafelski, S. M.; Gunawardane, R. N. Systematic Gene Tagging Using CRISPR/Cas9 in Human Stem Cells to Illuminate Cell Organization. Mol. Biol. Cell 2017, 28 (21), 2854–2874. [CrossRef]
- Tytgat, H. L. P.; Schoofs, G.; Driesen, M.; Proost, P.; Van Damme, E. J. M.; Vanderleyden, J.; Lebeer, S. Endogenous Biotin-Binding Proteins: An Overlooked Factor Causing False Positives in Streptavidin-Based Protein Detection. Microb. Biotechnol. 2015, 8 (1), 164–168. [CrossRef]
- Liu, Z.; Chen, O.; Wall, J. B. J.; Zheng, M.; Zhou, Y.; Wang, L.; Ruth Vaseghi, H.; Qian, L.; Liu, J. Systematic Comparison of 2A Peptides for Cloning Multi-Genes in a Polycistronic Vector. Sci. Rep. 2017, 7 (1), 1–9. [CrossRef]
- Gold, V. A.; Chroscicki, P.; Bragoszewski, P.; Chacinska, A. Visualization of Cytosolic Ribosomes on the Surface of Mitochondria by Electron Cryo-Tomography. EMBO Rep. 2017, 18 (10), 1786–1800. [CrossRef]
- Pla-Martín, D.; Schatton, D.; Wiederstein, J. L.; Marx, M.; Khiati, S.; Krüger, M.; Rugarli, E. I. CLUH Granules Coordinate Translation of Mitochondrial Proteins with MTORC1 Signaling and Mitophagy. EMBO J. 2020, 39 (9). [CrossRef]
- Youn, D. Y.; Xiaoli, A. M.; Pessin, J. E.; Yang, F. Regulation of Metabolism by the Mediator Complex. Biophys. Reports 2016, 2 (2–4), 69–77. [CrossRef]
- Yang, F.; Vought, B. W.; Satterlee, J. S.; Walker, A. K.; Jim Sun, Z. Y.; Watts, J. L.; DeBeaumont, R.; Mako Saito, R.; Hyberts, S. G.; Yang, S.; Macol, C.; Iyer, L.; Tjian, R.; Van Den Heuvel, S.; Hart, A. C.; Wagner, G.; Näär, A. M. An ARC/Mediator Subunit Required for SREBP Control of Cholesterol and Lipid Homeostasis. Nature 2006, 442 (7103), 700–704. [CrossRef]
- Tuttle, L. M.; Pacheco, D.; Warfield, L.; Wilburn, D. B.; Hahn, S.; Klevit, R. E. Mediator Subunit Med15 Dictates the Conserved “Fuzzy” Binding Mechanism of Yeast Transcription Activators Gal4 and Gcn4. Nat. Commun. 2021 121 2021, 12 (1), 1–11. [CrossRef]
- Zhang, Y.; Sun, Y.; Shi, Y.; Walz, T.; Tong, L. Structural Insights into the Human Pre-MRNA 3′-End Processing Machinery. Mol. Cell 2020, 77 (4), 800-809.e6. [CrossRef]
- Davis, M. R.; Delaleau, M.; Borden, K. L. B. Nuclear EIF4E Stimulates 3′-End Cleavage of Target RNAs. Cell Rep. 2019, 27 (5), 1397-1408.e4. [CrossRef]
- Hirawake-Mogi, H.; Thanh Nhan, N. T.; Okuwaki, M. G-Patch Domain-Containing Protein 4 Localizes to Both the Nucleoli and Cajal Bodies and Regulates Cell Growth and Nucleolar Structure. Biochem. Biophys. Res. Commun. 2021, 559, 99–105. [CrossRef]
- Morgenstern, M.; Peikert, C. D.; Lübbert, P.; Suppanz, I.; Klemm, C.; Alka, O.; Steiert, C.; Naumenko, N.; Schendzielorz, A.; Melchionda, L.; Mühlhäuser, W. W. D.; Knapp, B.; Busch, J. D.; Stiller, S. B.; Dannenmaier, S.; Lindau, C.; Licheva, M.; Eickhorst, C.; Galbusera, R.; Zerbes, R. M.; Ryan, M. T.; Kraft, C.; Kozjak-Pavlovic, V.; Drepper, F.; Dennerlein, S.; Oeljeklaus, S.; Pfanner, N.; Wiedemann, N.; Warscheid, B. Quantitative High-Confidence Human Mitochondrial Proteome and Its Dynamics in Cellular Context. Cell Metab. 2021, 33 (12). [CrossRef]
- Bogenhagen, D. F.; Haley, J. D. Pulse-Chase SILAC-Based Analyses Reveal Selective Oversynthesis and Rapid Turnover of Mitochondrial Protein Components of Respiratory Complexes. J. Biol. Chem. 2020, 295 (9), 2544–2554. [CrossRef]
- Bogenhagen, D. F.; Ostermeyer-Fay, A. G.; Haley, J. D.; Garcia-Diaz, M. Kinetics and Mechanism of Mammalian Mitochondrial Ribosome Assembly. Cell Rep. 2018, 22 (7), 1935–1944. [CrossRef]
- Tigges, J.; Weighardt, H.; Wolff, S.; Götz, C.; Förster, I.; Kohne, Z.; Huebenthal, U.; Merk, H. F.; Abel, J.; Haarmann-Stemmann, T.; Krutmann, J.; Fritsche, E. Aryl Hydrocarbon Receptor Repressor (AhRR) Function Revisited: Repression of CYP1 Activity in Human Skin Fibroblasts Is Not Related to AhRR Expression. J. Invest. Dermatol. 2013, 133 (1), 87–96. [CrossRef]
- Concordet, J. P.; Haeussler, M. CRISPOR: Intuitive Guide Selection for CRISPR/Cas9 Genome Editing Experiments and Screens. Nucleic Acids Res. 2018, 46 (W1), W242–W245. [CrossRef]
- Vandemoortele, G.; De Sutter, D.; Eyckerman, S. Robust Generation of Knock-in Cell Lines Using CRISPR-Cas9 and RAAV-Assisted Repair Template Delivery. BIO-PROTOCOL 2017, 7 (7). [CrossRef]
- Czlapinski, J. L.; Schelle, M. W.; Miller, L. W.; Laughlin, S. T.; Kohler, J. J.; Cornish, V. W.; Bertozzi, C. R. Conditional Glycosylate in Eukaryotic Cells Using a Biocompatible Chemical Inducer of Dimerization. J. Am. Chem. Soc. 2008, 130 (40), 13186–13187. [CrossRef]
- Le Sage, V.; Cinti, A.; Mouland, A. J. Proximity-Dependent Biotinylation for Identification of Interacting Proteins. Curr. Protoc. Cell Biol. 2016, 73 (1), 17.19.1-17.19.12. [CrossRef]
- Meier, F.; Brunner, A. D.; Koch, S.; Koch, H.; Lubeck, M.; Krause, M.; Goedecke, N.; Decker, J.; Kosinski, T.; Park, M. A.; Bache, N.; Hoerning, O.; Cox, J.; Räther, O.; Mann, M. Online Parallel Accumulation-Serial Fragmentation (PASEF) with a Novel Trapped Ion Mobility Mass Spectrometer. Mol. Cell. Proteomics 2018, 17 (12), 2534–2545. [CrossRef]
- Lin, H.; He, L.; Ma, B. A Combinatorial Approach to the Peptide Feature Matching Problem for Label-Free Quantification. Bioinformatics 2013, 29 (14), 1768–1775. [CrossRef]
- Blighe, K.; Rana, S.; Turkes, E.; Ostenforf, B.; Grioni, A.; Lewis, M. Publication-ready volcano plots with enhanced colouring and labeling. [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D. J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; Walzer, M.; Wang, S.; Brazma, A.; Vizcaíno, J. A. The PRIDE Database Resources in 2022: A Hub for Mass Spectrometry-Based Proteomics Evidences. Nucleic Acids Res. 2022, 50 (D1), D543–D552. [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; Tinevez, J. Y.; White, D. J.; Hartenstein, V.; Eliceiri, K.; Tomancak, P.; Cardona, A. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012 97 2012, 9 (7), 676–682. [CrossRef]
- Katrukha, E. Ekatrukha/ComDet: ComDet 0.5.3. 2020. [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; Fu, X.; Liu, S.; Bo, X.; Yu, G. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innov. 2021, 2 (3). [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




