1. Introduction
Soybean (Glycine max (L.) Merrill) is an economically important crop in Canada, with major production in Ontario, Quebec, and Manitoba. In the 2023/2024 season, Canadian soybean production reached approximately 7.57 million metric tons, contributing substantially to domestic feed, food, and export markets (Statistics Canada, 2024; Soy Canada, 2025 ).
Soybean is also a cornerstone of global food, feed, and biofuel production. In the 2023/2024 season, global soybean production reached approximately 397 million metric tons, with Brazil and the United States contributing 40% and 28%, respectively (USDA Foreign Agricultural Service, 2025).
Worldwide, and in Canada’s soybean-growing regions, fungal diseases represent a persistent and emerging threat to yield stability (Rossman, 2009; Udayanga et al., 2014). Annual losses in soybean due to fungal diseases are estimated to exceed 8 Million MTs (Bradley et al 2021) Species of the genus Diaporthe Nitschke (≡ Phomopsis (Sacc.) Bubak) typically cause pod and stem blight, stem canker, and seed decay, leading to yield reductions of 7.8 million MTs in the Northern USA and Canadian soybean producing regions(Kulik, 1984; Sinclair, 1993; Dorrance & Mills, 2009; Vidić et al., 2011, Bradley et al 2021, Rupe et al. 2025 ) .Despite being known to infect soybean roots and cause root rot, the impact of Diaporthe species on root-associated diseases remains largely overlooked (Ghissi et al., 2014), in comparison to more well studied root rot diseases (Wrather et al., 2001; Zhang et al., 2013; Abdelmagid et al., 2018; Hafez et al., 2021; Markell et al., 2024). Further investigation into Diaporthe spp. role in root disease complexes is required to make informed management strategies.
Diaporthe sojae, is a homothallic species characterized by abundant perithecia formation in culture, primarily causing soybeanpod and stem blights (Lehman, 1923; Santos et al., 2011; Udayanga et al., 2014). Direct confirmation of D. sojae as a root infecting pathogen leading to soybean root rot remains uncommon (Gerdemann, 1954; Ghissi et al., 2014). . Evidence indicates that D. sojae (syn D. phaseolorum var sojae)can colonize subterranean tissues at the stem base and upper taproot. Pedersen and Grau (2010) isolated D. sojae from the upper portion of soybean roots and with associated yield reductions in relation to its incidence. In Ontario, D.sojae was found as a component of the Diaporthe–Phomopsis complex in field-grown soybean (isolated from stems, pods, and seed) and reported that multiple seed treatments reduced lesion severity in the lower stem–taproot region scored as “root-rot severity,”(Xue et al 2007). .Accordingly, the isolation of D. sojae from soybean roots exhibiting root rot symptoms in Manitoba warrants evaluation of its epidemiological role and pathogenicity as a potential root rot pathogen or association with other pathogens as part of a complex. To our knowledge, there have been no previously documented cases of D. sojae as a confirmed a direct root rot pathogen in North America.
The objectives of the present study were to:
Document the first confirmed occurrence of D. sojae as a causal agent of soybean root rot in Western Canada and evaluate its pathogenicity using Koch’s postulates.
Determine the extent of virulence diversity among different D. sojae isolates.
These findings aim to broaden the known disease profile of D. sojae, shed light on its epidemiological significance, and underscore the critical need for accurate pathogen identification in developing effective disease management strategies.
2. MATERIALS AND METHODS
2.1. Plant Materials and Fungal Isolation
During the 2024 Manitoba soybean survey, plants showing root and crown decline symptoms were collected from 20 commercial fields. Affected plants exhibited noticeable thinning, stunted growth, and reduced pod set compared with healthy plants. Symptomatic roots were dark brown to black, with progressive taproot deterioration, poorly developed lateral roots, and reduced nodulation. Longitudinal sections of roots and lower stems revealed cortical decay and tissue maceration, consistent with advanced root rot (
Figure 1A, B). From each field, five symptomatic plants were sampled, resulting in a total of 100 plants processed for pathogen isolation.
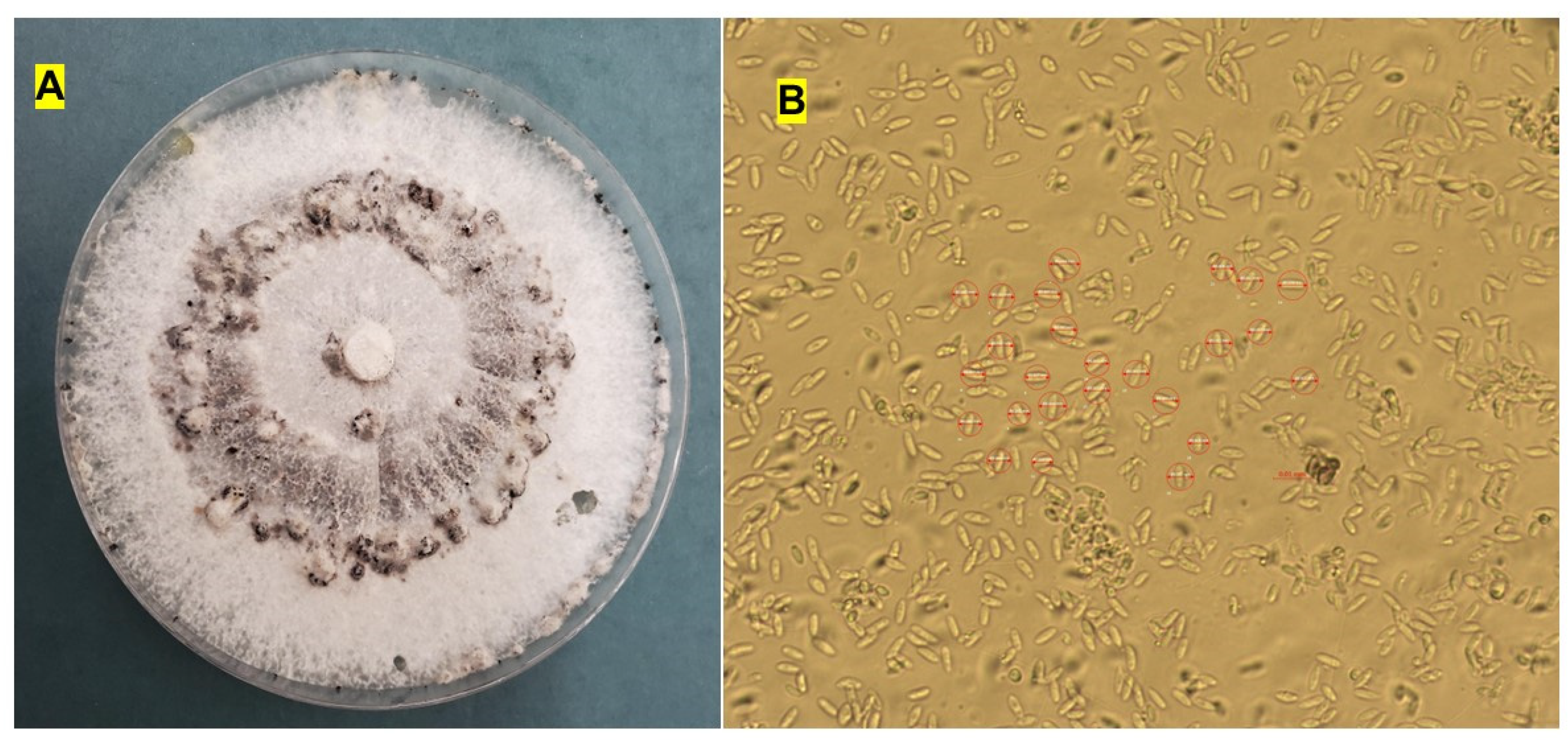
Symptomatic roots were cut into 1–2 cm segments, surface-sterilized in 1% sodium hypochlorite (NaOCl) for 2 min, rinsed twice in sterile distilled water, and air-dried on autoclaved filter paper. Sterilized root segments were plated onto potato dextrose agar (PDA; Becton, Dickinson and Company, Sparks, MD, USA) supplemented with 100 mg L⁻¹ streptomycin sulfate to suppress bacterial contamination. For each field, root segments from five plants were distributed across three PDA plates, with five tissue pieces per plate. Plates were incubated at 25 ± 2 °C under a 12 h light/12 h dark photoperiod for 48 h. Emerging fungal colonies were sub-cultured onto fresh PDA using the hyphal-tip method (Leyronas et al., 2012) and maintained under the same conditions. Pure cultures obtained by hyphal-tip isolation were subjected to preliminary morphological identification based on colony and microscopic characteristics. Colony morphology, including color, texture, and growth pattern, was assessed on PDA after incubation at 25 ± 2 °C for 3 weeks. Microscopic examination of conidial morphology was conducted using a compound light microscope, and conidial measurements were recorded to support species-level identification. All measurements and image rendering were performed using ZEN 3.1 (blue edition; Carl Zeiss Microscopy GmbH, Jena, Germany).
2.2. Molecular Identification and Phylogenetic Analysis
2.2.1. DNA Extraction, PCR Amplification, and Sequencing
Genomic DNA was extracted from 4-week-old pure fungal cultures using a DNeasy Plant DNA Extraction Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Species-level identification was performed by sequencing two conserved genetic regions: the translation elongation factor 1-alpha (EF1-α) gene and the nuclear ribosomal internal transcribed spacer (ITS) region. The EF1-α gene was amplified using primers EF1 (5'-ATG GGT AAG GAG GAC AAG AC-3') and EF2 (5'-GGA GGT ACC AGT GAT CAT G-3'), while the ITS region was amplified using primers ITS4B (5'-TTC CAC CGC TTA TTG ATA TGC-3') and BMBC-F (5'-GTA CAC ACC GCC CGT CG-3') (O’Donnell et al., 1998).
PCR amplifications were conducted under the following thermal cycling conditions: initial denaturation at 95 °C for 2 min; followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 40 s; with a final extension at 72 °C for 10 min. PCR products were resolved by electrophoresis on 1.5% agarose gels stained with ethidium bromide (0.5 μg mL⁻¹) and visualized using a Gel Doc-It™ Imaging System (UVP-LLC, Upland, CA, USA). Amplicons were purified using the ExoSAP-IT Express PCR Product Cleanup Kit (Applied Biosystems) following the manufacturer’s protocol. Purified amplicons were subjected to Sanger sequencing at Génome Québec (Montréal, QC, Canada) using the same primers (EF1/EF2 and ITS4B/BMBC-F) as for PCR amplification. The resulting consensus sequences were deposited in GenBank (
Table 2).
2.2.2. Phylogenetic Analysis
To ensure accurate identification and overcome potential limitations of partial sequences in public databases, a comprehensive reference dataset was compiled. This included EF1-α and ITS sequences extracted from all available complete
Diaporthe genomes in NCBI, along with sequences from 24 complete genomes representing 22
Diaporthe species associated with soybean and other crops worldwide (
Supplementary Table S1).
Sequence similarity searches were performed using a combination of MEGABLAST and discontiguous MEGABLAST algorithms within Geneious Prime® (v2022.1.1). Multiple sequence alignments for both loci were generated using MAFFT within Geneious Prime. Phylogenetic trees were constructed using the Neighbor-Joining method with the Jukes–Cantor genetic distance model. Nodal support was assessed via bootstrap analysis with 1,000 replicates. Trees were rooted with Diaporthella corylina to serve as an outgroup. Final consensus trees were visualized and annotated using iTOL v3 (Letunic and Bork, 2016).
2.3. Pathogenicity Assays
The pathogenicity of 19 Diaporthe sojae isolates (SB24-114, SB24-117, SB24-131, SB24-136, SB24-140, SB24-143, SB24-144, SB24-300, SB24-311, SB24-312, SB24-315, SB24-316, SB24-317, SB24-318, SB24-322, SB24-331, SB24-332, SB24-339, and SB24-346) was evaluated on soybean. Inoculum was prepared by inoculating sterilized wheat kernels with each isolate following the method of Luckew et al. (2012). Wheat (Triticum aestivum L.) kernels (cv. Brandon) were soaked overnight in distilled water, drained, and transferred into 1-L stainless steel boxes (~500 g per box). Kernels were autoclaved at 121 °C for 1 h on two consecutive days to ensure sterility. After cooling, each box was inoculated under aseptic conditions with six 6-mm mycelial plugs cut from 5- to 7-day-old PDA cultures of the respective isolate. Boxes were incubated at 25 °C in darkness, shaken on days 3 and 6 to promote uniform colonization, and maintained until full colonization of the wheat kernels was achieved after 14 days. Control inoculum consisted of autoclaved wheat kernels inoculated with sterile PDA plugs only.
For pathogenicity testing, 25 g of wheat kernels were thoroughly mixed with 1 kg of pasteurized potting substrate (Sunshine® Mix #4, Sungro, Maryland, USA) and placed into disinfected 6-inch pots. Four isolates included in the phylogenetic analysis (SB24-139, SB24-148, SB24-313, and SB24-320) were omitted from the pathogenicity assay due to insufficient biomass production during the inoculum preparation stage. Each treatment included four replicate pots, with four seeds of soybean cultivar Sperling sown per pot. Each pot was considered a biological replicate. Pots were arranged in a randomized complete block design (RCBD) and maintained in a greenhouse at 24/16 °C day/night temperature, 13/11 h light/dark cycle, and 70–80% relative humidity. Plants were irrigated three times per week with tap water and received no additional fertilizer. At the V5 growth stage (approximately 28–32 days after inoculation), plants were carefully uprooted, roots were gently washed, and disease severity was visually scored using the 0–4 root rot rating scale described by Taheri et al. (2011) (
Table 1). To confirm pathogenicity, symptomatic root tissues were surface-sterilized and plated on PDA, and the recovered fungi were reidentified morphologically and molecularly as described above. The entire experiment was repeated twice.
2.4. Statistical Analysis
Statistical analysis was performed using Python 3.11.0 with pandas, NumPy, SciPy, and statsmodels libraries. To account for subsampling variability and avoid pseudoreplication, the disease severity ratings of the four individual plants within each pot were averaged to generate a single value for each biological replicate. Data from the two experimental repetitions were pooled for analysis after confirming homogeneity of variance (Levene’s test, P > 0.05), resulting in a total sample size of n = 80 independent observations (20 treatments × 4 replicate pots). Prior to analysis, residuals were assessed for normality using the Shapiro-Wilk test.
A Disease Severity Index (DSI) was calculated for each treatment using the formula:
DSI (%) = (Mean Severity / 4) × 100
In addition, Mortality Rate (%) was calculated as the percentage of plants receiving a score of 4 (dead). A one-way Analysis of Variance (ANOVA) was performed to test for treatment differences, followed by Tukey's Honestly Significant Difference (HSD) post-hoc test for pairwise comparisons. Statistical significance was set at α = 0.05.
3. RESULTS
3.1. Fungal Isolation and Morphological Identification
From the 2024 Manitoba soybean survey, a total of 287 fungal isolates were recovered from symptomatic roots. Based on initial ITS sequencing, 49 isolates were identified as Diaporthe sojae. To further validate species identity, 23 representative isolates were selected for detailed characterization (
Table 2). These isolates were selected to maximize geographic and population diversity across Manitoba; selection ensured representation from each field where D. sojae was detected, with additional isolates included from fields yielding multiple isolates to capture within-field variability.
On PDA, colonies exhibited rapid growth, producing abundant white aerial mycelium with a yellowish reverse that darkened with senescence. Pycnidia developed after approximately 5 weeks of incubation in the dark at 25 °C. These structures were black, globose to subglobose, occasionally flask-shaped, and generally immersed to partially erumpent within the agar surface. Alpha conidia were hyaline, aseptate, ellipsoid to clavate, tapering at both ends, and biguttulate (containing two distinct oil droplets), measuring 6.0–8.0 µm (n = 25). Beta conidia were not observed under the culture conditions used in this study (
Figure 2A, B).
3.2. Phylogenetic Analysis
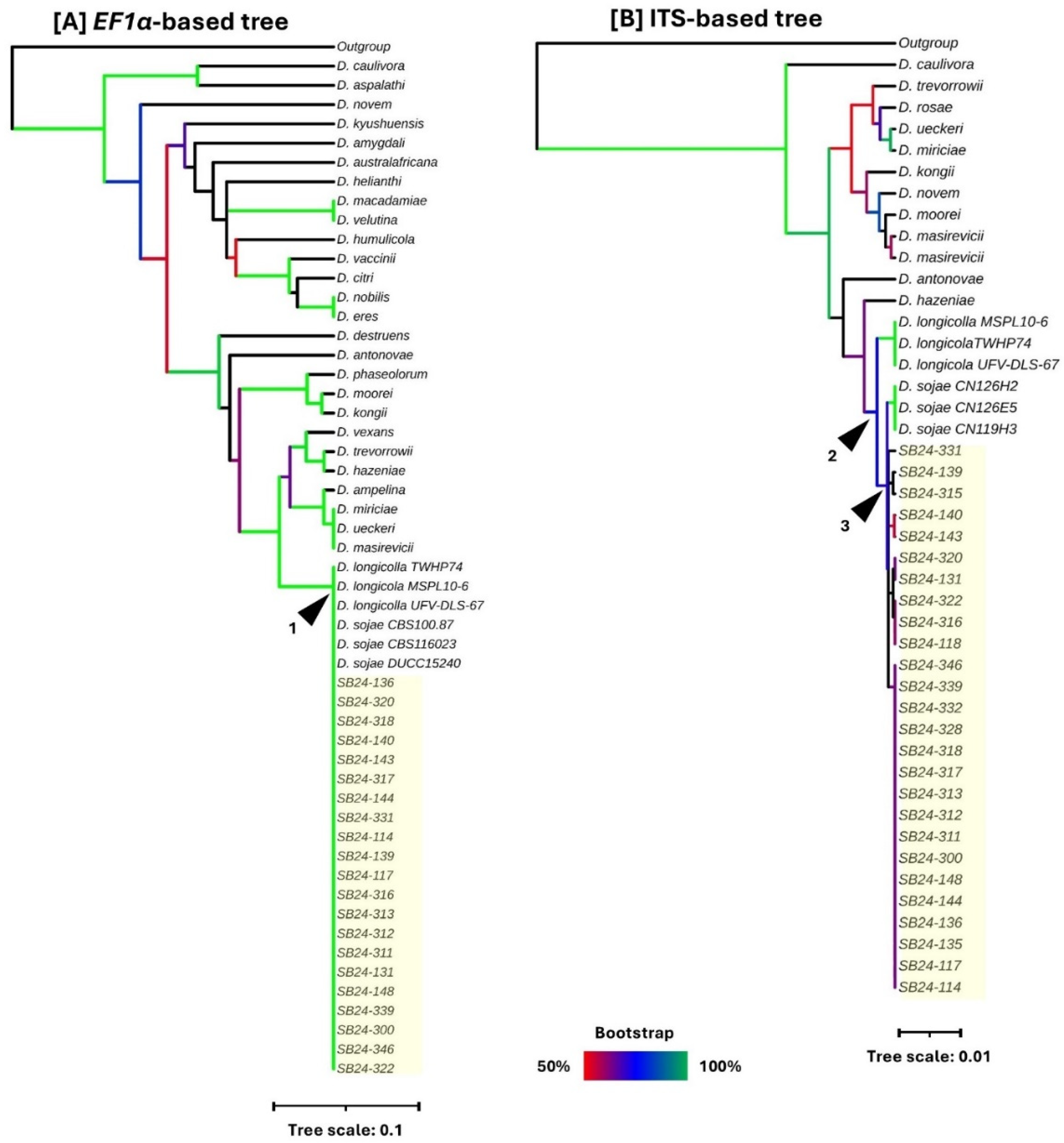
Phylogenetic analyses based on EF1-α and ITS sequences were performed to investigate the evolutionary relationships of the Diaporthe isolates recovered from soybean roots (
Figure 3). The analyses included all available Diaporthe species sequences for the respective loci retrieved from GenBank and extracted from complete genomes (
Supplementary Table S1).
The EF1-α-based phylogenetic tree (
Figure 3A) revealed that the soybean isolates clustered with both D. longicolla and
D. sojae reference isolates within a strongly supported monophyletic clade (Node 1; 100% bootstrap support). This indicates that the EF1-α locus alone provided insufficient resolution to distinguish the soybean isolates from
D. longicolla, as all isolates within this clade shared a close evolutionary relationship.
In contrast, the ITS-based phylogenetic tree (
Figure 3B) demonstrated improved resolution among closely related species. The soybean isolates formed a distinct monophyletic group with
D. sojae reference strains (Node 3; 75% bootstrap support), which was clearly separated from the closely related
D. longicolla cluster (Node 2; 72% bootstrap support). This confirms that the ITS region is a more informative marker for discriminating
D. sojae from
D. longicolla in this context. Consequently, the combined morphological, molecular, and phylogenetic data confirmed the identity of the 23 representative isolates as
D. sojae. Sequences from these confirmed isolates were deposited in GenBank (
Table 2).
3.3. Pathogenicity Test
Inoculation of soybean cv. Sperling with the 19 D. sojae isolates produced dark brown lesions on the hypocotyl and taproot, leading to significant root necrosis and seedling stunting compared with the asymptomatic, healthy roots of the control plants (
Figure 1). These symptoms were morphologically identical to the root rot symptoms observed in the field.
One-way ANOVA revealed a highly significant treatment effect on root rot severity among the 20 treatments (19
D. sojae isolates plus the non-inoculated control) (
,
) (
Table 3). Tukey’s HSD test separated all 19 isolates from the control (
), confirming their pathogenic status. Furthermore, the analysis revealed significant virulence diversity within the population, separating the isolates into overlapping virulence groups.
Across isolates, the Disease Severity Index (DSI) ranged from 68.8% for the least aggressive isolates (SB24-131, SB24-136) to 100% for the highly virulent isolates (SB24-315, SB24-322) (
Table 4). Based on Tukey’s HSD test, three statistically distinct yet overlapping virulence groups were identified:
High Virulence (Group a): Seven isolates (SB24-322, SB24-315, SB24-311, SB24-317, SB24-318, SB24-300, and SB24-143) belonged exclusively to Group ‘a’ (DSI 89.0–100%; mortality 62.5–100%). These isolates were acutely aggressive, causing rapid damping-off and resulting in high seedling mortality within the experimental period.
Intermediate Virulence (Groups ab and bc): Ten isolates clustered within the overlapping groups ‘ab’ and ‘bc’ (
Table 4). These isolates exhibited a broad range of disease severity (DSI 70.3–87.5%) and moderate mortality (18.8–56.3%), representing transitional phenotypes that caused substantial root necrosis and stunting, with variable progression to plant death.
Lower Lethality / Chronic Disease Expression (Group c): Two isolates (SB24-131 and SB24-136) were classified in Group ‘c’ (DSI = 68.8%; mortality 25.0–37.5%). These isolates caused pronounced root rot symptoms, including extended lesions, but significantly lower mortality compared with Group ‘a’, resulting in persistent disease and survival of plants with compromised root systems.
4. Discussion
Soybean root rot is a destructive and complex disease syndrome involving a diverse assemblage of soilborne fungal pathogens that negatively affect seedling emergence, vigor, and ultimately yield stability (Dixon & Tilston 2010). Globally, Fusarium spp., Phytophthora sojae, Pythium spp., and Rhizoctonia solani (Hafez et al. 2021; Tyler 2007; Zhang et al. 2013) are considered the predominant root rot pathogens in soybean. The fungal community associated with soybean roots is highly diverse (Impullitti & Malvick 2013). Diaporthe spp. (including their Phomopsis anamorphs) have received comparatively less attention as causal agents of soybean root rot; however, studies indicate that Diaporthe spp. can be an important and under-recognized contributor to root decay, crown rot, and seedling blight, particularly in mixed-pathogen environments where multiple soilborne fungi coexist (Hosseini et al. 2020; Petrović et al. 2021; Zhao et al. 2021).
Diaporthe sojae is known to cause pod and stem blight, stem canker, and Phomopsis seed decay, typically observed late in the season under high humidity. It is frequently isolated from aerial portions of soybean and is considered one of the most important fungal pathogens impacting seed quality (Petrović et al. 2021; Zhao et al. 2022; DEFRA 2024). Although D. sojae has previously been recovered from soybean roots (Pedersen and Grau 2010), reports of it causing root rot are lacking. The present study provides the first confirmed evidence that D. sojae can act as a causal agent of soybean root rot, expanding the known pathogenic profile of this species. Diaporthe sojae was isolated from field-grown soybean displaying root rot symptoms and was shown to be capable of colonizing and aggressively damaging soybean root tissues in controlled studies.
The taxonomic history of
D. sojae has long been complicated, reflecting early reliance on morphology and later refinement through molecular phylogenetics. Initially described by Lehman (1922, 1923) as the cause of pod and stem blight, the species was later grouped within
D. phaseolorum due to morphological similarity (Wehmeyer 1933). Kulik (1984) proposed merging the taxa, while Hobbs et al. (1985) described a new
Phomopsis species from soybean, and Morgan-Jones (1989) introduced
D. phaseolorum f. sp.
sojae. Fernández and Hanlin (1996) recommended varietal designation, whereas molecular studies subsequently revealed substantial genetic divergence within what had been broadly referred to as
D. phaseolorum var.
sojae (Zhang et al. 1998). Historically, misconceptions regarding heterothallism were corrected when later research demonstrated a homothallic reproductive strategy, with morphological diversity reflecting species-level variation rather than reproductive mode (Jensen 1983; Santos et al. 2011; Udayanga et al. 2015). The application of multi-locus phylogenetics—including ITS, EF1-α, β-tubulin, histone, calmodulin, and actin—greatly improved species delimitation (Gomes et al. 2013; Udayanga et al. 2015; Petrović et al. 2018, 2021). The morphological features observed in our isolates, including the size and shape of the biguttulate alpha conidia, were consistent with published descriptions of
D. sojae (Lehman 1923; Fernández and Hanlin 1996; Dissanayake et al. 2015; Udayanga et al. 2015; Du et al. 2021; Petrović et al. 2021) (
Figure 2A, B), thereby supporting our initial identification. Although early analyses suggested
D. sojae and
D. longicolla might be synonymous (Gomes et al. 2013), a more comprehensive multi-locus phylogeny incorporating authenticated reference isolates unambiguously resolved
D. sojae,
D. longicolla, and
D. phaseolorum as distinct species (Udayanga et al. 2015).
The multilocus phylogenetic analysis performed in this study aimed to accurately identify the Diaporthe isolates associated with symptomatic soybean roots. By analyzing the ITS and EF1-α loci and integrating all available sequences from NCBI GenBank as well as curated sequences extracted from 24 complete Diaporthe genomes representing 33 species, we achieved a robust species-level placement of the isolates. Notably, no complete genome sequence yet exists for D. sojae, necessitating comparative inference from related species. The EF1-α–based phylogeny provided strong clustering but lacked sufficient resolution to discriminate D. sojae from D. longicolla, consistent with previous findings that EF1-α alone cannot reliably separate these closely related species (Udayanga et al. 2015; Dissanayake et al. 2024; Mena et al. 2024). Conversely, the ITS marker yielded clearer resolution, placing the soybean isolates within a well-supported D. sojae clade, fully separated from D. longicolla. This aligns with prior evidence that ITS, while insufficient in some fungal genera, retains discriminatory power within the D. sojae–D. longicolla species complex.
Our comprehensive phylogenetic analysis also included multiple Diaporthe species known to infect soybean or other legumes, such as D. aspalathi, D. caulivora, D. longicolla, D. miriciae, D. novem, and D. ueckeri (Petrović et al. 2016). These species form part of the broader Diaporthe/Phomopsis disease complex (Zhao et al. 2022), which collectively leads to substantial yield losses. Diaporthe longicolla is recognized as the predominant pathogen of Diaporthe seed decay and a major contributor to pod and stem blight (Abdelmagid et al. 2022; López-Cardona et al. 2022). Diaporthe aspalathi and D. caulivora cause southern and northern stem canker, respectively (Ghimire et al. 2019; Mena et al. 2020). Several Diaporthe species have recently been linked to root diseases in non-soybean hosts, such as D. ueckeri on cassava (da Silva et al. 2024), D. eres on Chinese goldthread (Mei et al. 2021), and D. betae on sugar beet (Shao et al. 2025). These reports highlight the pathogenic plasticity of the genus and demonstrate that some species can cause severe disease in both above- and below-ground tissues.
The present study demonstrates that D. sojae can act as a damaging root rot pathogen in soybean. Pathogenicity assays confirmed that all 19 isolates tested can cause root rot in controlled assays. Analyses of virulence revealed two distinct pathogenicity categories among the isolates tested: (i) a lethal, acute phenotype characterized by rapid damping-off and seedling mortality (e.g., isolates SB24-322 and SB24-315), and (ii) a chronic phenotype, in which isolates caused severe root lesions but lower mortality levels (<40%), enabling plants to survive with compromised root systems. This phenotypic dichotomy is epidemiologically significant. Lethal isolates threaten stand establishment, while chronic isolates may enable infected plants to persist, leading to extensive pycnidial development on senescent stems and roots, thereby increasing overwintering inoculum and promoting long-term disease persistence.
The identification of D. sojae as a root rot pathogen further complicates the model of soybean root rot etiology, which has been largely attributed to complexes of pathogens previously driven by Fusarium, Rhizoctonia, Pythium, and Phytophthora species (Wrather et al. 2001; Abdelmagid et al. 2018). The addition of D. sojae as a potential driver of root rot complexes demonstrates the importance of improved diagnostic methods, as morphological features alone are insufficient to discriminate the causal pathogens. The combined use of morphological and multilocus molecular analysis leads to more accurate pathogen identification.
From an epidemiological perspective, the recovery of D. sojae from multiple fields in Manitoba suggests that this pathogen may be more widespread in soybean-producing regions than previously assumed. Its ability to colonize roots, stems, pods, and seeds positions it as a multi-tissue pathogen that can cause losses throughout the entire soybean growth cycle (Petrović et al. 2021; Udayanga et al. 2015). Moreover, early-season root infections may interact synergistically with other soilborne pathogens, potentially exacerbating disease severity under conducive environmental conditions (Hafez et al. 2021; Datnoff and Sinclair 1988).
The emergence of D. sojae as a root rot pathogen in Manitoba is driven by the region's unique agroclimatic and agronomic conditions. Early-season planting into cold, wet soils is a common challenge in the Canadian prairies, which slows seedling growth, weakens root development, and increases susceptibility to opportunistic pathogens (Batista et al. 2024; Comas et al. 2010; Sebastian et al. 2016). Furthermore, the widely adopted practice of conservation tillage across Manitoba (Awada et al. 2014; Khakbazan and Hamilton 2012) is used to improve soil health (Mitchell et al. 2017), but it also promotes the accumulation of infected residue at the soil surface, serving as a reservoir for D. sojae inoculum (Almeida et al. 2001; Whalen et al. 2007). This combination of a favorable disease environment and high potential inoculum pressure creates a situation conducive to aggressive early-season infections by opportunistic fungi, including D. sojae. This pattern mirrors the first worldwide report of root rot caused by Fusarium cerealis in Manitoba (Abdelmagid et al. 2018), where cold, wet soils and heavy residues facilitated cross-host infection by a traditionally cereal-associated pathogen.
In summary, this study expands the known pathological profile of D. sojae and establishes it as a causal agent of soybean root rot in Western Canada. While all isolates exhibited pathogenicity, virulence varied significantly, forming two epidemiologically important groups: acute, lethal isolates that eliminate seedlings rapidly, and chronic isolates that compromise root systems while allowing plant survival. The emergence of D. sojae as a root rot pathogen highlights both the pathogen’s biological versatility and the influence of regional environmental factors that favor opportunistic infection. These findings underscore the importance of including D. sojae in future root rot surveys, diagnostic protocols, and breeding programs. The high-virulence isolates identified here (e.g., SB24-322) should be incorporated into screening programs to avoid false resistance. Future research should focus on population-level surveys to determine the prevalence of the identified virulence phenotypes, genomic analyses to uncover determinants of aggressiveness, and the investigation of interactions with other soilborne pathogens to inform integrated disease management strategies.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Acknowledgments
This work was supported by the Manitoba SCAP program, the Manitoba Pulse & SoybeanGrowers Association, and Agriculture and Agri-Food Canada through project J-003419 (Staying on Top of Soybean Root Diseases under Climate Change in Manitoba).
References
- Abdelmagid, A., Hafez, M., Lawley, Y., Adam, L. R., and Daayf, F. 2018. First report of Fusarium cerealis causing root rot on soybean. Plant Dis. 102:1225. [CrossRef]
- Abdelmagid, A., Hafez, M., Lawley, Y., Rehal, P. K., and Daayf, F. 2022. First report of pod and stem blight and seed decay caused by Diaporthe longicolla on soybean in Western Canada. Plant Dis. 106:1061. [CrossRef]
- Almeida, Á. M. R., Saraiva, O. F., Farias, J. R. B., Gaudêncio, C. A., and Torres, E. 2001. Survival of pathogens on soybean debris under no-tillage and conventional tillage systems. Pesqui. Agropecu. Bras. 36:1231–1238. [CrossRef]
- Awada, L., Lindwall, C. W., and Sonntag, B. 2014. The development and adoption of conservation tillage systems on the Canadian Prairies. Int. Soil Water Conserv. Res. 2:47–65. [CrossRef]
- Batista, B. D., Wang, J., Liu, H., Kaur, S., Macdonald, C. A., Qiu, Z., Trivedi, P., Delgado-Baquerizo, M., Xiong, C., Liang, J., and Bange, M. 2024. Biotic and abiotic responses to soilborne pathogens and environmental predictors of soil health. Soil Biol. Biochem. 189:109246. [CrossRef]
- Bradley, C. A., Allen, T. W., Saulters, B., et al. 2021. Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2015 to 2019. Plant Health Prog. 22:483–495. (Note: This author list is extremely long; verify if the journal requires all 20+ names or allows et al. for this specific type of report). [CrossRef]
- Comas, L. H., Bauerle, T. L., and Eissenstat, D. M. 2010. Biological and environmental factors controlling root dynamics and function: Effects of root ageing and soil moisture. Aust. J. Grape Wine Res. 16:131–137. [CrossRef]
- da Silva, J. S. A., Alves, V. C. S., da Silva, S. F., do Nascimento Barbosa, R., de Souza, C. A. F., da Costa, D. P., Machado, A. R., de Medeiros, E. V., and de Souza-Motta, C. M. 2024. Diaporthe ueckeri causing cassava root rot in Pernambuco, Brazil. Crop Prot. 184:106811. [CrossRef]
- Datnoff, L. E., and Sinclair, J. B. 1988. The interaction of Fusarium oxysporum and Rhizoctonia solani in causing root rot of soybeans. Phytopathology 78:771–777. [CrossRef]
- Department for Environment, Food & Rural Affairs (DEFRA). 2024. Rapid Pest Risk Analysis (PRA) for Diaporthe sojae. Online. https://planthealthportal.defra.gov.uk/assets/Diaporthe_sojae_PRA_draft.pdf.
- Dissanayake, A. J., Liu, M., Zhang, W., Chen, Z., Udayanga, D., Chukeatirote, E., Li, X., Yan, J., and Hyde, K. D. 2015. Morphological and molecular characterisation of Diaporthe species associated with grapevine trunk disease in China. Fungal Biol. 119:283–294. [CrossRef]
- Dissanayake, A. J., Zhu, J. T., Chen, Y. Y., Maharachchikumbura, S. S., Hyde, K. D., and Liu, J. K. 2024. A re-evaluation of Diaporthe: Refining the boundaries of species and species complexes. Fungal Divers. 126:1–125. [CrossRef]
- Dixon, G. R., and Tilston, E. L. 2010. Soil-borne pathogens and their interactions with the soil environment. Pages 197–271 in: Soil Microbiology and Sustainable Crop Production. Springer.
- Dorrance, A. E., and Mills, D. 2009. Phomopsis seed decay. Plant Health Instr. DOI: 10.1094/PHI-I-2009-0129-01. [CrossRef]
- dos Santos, G. C., Lima Horn, L. M., Trezzi Casa, R., Soardi, K., Lopes, M. A., Nascimento, S. C. D., Santi, V. M., Nascimento da Silva, F., Kór, D. G., Gonçalves, M. J., and Gorayeb, E. S. 2024. First report of seed decay caused by Diaporthe ueckeri on soybean in Brazil. Plant Dis. 108:2925. [CrossRef]
- Du, Y., Wang, X., Guo, Y., Xiao, F., Peng, Y., Hong, N., and Wang, G. 2021. Biological and molecular characterization of seven Diaporthe species associated with kiwifruit shoot blight and leaf spot in China. Phytopathol. Mediterr. 60:177–198. [CrossRef]
- Esmaeili Taheri, A., Chatterton, S., Foroud, N. A., Gossen, B. D., and McLaren, D. L. 2017. Identification and community dynamics of fungi associated with root, crown, and foot rot of field pea in western Canada. Eur. J. Plant Pathol. 147:489–500. [CrossRef]
- Fernández, F. A., and Hanlin, R. T. 1996. Morphological and RAPD analyses of Diaporthe phaseolorum from soybean. Mycologia 88:425–440. [CrossRef]
- Gerdemann, J. W. 1954. The association of Diaporthe phaseolorum var. sojae with root and basal stem rot of soybean. Plant Dis. Rep. 38:742–743.
- Ghimire, K., Petrović, K., Kontz, B. J., Bradley, C. A., Chilvers, M. I., Mueller, D. S., Smith, D. L., Wise, K. A., and Mathew, F. M. 2019. Inoculation method impacts symptom development associated with Diaporthe aspalathi, D. caulivora, and D. longicolla on soybean (Glycine max). Plant Dis. 103:677–684. [CrossRef]
- Ghissi, V. V., Reis, E. M., and Deuner, C. C. 2014. Etiology of phomopsis root rot in soybean. Summa Phytopathol. 40:270–272. [CrossRef]
- Gomes, R. R., Glienke, C., Videira, S. I. R., Lombard, L., Groenewald, J. Z., and Crous, P. W. 2013. Diaporthe: A genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31:1–41. [CrossRef]
- Hafez, M., Abdelmagid, A., Aboukhaddour, R., Adam, L. R., and Daayf, F. 2021. Fusarium root rot complex in soybean: Molecular characterization, trichothecene formation, and cross-pathogenicity. Phytopathology 111:2287–2302. [CrossRef]
- Hobbs, T. W., Schmitthenner, A. F., and Kuter, G. A. 1985. A new Phomopsis species from soybean. Mycologia 77:535–544. [CrossRef]
- Hosseini, B., El-Hasan, A., Link, T., and Voegele, R. T. 2020. Analysis of the species spectrum of the Diaporthe/Phomopsis complex in European soybean seeds. Mycol. Prog. 19:455–469. [CrossRef]
- Impullitti, A. E., and Malvick, D. K. 2013. Fungal endophyte diversity in soybean. J. Appl. Microbiol. 114:1500–1506. [CrossRef]
- Jensen, J. D. 1983. The development of Diaporthe phaseolorum var. sojae in culture. Mycologia 75:1074–1091. [CrossRef]
- Khakbazan, M., and Hamilton, C. 2012. Economic evaluation of tillage management systems for spring wheat production in the Canadian Prairies. Soil Tillage Res. 118:40–51. [CrossRef]
- Kulik, M. M. 1984. Symptomless infection, persistence, and production of pycnidia in host and non-host plants by Phomopsis batatas, Phomopsis phaseoli, and Phomopsis sojae. Mycologia 76:274–291. [CrossRef]
- Lehman, S. G. 1922. Pod and stem blight of the soybean. J. Elisha Mitchell Sci. Soc. 38:13. [CrossRef]
- Lehman, S. G. 1923. Pod and stem blight of the soybean. Ann. Mo. Bot. Gard. 10:111–178. [CrossRef]
- Letunic, I., and Bork, P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44:W242–W245. [CrossRef]
- Leyronas, C., Morris, C. E., and Nicot, P. C. 2012. Optimizing the sampling of Fusarium populations on wheat spikes to estimate the genetic diversity. Mycology 103:1269–1280.
- López-Cardona, N., Guevara-Castro, A., López-Casallas, M., and Gañán-Betancur, L. 2022. Occurrence of seed decay caused by Diaporthe longicolla on soybean in Colombia. Summa Phytopathol. 48:139–140. [CrossRef]
- Luckew, A. S., Cianzio, S. R., and Leandro, L. F. 2012. Screening method for distinguishing soybean resistance to Fusarium virguliforme. Crop Sci. 52:2215–2223. [CrossRef]
- Markell, S., Malvick, D., and Nelson, B. 2024. Soybean disease diagnostic series. PP1867. North Dakota State University Extension.
- Mei, P., Song, X. H., Zhu, Z. Y., and Li, L. Y. 2021. First report of Diaporthe eres causing root rot of Coptis chinensis in China. Plant Dis. 105:1854. [CrossRef]
- Mena, E., Stewart, S., Montesano, M., and Ponce de León, I. 2020. Soybean stem canker caused by Diaporthe caulivora: Pathogenicity and fungus–plant interactions. Front. Plant Sci. 10:1733. [CrossRef]
- Mena, E., Stewart, S., Montesano, M., and Ponce de León, I. 2024. Current understanding of the Diaporthe/Phomopsis complex in soybean. Plant Pathol. 73:31–46. [CrossRef]
- Mitchell, J. P., Shrestha, A., Mathesius, K., Scow, K. M., Southard, R. J., Haney, R. L., Schmidt, R., Munk, D. S., and Horwath, W. R. 2017. Cover cropping and no-tillage improve soil health in an arid irrigated cropping system in California’s San Joaquin Valley. Soil Tillage Res. 165:325–335. [CrossRef]
- Morgan-Jones, G. 1989. The Diaporthe/Phomopsis complex: Taxonomy, biology, pathogenicity. Mycotaxon 34:279–290.
- Nascimento, C. R., et al. 2025. First report of Diaporthe ueckeri and D. longicolla causing stem canker on soybean in Brazil. Plant Dis. 109:506. [NOTE: You MUST check the PDF of this paper to list all authors. "et al." is not allowed.]. [CrossRef]
- O’Donnell, K., Kistler, H. C., Cigelnik, E., and Ploetz, R. C. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. U.S.A. 95:2044–2049. [CrossRef]
- Pedersen, P., and Grau, C. R. 2010. Ecology and epidemiology of Diaporthe/Phomopsis species complex in soybean. Compendium of Soybean Diseases. 5th ed. APS Press, St. Paul, MN.
- Petrović, K., Skaltsas, D., Castlebury, L. A., Kontz, B., Allen, T. W., Chilvers, M. I., Gregory, N., Kelly, H. M., Robertson, A. E., and Mathew, F. M. 2016. First report of Diaporthe novem, D. foeniculina, and D. rudis associated with soybean seed decay in Serbia. Plant Dis. 100:2324. [CrossRef]
- Petrović, K., Vidić, M., Riccioni, L., Đorđević, V., and Rajković, D. 2018. Diaporthe pseudolongicolla: A new pathogen on soybean seed in Serbia. Field Veg. Crops Res. 55:103–109. [CrossRef]
- Petrović, K., Zdravković, J., Jelovac, D., and Vidić, M. 2021. Diaporthe seed decay of soybean. Plant Dis. 105:1621–1629. [CrossRef]
- Rossman, A. Y. 2009. The impact of invasive fungi on agricultural ecosystems in the United States. Biol. Invasions 11:97–107. [CrossRef]
- Rupe, J. C., da Silva, M. P., Rojas, J. A., Holland, R., and Zaia, R. 2025. Estimating the potential for soybean seed infection by Diaporthe spp. using a weather-based model. Plant Health Prog. 26:145–156.
- Santos, J. M., Vrandečić, K., Ćosić, J., Duvnjak, T., and Phillips, A. J. L. 2011. Resolving Diaporthe species occurring on soybean in Croatia. Persoonia 27:9–19. [CrossRef]
- Sebastian, N., Erika, H., and Christian, K. 2016. Critically low soil temperatures for root growth. Alpine Bot. 126:11–21. [CrossRef]
- Shao, H., Ma, C., Yu, B., Chen, S., and Li, H. 2025. Diaporthe betae sp. nov. causing sugar beet root rot. Front. Microbiol. 16:1453460. [CrossRef]
- Sinclair, J. B. 1993. Phomopsis seed decay of soybean. APS Press, St. Paul, MN.
- Soy Canada. 2025. Canadian soybean production statistics. https://soycanada.ca. Accessed 13 Feb 2025.
- Statistics Canada. 2024. Estimated area, yield, production of principal field crops. https://www150.statcan.gc.ca. Accessed 13 Feb 2025.
- Statistics Canada. 2025. Estimated areas, yield, production of principal field crops. https://www150.statcan.gc.ca. Accessed 13 Feb 2025.
- Taheri, P., Gnanamanickam, S. S., and Höfte, M. 2011. Characterization of Phytophthora and Pythium species associated with root rot of cocoyam (Xanthosoma sagittifolium) in Nicaragua and Costa Rica. Plant Pathol. 60:469–480.
- Tyler, B. M. 2007. Phytophthora sojae: Root rot pathogen of soybean. Mol. Plant Pathol. 8:1–8.
- Udayanga, D., Liu, X., McKenzie, E. H. C., Chukeatirote, E., Bahkali, A. H., and Hyde, K. D. 2011. The genus Phomopsis: Biology, applications, species concepts and names of common phytopathogens. Fungal Divers. 50:189–225. [CrossRef]
- Udayanga, D., Castlebury, L. A., Rossman, A. Y., Chukeatirote, E., and Hyde, K. D. 2015. The Diaporthe sojae species complex: Phylogenetic re-assessment of pathogens associated with soybean, cucurbits and other field crops. Fungal Biol. 119:383–407. [CrossRef]
- USDA Foreign Agricultural Service. 2025. Soybeans – Global production statistics (2023/2024). https://www.fas.usda.gov. Accessed 13 Feb 2025.
- Vidić, M., Jasnić, S., and Škrbić, B. 2011. Phomopsis seed decay in soybean. Zb. Matice Srp. Prir. Nauke 120:75–84.
- Whalen, J. K., Prasher, S. O., and Benslim, H. 2007. Monitoring corn and soybean agroecosystems in Quebec, Canada. Can. J. Plant Sci. 87:841–849. [CrossRef]
- Wrather, J. A., Stienstra, W. C., and Koenning, S. R. 2001. Soybean disease loss estimates for the United States from 1996 to 1998. Can. J. Plant Pathol. 23:122–131. [CrossRef]
- Xue, A. G., Cober, E., Voldeng, H. D., and Babcock, C. 2007. Prevalence and pathogenicity of Diaporthe/Phomopsis complex on soybean in Ontario. Can. J. Plant Pathol. 29:442.
- Zhang, A. W., Riccioni, L., Pedersen, W. L., Kollipara, K. P., and Hartman, G. L. 1998. Molecular identification and grouping of Diaporthe phaseolorum and Phomopsis longicolla from soybean. Phytopathology 88:1306–1314. [CrossRef]
- Zhang, J. X., Xue, A. G., Cober, E. R., Morrison, M. J., Zhang, H. J., Zhang, S. Z., and Gregorich, E. 2013. Prevalence, pathogenicity and cultivar resistance of Fusarium and Rhizoctonia species causing soybean root rot. Can. J. Plant Sci. 93:221–236. [CrossRef]
- Zhao, X., Li, K., Zheng, S., Yang, J., Chen, C., Zheng, X., Wang, Y., and Ye, W. 2022. Diaporthe diversity and pathogenicity in soybean stem blight. Plant Dis. 106:2892–2903. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).