Submitted:
16 December 2025
Posted:
17 December 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
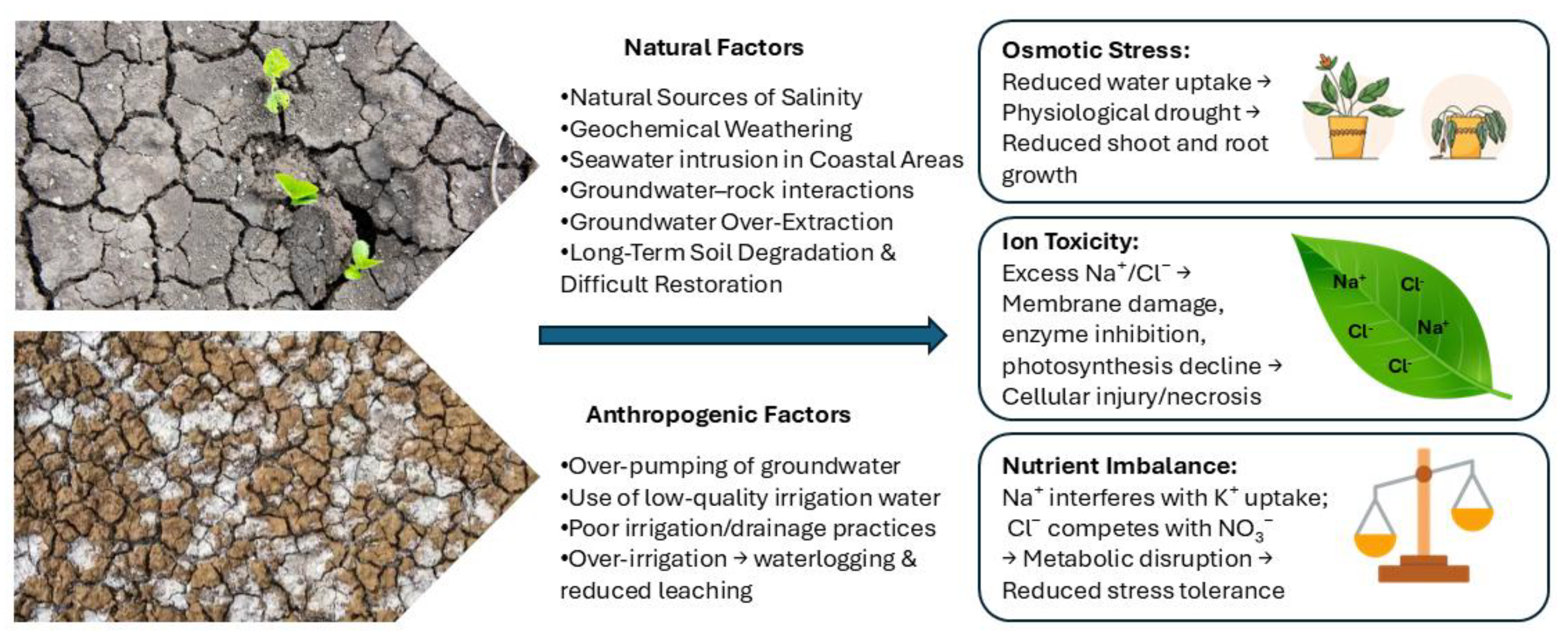
1.1. Soil Salinity and Its Impact in Agriculture
1.2. Natural and Human-Induced Causes of Soil Salinity
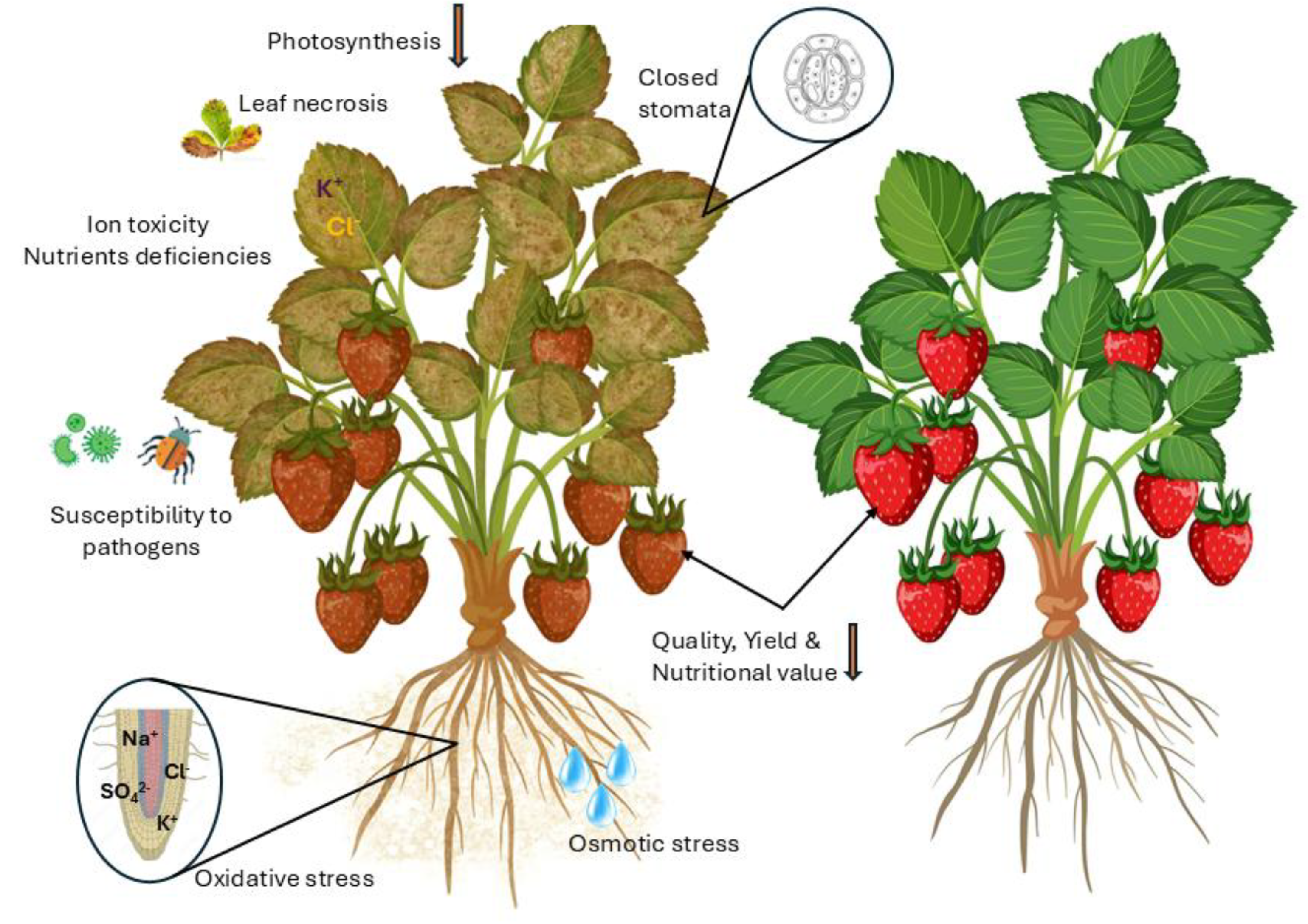
2. Salinity Stress and Strawberry Cultivation
2.1. Strawberry Cultivation and Salinity Stress
2.2. Variation in Salinity Responses Across Different Strawberry Species and Cultivars
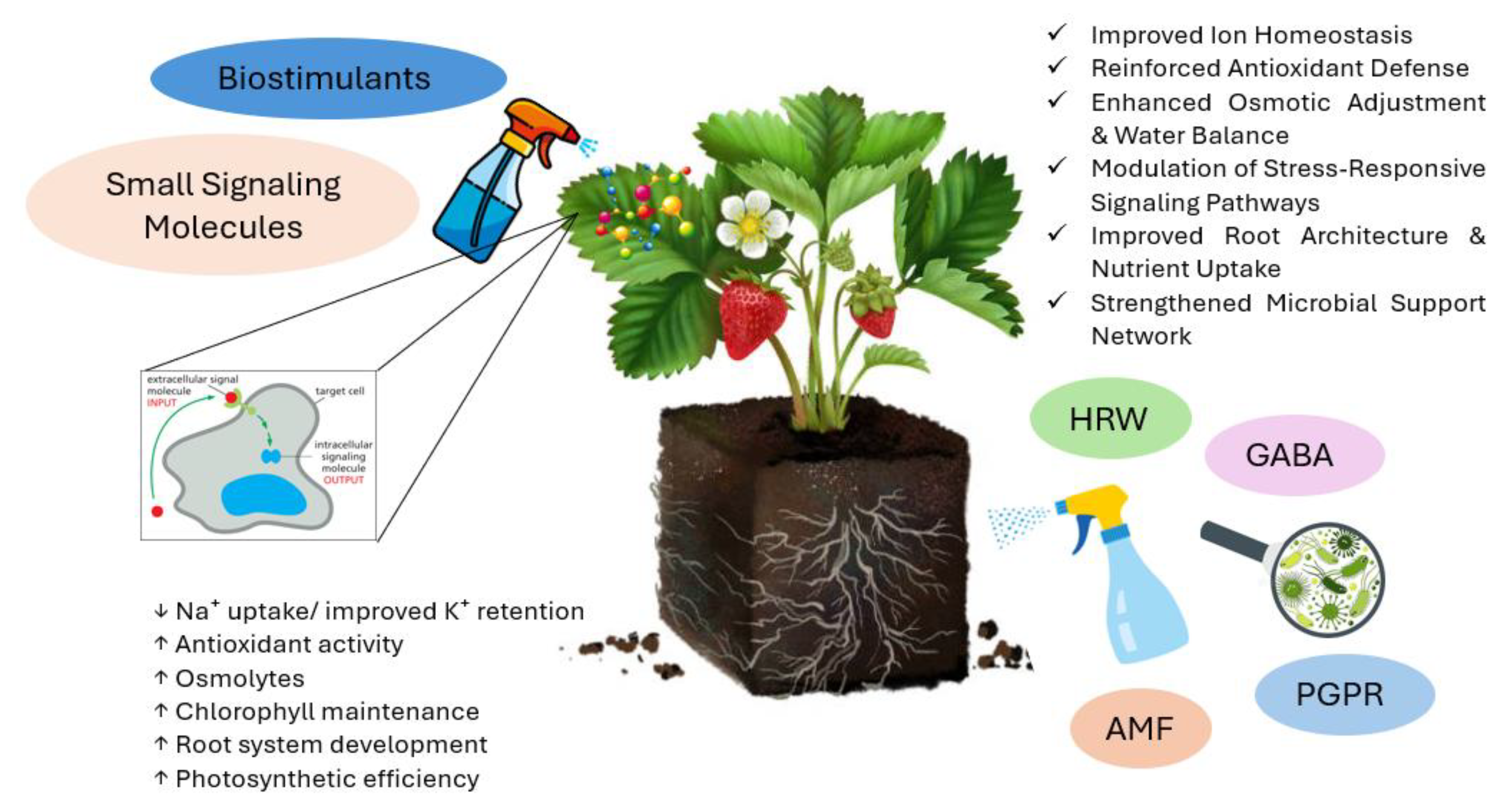
3. Mitigation Mechanisms in Strawberry under Salinity Stress
3.1. The Role of Small Signaling Molecules and Biostimulants in Mitigating Salinity Stress in Strawberry
3.2. Microorganisms and Plant-Microbe Interactions under Salinity
4. Genetic and Molecular Approaches for Improving Salinity Tolerance in Strawberry
Gene Identification and Functional Annotation for Breeding Targets
| Gene / Protein | Function / Role In SalinityTolerance | Mechanism / Pathway | Reference |
|---|---|---|---|
| SOS1 | Plasma membrane Na⁺/H⁺ antiporter; removes Na⁺ from cytoplasm | Na⁺ transport / ionhomeostasis | [96,120] |
| SOS2 | Part of SOS pathway; regulates Na⁺ homeostasis | Na⁺ transport / signaltransduction | [96] |
| SOS3 | Part of SOS pathway; regulates Na⁺ homeostasis | Na⁺ transport / signaltransduction | [96] |
| NHX1 | Vacuolar Na⁺/H⁺ antiporter; sequesters Na⁺ into vacuoles | Na⁺ compartmentalization / ion homeostasis | [96,120] |
| NHX2 | Vacuolar Na⁺/H⁺ antiporter; ion compartmentalization | Na⁺ compartmentalization / ion homeostasis | [96] |
| HKT1 | Na⁺ transporter; regulates Na⁺ uptake and distribution | Na⁺ transport / ionhomeostasis | [96,120] |
| AVP1 | Vacuolar H⁺-PPase; generates proton gradient for ion sequestration | Proton gradient / ioncompartmentalization | [120] |
| Osmotin | Enhances proline, total soluble protein, chlorophyllcontent; improves salinitytolerance | Osmotic adjustment / stress-responsive protein | [98,121] |
| FvGRF3 | Growth-Regulating Factor; involved in chromatin remodeling and ABA-mediated stress response | Transcriptional regulation / ABA signaling | [97] |
| FvGRF6 | Growth-Regulating Factor; involved in chromatin remodeling and ABA-mediated stress response | Transcriptional regulation / ABA signaling | [97] |
| FvGRF8 | Growth-Regulating Factor; involved in chromatin remodeling and ABA-mediated stress response | Transcriptional regulation / ABA signaling | [97] |
| FvMYB82 | MYB transcription factor; increases proline, chlorophyll, and antioxidant enzyme activity | Transcriptional regulation / antioxidant defense | [110] |
| FaNAC2 | NAC transcription factor; regulates proline and ABA biosynthesis under stress | Transcriptional regulation / osmotic adjustment | [104] |
| FvNAC29 | NAC transcription factor; reduces ROS and MDA, enhances antioxidant enzymes and osmotic adjustment | Transcriptional regulation / oxidative stress & osmoprotection | [105] |
| FaTINY2 | AP2/ERF transcription factor; coordinates osmotic adjustment and antioxidant defense | Transcriptional regulation / stress signaling | [107] |
| FaTEDT1L | HDG11-like transcriptionfactor; enhances root growth, reduces waterloss, upregulates SOS genes | Transcriptional regulation / osmotic stress response | [113] |
| G6PDH gene family | Redox regulation via pentosephosphate pathway; protects cells from oxidative stress and contributes to NADPH production under salinity | Redox homeostasis / ROS detoxification | [115] |
| AKT1 | K⁺ channel; maintains ionbalance under salinity stress | K⁺ transport / ionhomeostasis | [96] |
| SAL1 | Phosphatase involved in stress signaling and ionhomeostasis | Signal transduction / stress signaling | [96] |
| CLC_G | Chloride channel; regulates Cl⁻ transport and homeostasis | Cl⁻ transport / ionhomeostasis | [96] |
| CLC_C | Chloride channel; regulates Cl⁻ transport and homeostasis | Cl⁻ transport / ionhomeostasis | [96] |
| SLAH3 | Anion channel; contributesto Cl⁻ homeostasis under salt stress | Cl⁻ transport / ionhomeostasis | [96] |
| ALMT12 | Malate transporter; contributes to ion balance and stress response | Organic acid transport / ion balance | [96] |
5. Future Prospects
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pisinaras, V.; Tsihrintzis, V.A.; Petalas, C.; Ouzounis, K. Soil Salinization in the Agricultural Lands of Rhodope District, Northeastern Greece. Environmental monitoring and assessment 2010, 166, 79–94. [Google Scholar] [CrossRef]
- Hardie, M.; Doyle, R. Measuring Soil Salinity. In Plant Salt Tolerance;Methods in Molecular Biology; Shabala, S., Cuin, T.A., Eds.; Humana Press: Totowa, NJ, 2012; Vol. 913, pp. 415–425. ISBN 978-1-61779-985-3. [Google Scholar]
- Daliakopoulos, I.N.; Tsanis, I.K.; Koutroulis, A.; Kourgialas, N.N.; Varouchakis, A.E.; Karatzas, G.P.; Ritsema, C.J. The Threat of Soil Salinity: A European Scale Review. Science of the total environment 2016, 573, 727–739. [Google Scholar] [CrossRef]
- FAO Global Map of Salt-Affected Soils 2021.
- Tóth, T. Classification and Mitigation of Soil Salinization. In Oxford Research Encyclopedia of Environmental Science; 2017. [Google Scholar]
- Singh, M.; Nara, U.; Kumar, A.; Choudhary, A.; Singh, H.; Thapa, S. Salinity Tolerance Mechanisms and Their Breeding Implications. Journal of Genetic Engineering and Biotechnology 2021, 19, 173. [Google Scholar] [CrossRef] [PubMed]
- Pessarakli, M.; Szabolcs, I. Soil Salinity and Sodicity as Particular Plant/Crop Stress Factors. In Handbook of Plant and Crop Stress, Fourth Edition ed; CRC press, 2019; pp. 3–21. [Google Scholar]
- Shokat, S.; Großkinsky, D.K. Tackling Salinity in Sustainable Agriculture—What Developing Countries May Learn from Approaches of the Developed World. Sustainability 2019, 11, 4558. [Google Scholar] [CrossRef]
- Petalas, C.; Pisinaras, V.; Gemitzi, A.; Tsihrintzis, V.A.; Ouzounis, K. Current Conditions of Saltwater Intrusion in the Coastal Rhodope Aquifer System, Northeastern Greece. Desalination 2009, 237, 22–41. [Google Scholar] [CrossRef]
- Karamanos, A.; Aggelides, S.; Londra, P. Water Use Efficiency and Water Productivity in Greece. Options Méditerranéennes 2005, 57, 92–100. [Google Scholar]
- Yuan, X.; Li, S.; Chen, J.; Yu, H.; Yang, T.; Wang, C.; Huang, S.; Chen, H.; Ao, X. Impacts of Global Climate Change on Agricultural Production: A Comprehensive Review. Agronomy 2024, 14, 1360. [Google Scholar] [CrossRef]
- Terán, F.; Vives-Peris, V.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Facing Climate Change: Plant Stress Mitigation Strategies in Agriculture. Physiologia Plantarum 2024, 176, e14484. [Google Scholar] [CrossRef]
- Zaman, M.; Shahid, S.A.; Heng, L. Guideline for Salinity Assessment, Mitigation and Adaptation Using Nuclear and Related Techniques; Springer Nature, 2018. [Google Scholar]
- Negrão, S.; Schmöckel, S.M.; Tester, M. Evaluating Physiological Responses of Plants to Salinity Stress. Annals of botany 2017, 119, 1–11. [Google Scholar] [CrossRef]
- Xiong, W.; Reynolds, M.; Xu, Y. Climate Change Challenges Plant Breeding. Current Opinion in Plant Biology 2022, 70, 102308. [Google Scholar] [CrossRef]
- Grieve, C.M.; Grattan, S.R.; Maas, E.V. Plant Salt Tolerance. ASCE manual and reports on engineering practice 2012, 71, 405–459. [Google Scholar]
- Maas, E.V.; Grattan, S.R. Crop Yields as Affected by Salinity. In Agronomy Monographs; Skaggs, R.W., Schilfgaarde, J., Eds.; Wiley, 1999; Vol. 38, pp. 55–108. ISBN 978-0-89118-141-5. [Google Scholar]
- Singh, A. Salinization and Drainage Problems of Agricultural Land. Irrigation and Drainage 2020, 69, 844–853. [Google Scholar] [CrossRef]
- Endo, T.; Yamamoto, S.; Larrinaga, J.A.; Fujiyama, H.; Honna, T. Status and Causes of Soil Salinization of Irrigated Agricultural Lands in Southern Baja California, Mexico. Applied and Environmental Soil Science 2011, 2011, 1–12. [Google Scholar] [CrossRef]
- Rengasamy, P. World Salinization with Emphasis on Australia. J Exp Bot 2006, 57, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Bouzourra, H.; Bouhlila, R.; Elango, L.; Slama, F.; Ouslati, N. Characterization of Mechanisms and Processes of Groundwater Salinization in Irrigated Coastal Area Using Statistics, GIS, and Hydrogeochemical Investigations. Environmental Science and Pollution Research 2015, 22, 2643–2660. [Google Scholar] [CrossRef]
- Nabi, F.; Sarfaraz, A.; Kama, R.; Kanwal, R.; Li, H. Structure-Based Function of Humic Acid in Abiotic Stress Alleviation in Plants: A Review. Plants 2025, 14, 1916. [Google Scholar] [CrossRef]
- Gkiougkis, I.; Kallioras, A.; Pliakas, F.; Pechtelidis, A.; Diamantis, V.; Diamantis, I.; Ziogas, A.; Dafnis, I. Assessment of Soil Salinization at the Eastern Nestos River Delta, NE Greece. Catena 2015, 128, 238–251. [Google Scholar] [CrossRef]
- Hancock, J.F. Temperate Fruit Crop Breeding: Germplasm to Genomics; Springer Science & Business Media, 2008. [Google Scholar]
- Ferreira, J.F.; Liu, X.; Suarez, D.L. Fruit Yield and Survival of Five Commercial Strawberry Cultivars under Field Cultivation and Salinity Stress. Scientia Horticulturae 2019, 243, 401–410. [Google Scholar] [CrossRef]
- Mazzoni, L.; Perez-Lopez, P.; Giampieri, F.; Alvarez-Suarez, J.M.; Gasparrini, M.; Forbes-Hernandez, T.Y.; Quiles, J.L.; Mezzetti, B.; Battino, M. The Genetic Aspects of Berries: From Field to Health. J Sci Food Agric 2016, 96, 365–371. [Google Scholar] [CrossRef]
- Giampieri, F.; Alvarez-Suarez, J.M.; Battino, M. Strawberry and Human Health: Effects beyond Antioxidant Activity. J. Agric. Food Chem. 2014, 62, 3867–3876. [Google Scholar] [CrossRef]
- Galli, V.; da Silva Messias, R.; Perin, E.C.; Borowski, J.M.; Bamberg, A.L.; Rombaldi, C.V. Mild Salt Stress Improves Strawberry Fruit Quality. Lwt 2016, 73, 693–699. [Google Scholar] [CrossRef]
- Wu, Y.; Li, L.; Li, M.; Zhang, M.; Sun, H.; Sigrimis, N. Optimal Fertigation for High Yield and Fruit Quality of Greenhouse Strawberry. PLoS One 2020, 15, e0224588. [Google Scholar] [CrossRef] [PubMed]
- Shannon, M.C.; Grieve, C.M. Tolerance of Vegetable Crops to Salinity. Scientia horticulturae 1998, 78, 5–38. [Google Scholar] [CrossRef]
- Keutgen, A.J.; Pawelzik, E. Quality and Nutritional Value of Strawberry Fruit under Long Term Salt Stress. Food chemistry 2008, 107, 1413–1420. [Google Scholar] [CrossRef]
- Keutgen, A.J.; Pawelzik, E. Impacts of NaCl Stress on Plant Growth and Mineral Nutrient Assimilation in Two Cultivars of Strawberry. Environmental and experimental botany 2009, 65, 170–176. [Google Scholar] [CrossRef]
- Singh, A. Soil Salinization and Waterlogging: A Threat to Environment and Agricultural Sustainability. Ecological indicators 2015, 57, 128–130. [Google Scholar] [CrossRef]
- Denaxa, N.-K.; Nomikou, A.; Malamos, N.; Liveri, E.; Roussos, P.A.; Papasotiropoulos, V. Salinity Effect on Plant Growth Parameters and Fruit Bioactive Compounds of Two Strawberry Cultivars, Coupled with Environmental Conditions Monitoring. Agronomy 2022, 12, 2279. [Google Scholar] [CrossRef]
- Ghaderi, N.; Hatami, M.R.; Mozafari, A.; Siosehmardeh, A. Change in Antioxidant Enzymes Activity and Some Morpho-Physiological Characteristics of Strawberry under Long-Term Salt Stress. Physiology and molecular biology of plants 2018, 24, 833–843. [Google Scholar] [CrossRef]
- Mozafari, A.; Ghaderi, N.; Havas, F.; Dedejani, S. Comparative Investigation of Structural Relationships among Morpho-Physiological and Biochemical Properties of Strawberry (Fragaria\times Ananassa Duch.) under Drought and Salinity Stresses: A Study Based on in Vitro Culture. Scientia Horticulturae 2019, 256, 108601. [Google Scholar] [CrossRef]
- Hussein, A.E.; El-Kerdany, A.Y.; Afifi, K.M. Effect of Drought and Salinity Stresses on Two Strawberry Cultivars during Their Regeneration in Vitro. International Journal of Innovative Science Engineering and Technology 2016, 4, 83–93. [Google Scholar]
- Akbari, M.; Mahna, N.; Ramesh, K.; Bandehagh, A.; Mazzuca, S. Ion Homeostasis, Osmoregulation, and Physiological Changes in the Roots and Leaves of Pistachio Rootstocks in Response to Salinity. Protoplasma 2018, 255, 1349–1362. [Google Scholar] [CrossRef] [PubMed]
- Sikder, R.K.; Wang, X.; Zhang, H.; Gui, H.; Dong, Q.; Jin, D.; Song, M. Nitrogen Enhances Salt Tolerance by Modulating the Antioxidant Defense System and Osmoregulation Substance Content in Gossypium Hirsutum. Plants 2020, 9, 450. [Google Scholar] [CrossRef] [PubMed]
- Asghar, R.; Biglarifard, A.; Mirdehghan, H.; Borghei, S.F. Influence of NaCl Salinity on Growth Analysis of Strawberry Cv. Camarosa. Journal of Stress Physiology & Biochemistry 2011, 7, 145–156. [Google Scholar]
- Moradi, P.; Vafaee, Y.; Mozafari, A.A.; Tahir, N.A. Silicon Nanoparticles and Methyl Jasmonate Improve Physiological Response and Increase Expression of Stress-Related Genes in Strawberry Cv. Paros under Salinity Stress. Silicon 2022, 14, 10559–10569. [Google Scholar] [CrossRef]
- Zhang, Y.; Deng, M.; Lin, B.; Tian, S.; Chen, Y.; Huang, S.; Lin, Y.; Li, M.; He, W.; Wang, Y.; et al. Physiological and Transcriptomic Evidence Revealed the Role of Exogenous GABA in Enhancing Salt Tolerance in Strawberry Seedlings. BMC Genomics 2025, 26, 196. [Google Scholar] [CrossRef]
- Golnari, S.; Vafaee, Y.; Nazari, F.; Ghaderi, N. Gamma-Aminobutyric Acid (GABA) and Salinity Impacts Antioxidative Response and Expression of Stress-Related Genes in Strawberry Cv. Aromas. Brazilian Journal of Botany 2021, 44, 639–651. [Google Scholar] [CrossRef]
- Keutgen, A.; Pawelzik, E. Modifications of Taste-Relevant Compounds in Strawberry Fruit under NaCl Salinity. Food chemistry 2007, 105, 1487–1494. [Google Scholar] [CrossRef]
- Sangiorgio, D.; Cellini, A.; Spinelli, F.; Donati, I. Promoting Strawberry (Fragaria\times Ananassa) Stress Resistance, Growth, and Yield Using Native Bacterial Biostimulants. Agronomy 2023, 13, 529. [Google Scholar] [CrossRef]
- Faghih, S.; Ghobadi, C.; Zarei, A. Response of Strawberry Plant Cv.‘Camarosa’to Salicylic Acid and Methyl Jasmonate Application under Salt Stress Condition. Journal of Plant Growth Regulation 2017, 36, 651–659. [Google Scholar] [CrossRef]
- Zahedi, S.M.; Hosseini, M.S.; Abadía, J.; Marjani, M. Melatonin Foliar Sprays Elicit Salinity Stress Tolerance and Enhance Fruit Yield and Quality in Strawberry (Fragaria\times Ananassa Duch.). Plant Physiology and Biochemistry 2020, 149, 313–323. [Google Scholar] [CrossRef]
- Haque, S.I.; Matsubara, Y. Salinity Tolerance and Sodium Localization in Mycorrhizal Strawberry Plants. Communications in Soil Science and Plant Analysis 2018, 49, 2782–2792. [Google Scholar] [CrossRef]
- Zeid, I.M.A.; Mohamed, F.H.; Metwali, E.M. Responses of Two Strawberry Cultivars to NaCl-Induced Salt Stress under the Influence of ZnO Nanoparticles. Saudi Journal of Biological Sciences 2023, 30, 103623. [Google Scholar] [CrossRef] [PubMed]
- Saidimoradi, D.; Ghaderi, N.; Javadi, T. Salinity Stress Mitigation by Humic Acid Application in Strawberry (Fragaria x Ananassa Duch.). Scientia Horticulturae 2019, 256, 108594. [Google Scholar] [CrossRef]
- Karlidag, H.; Yildirim, E.; Turan, M.; Pehluvan, M.; Donmez, F. Plant Growth-Promoting Rhizobacteria Mitigate Deleterious Effects of Salt Stress on Strawberry Plants (Fragaria\times Ananassa). HortScience 2013, 48, 563–567. [Google Scholar] [CrossRef]
- Yavuz, A.; Erdogan, U.; Turan, M.; Argın, S.; Kocaman, A. Synergistic Strategies for Overcoming Salt Stress in Strawberry Farming: The Use of Organic Fertilizers and Plant Growth Promoting Rhizobacteria (PGPR). Applied Fruit Science 2024, 66, 1787–1797. [Google Scholar] [CrossRef]
- Malekzadeh Shamsabad, M.R.; Esmaeilizadeh, M.; Roosta, H.R.; Dąbrowski, P.; Telesiński, A.; Kalaji, H.M. Supplemental Light Application Can Improve the Growth and Development of Strawberry Plants under Salinity and Alkalinity Stress Conditions. Scientific Reports 2022, 12, 9272. [Google Scholar] [CrossRef]
- Hu, Y.; Feng, C.; Yang, L.; Edger, P.P.; Kang, M. Genomic Population Structure and Local Adaptation of the Wild Strawberry Fragaria Nilgerrensis. Horticulture Research 2022, 9, uhab059. [Google Scholar] [CrossRef]
- Bringhurst, R.S.; Hancock, J.F.; Voth, V. The Beach Strawberry, an Important Natural Resource; 1977. [Google Scholar]
- VanDerZanden, A.M.; Cameron, J.S. Effect of Water Deficit Stress on 11 Native Fragaria Chiloensis Clones Selected as Ornamental Groundcovers. Scientia horticulturae 1996, 66, 241–253. [Google Scholar] [CrossRef]
- Carrasco, B.; Garcés, M.; Rojas, P.; Saud, G.; Herrera, R.; Retamales, J.B.; Caligari, P.D. The Chilean Strawberry [Fragaria Chiloensis (L.) Duch.]: Genetic Diversity and Structure. Journal of the American Society for Horticultural Science 2007, 132, 501–506. [Google Scholar] [CrossRef]
- Nikoloudi, A. Salinity Tolerance in Strawberry (Fragaria Spp) as Influenced by Genotype; Michigan State University, 2003. [Google Scholar]
- Garriga, M.; Muñoz, C.A.; Caligari, P.D.; Retamales, J.B. Effect of Salt Stress on Genotypes of Commercial (Fragaria x Ananassa) and Chilean Strawberry (F. Chiloensis). Scientia Horticulturae 2015, 195, 37–47. [Google Scholar] [CrossRef]
- Suarez, D.L.; Grieve, C.M. GROWTH, YIELD, AND ION RELATIONS OF STRAWBERRY IN RESPONSE TO IRRIGATION WITH CHLORIDE-DOMINATED WATERS. Journal of Plant Nutrition 2013, 36, 1963–1981. [Google Scholar] [CrossRef]
- Demiral, M.A. Effect of Salt Stress on Concentration of Nitrogen and Phosphorus in Root and Leaf of Strawberry Plant. Eurasian Journal of Soil Science 2017, 6, 357–364. [Google Scholar] [CrossRef]
- Orsini, F.; Alnayef, M.; Bona, S.; Maggio, A.; Gianquinto, G. Low Stomatal Density and Reduced Transpiration Facilitate Strawberry Adaptation to Salinity. Environmental and Experimental Botany 2012, 81, 1–10. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Zhou, M.; Shabala, S. How Does Stomatal Density and Residual Transpiration Contribute to Osmotic Stress Tolerance? Plants 2023, 12, 494. [Google Scholar] [CrossRef]
- Turhan, E.; Eris, A. Changes of Growth, Amino Acids, and Ionic Composition in Strawberry Plants under Salt Stress Conditions. Communications in Soil Science and Plant Analysis 2009, 40, 3308–3322. [Google Scholar] [CrossRef]
- Turhan, E.; Eris, A. Changes of Micronutrients, Dry Weight, and Chlorophyll Contents in Strawberry Plants Under Salt Stress Conditions*. Communications in Soil Science and Plant Analysis 2005, 36, 1021–1028. [Google Scholar] [CrossRef]
- Turhan, E.; Eris, A. Effects of Sodium Chloride Applications and Different Growth Media on Ionic Composition in Strawberry Plant. Journal of Plant Nutrition 2005, 27, 1653–1665. [Google Scholar] [CrossRef]
- Gulen, H.; Turhan, E.; Eris, A. Changes in Peroxidase Activities and Soluble Proteins in Strawberry Varieties under Salt-Stress. Acta Physiologiae Plantarum 2006, 28, 109–116. [Google Scholar] [CrossRef]
- Kortekamp, T.; Chen, T.-W.; Wagner, H.; Olbricht, K. Evaluation of Salinity Tolerance in a Fragaria F2 Population. In Proceedings of the XVI EUCARPIA Symposium on Fruit Breeding and Genetics 1412, 2023; pp. 111–116. [Google Scholar]
- Alnayef, M.; Hartley, J.; Orsini, F.; Di Silvestro, R.; Sanoubar, R.; Marotti, I.; Gianquinto, G.; Dinelli, G.; Puniran-Hartley, N. Using Salinity to Improve Nutritional and Market Value of Strawberries. Australian Journal of Crop Science 2022, 16, 7–S3. [Google Scholar] [CrossRef]
- Pirlak, L.; Eşitken, A. Salinity Effects on Growth, Proline and Ion Accumulation in Strawberry Plants. Acta Agriculturae Scandinavica, Section B - Soil & Plant Science 2004, 54, 189–192. [Google Scholar] [CrossRef]
- Rahimi, A.; Biglarifard, A. Influence of NaCl Salinity and Different Substracts on Plant Growth, Mineral Nutrient Assimilation and Fruit Yield of Strawberry. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 2011, 39, 219–226. [Google Scholar] [CrossRef]
- Ondrašek, G.; Romić, D.; Romić, M.; Duralija, B.; Mustač, I. Strawberry Growth and Fruit Yield in a Saline Environment. Agriculturae conspectus scientificus 2006, 71, 155–158. [Google Scholar]
- Chan, Z.; Shi, H. Improved Abiotic Stress Tolerance of Bermudagrass by Exogenous Small Molecules. Plant Signaling & Behavior 2015, 10, e991577. [Google Scholar] [CrossRef] [PubMed]
- Averina, N.G.; Gritskevich, E.R.; Vershilovskaya, I.V.; Usatov, A.V.; Yaronskaya, E.B. Mechanisms of Salt Stress Tolerance Development in Barley Plants under the Influence of 5-Aminolevulinic Acid. Russian Journal of Plant Physiology 2010, 57, 792–798. [Google Scholar] [CrossRef]
- Wu, Y.; Liao, W.; Dawuda, M.M.; Hu, L.; Yu, J. 5-Aminolevulinic Acid (ALA) Biosynthetic and Metabolic Pathways and Its Role in Higher Plants: A Review. Plant Growth Regul 2019, 87, 357–374. [Google Scholar] [CrossRef]
- Wu, Y.; Jin, X.; Liao, W.; Hu, L.; Dawuda, M.M.; Zhao, X.; Tang, Z.; Gong, T.; Yu, J. 5-Aminolevulinic Acid (ALA) Alleviated Salinity Stress in Cucumber Seedlings by Enhancing Chlorophyll Synthesis Pathway. Frontiers in plant science 2018, 9, 635. [Google Scholar] [CrossRef]
- Wei, W.; Li, Q.-T.; Chu, Y.-N.; Reiter, R.J.; Yu, X.-M.; Zhu, D.-H.; Zhang, W.-K.; Ma, B.; Lin, Q.; Zhang, J.-S. Melatonin Enhances Plant Growth and Abiotic Stress Tolerance in Soybean Plants. Journal of experimental botany 2015, 66, 695–707. [Google Scholar] [CrossRef]
- Li, J.; Liu, J.; Zhu, T.; Zhao, C.; Li, L.; Chen, M. The Role of Melatonin in Salt Stress Responses. International journal of molecular sciences 2019, 20, 1735. [Google Scholar] [CrossRef]
- Pardo-Hernández, M.; López-Delacalle, M.; Rivero, R.M. ROS and NO Regulation by Melatonin under Abiotic Stress in Plants. Antioxidants 2020, 9, 1078. [Google Scholar] [CrossRef]
- Georgiadou, E.C.; García, C.J.; Taliadorou, A.M.; Gedeon, S.; Valanides, N.; Varaldo, A.; Gohari, G.; Balsells-Llauradó, M.; Alcázar, R.; Hertog, M.L. Pre-Harvest Application of Sodium Alginate Functionalized with Melatonin Enhances Secondary Metabolism in Strawberry Fruit. Current Plant Biology 2025, 43, 100515. [Google Scholar] [CrossRef]
- Bandurska, H. Salicylic Acid: An Update on Biosynthesis and Action in Plant Response to Water Deficit and Performance Under Drought. In SALICYLIC ACID; Hayat, S., Ahmad, A., Alyemeni, M.N., Eds.; Springer Netherlands: Dordrecht, 2013; pp. 1–14. ISBN 978-94-007-6427-9. [Google Scholar]
- Karlidag, H.; Yildirim, E.; Turan, M. Melhora Do Efeito de Estresse Salino Em Morango Por Adição de Ácido Salicílico. Scientia Agricola 2009, 66, 180–187. [Google Scholar] [CrossRef]
- Jamali, B.; Eshghi, S. Salicylic Acid–Induced Salinity Redressal in Hydroponically Grown Strawberry. Communications in Soil Science and Plant Analysis 2015, 46, 1482–1493. [Google Scholar] [CrossRef]
- Samadi, S.; Habibi, G.; Vaziri, A. Effects of Exogenous Salicylic Acid on Antioxidative Responses, Phenolic Metabolism and Photochemical Activity of Strawberry under Salt Stress. Iranian Journal of Plant Physiology 2019, 9, 2685–2694. [Google Scholar]
- Roshdy, A.E.-D.; Alebidi, A.; Almutairi, K.; Al-Obeed, R.; Elsabagh, A. The Effect of Salicylic Acid on the Performances of Salt Stressed Strawberry Plants, Enzymes Activity, and Salt Tolerance Index. Agronomy 2021, 11, 775. [Google Scholar] [CrossRef]
- Canellas, L.P.; Da Silva, R.M.; Busato, J.G.; Olivares, F.L. Humic Substances and Plant Abiotic Stress Adaptation. Chem. Biol. Technol. Agric. 2024, 11, 66. [Google Scholar] [CrossRef]
- Mora, V.; Olaetxea, M.; Bacaicoa, E.; Baigorri, R.; Fuentes, M.; Zamarreño, A.M.; Garcia-Mina, J.M. Abiotic Stress Tolerance in Plants: Exploring the Role of Nitric Oxide and Humic Substances. In Nitric Oxide in Plants: Metabolism and Role in Stress Physiology; Khan, M.N., Mobin, M., Mohammad, F., Corpas, F.J., Eds.; Springer International Publishing: Cham, 2014; pp. 243–264. ISBN 978-3-319-06709-4. [Google Scholar]
- Mirfattahi, Z.; Eshghi, S.; Gharaghani, A.; Etemadi, M.; Moghadam, A. Time and Rate of Acetate Foliar Spray Can Ameliorate Adverse Effect of NaCl Stress on Strawberry. South African Journal of Botany 2022, 150, 797–805. [Google Scholar] [CrossRef]
- Rahman, Md.M.; Keya, S.S.; Sahu, A.; Gupta, A.; Dhingra, A.; Tran, L.-S.P.; Mostofa, M.G. Acetic Acid: A Cheap but Chief Metabolic Regulator for Abiotic Stress Tolerance in Plants. Stress Biology 2024, 4, 34. [Google Scholar] [CrossRef] [PubMed]
- Evelin, H.; Kapoor, R.; Giri, B. Arbuscular Mycorrhizal Fungi in Alleviation of Salt Stress: A Review. Annals of botany 2009, 104, 1263–1280. [Google Scholar] [CrossRef] [PubMed]
- Boorboori, M.R.; Lackóová, L. Arbuscular Mycorrhizal Fungi and Salinity Stress Mitigation in Plants. Frontiers in Plant Science 2025, 15, 1504970. [Google Scholar] [CrossRef]
- Redondo-Gómez, S.; Mesa-Marín, J.; Pérez-Romero, J.A.; López-Jurado, J.; García-López, J.V.; Mariscal, V.; Molina-Heredia, F.P.; Pajuelo, E.; Rodríguez-Llorente, I.D.; Flowers, T.J. Consortia of Plant-Growth-Promoting Rhizobacteria Isolated from Halophytes Improve Response of Eight Crops to Soil Salinization and Climate Change Conditions. Agronomy 2021, 11, 1609. [Google Scholar] [CrossRef]
- Wang, R.; Chu, S.; Zhang, D.; Hayat, K.; Zhang, X.; Chi, Y.; Ma, X.; Chen, X.; Yang, H.; Ding, W.; et al. Alleviation of Salt Stress in Strawberries by Hydrogen-rich Water: Physiological, Transcriptomic and Metabolomic Responses. Physiologia Plantarum 2025, 177, e70151. [Google Scholar] [CrossRef]
- Ullah, I.; Toor, M.D.; Yerlikaya, B.A.; Mohamed, H.I.; Yerlikaya, S.; Basit, A.; Rehman, A. High-Temperature Stress in Strawberry: Understanding Physiological, Biochemical and Molecular Responses. Planta 2024, 260, 118. [Google Scholar] [CrossRef] [PubMed]
- Roychowdhury, R.; Das, S.P.; Gupta, A.; Parihar, P.; Chandrasekhar, K.; Sarker, U.; Kumar, A.; Ramrao, D.P.; Sudhakar, C. Multi-Omics Pipeline and Omics-Integration Approach to Decipher Plant’s Abiotic Stress Tolerance Responses. Genes 2023, 14, 1281. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, D.; Pudussery, M.V.; Ferreira, J.F.; Liu, X.; Pallete, A.; Grover, K.K.; Hummer, K. Variable Salinity Responses and Comparative Gene Expression in Woodland Strawberry Genotypes. Scientia Horticulturae 2019, 254, 61–69. [Google Scholar] [CrossRef]
- Li, Z.; Xie, Q.; Yan, J.; Chen, J.; Chen, Q. Genome-Wide Identification and Characterization of the Abiotic-Stress-Responsive GRF Gene Family in Diploid Woodland Strawberry (Fragaria Vesca). Plants 2021, 10, 1916. [Google Scholar] [CrossRef]
- Husaini, A.M.; Abdin, M.Z. Development of Transgenic Strawberry (Fragaria x Ananassa Duch.) Plants Tolerant to Salt Stress. Plant Science 2008, 174, 446–455. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Y.; Gao, W.; Tian, S.; Lin, B.; Gu, X.; Lin, Y.; Li, M.; Wang, Y.; He, W. Characterization of Superoxide Dismutase (SOD) Gene Family and Their Responses to Salinity Stress and Fruit Development in Octoploid Strawberry. Agronomy 2024, 14, 2514. [Google Scholar] [CrossRef]
- Kong, J.; Xiong, R.; Qiu, K.; Lin, X.; Li, D.; Lu, L.; Zhou, J.; Zhu, S.; Liu, M.; Sun, Q. Genome-Wide Identification and Characterization of the Laccase Gene Family in Fragaria Vesca and Its Potential Roles in Response to Salt and Drought Stresses. Plants 2024, 13, 3366. [Google Scholar] [CrossRef]
- Fernando, V.D. Major Transcription Factor Families Involved in Salinity Stress Tolerance in Plants. Transcription factors for abiotic stress tolerance in plants 2020, 99–109. [Google Scholar]
- Shao, H.; Wang, H.; Tang, X. NAC Transcription Factors in Plant Multiple Abiotic Stress Responses: Progress and Prospects. Frontiers in plant science 2015, 6, 902. [Google Scholar] [CrossRef]
- Zhang, H.; Kang, H.; Su, C.; Qi, Y.; Liu, X.; Pu, J. Genome-Wide Identification and Expression Profile Analysis of the NAC Transcription Factor Family during Abiotic and Biotic Stress in Woodland Strawberry. PLoS One 2018, 13, e0197892. [Google Scholar] [CrossRef]
- Liang, J.; Zheng, J.; Wu, Z.; Wang, H. Strawberry FaNAC2 Enhances Tolerance to Abiotic Stress by Regulating Proline Metabolism. Plants 2020, 9, 1417. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, H.; Wei, Y.; Han, J.; Wang, Y.; Li, X.; Zhang, L.; Han, D. Overexpression of a Fragaria Vesca NAM, ATAF, and CUC (NAC) Transcription Factor Gene (FvNAC29) Increases Salt and Cold Tolerance in Arabidopsis Thaliana. International Journal of Molecular Sciences 2024, 25, 4088. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA 2/Ethylene Responsive Factor ( AP 2/ ERF ) Transcription Factors: Mediators of Stress Responses and Developmental Programs. New Phytologist 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, W.; Li, H.; Yao, A.; Ma, Z.; Kang, R.; Guo, Y.; Li, X.; Yu, W.; Han, D. Overexpression of a Fragaria\times Ananassa AP2/ERF Transcription Factor Gene (FaTINY2) Increases Cold and Salt Tolerance in Arabidopsis Thaliana. International Journal of Molecular Sciences 2025, 26, 2109. [Google Scholar]
- Li, S.; Chang, L.; Sun, R.; Dong, J.; Zhong, C.; Gao, Y.; Zhang, H.; Wei, L.; Wei, Y.; Zhang, Y. Combined Transcriptomic and Metabolomic Analysis Reveals a Role for Adenosine Triphosphate-Binding Cassette Transporters and Cell Wall Remodeling in Response to Salt Stress in Strawberry. Frontiers in Plant Science 2022, 13, 996765. [Google Scholar] [CrossRef]
- Wang, S.; Shi, M.; Zhang, Y.; Xie, X.; Sun, P.; Fang, C.; Zhao, J. FvMYB24, a Strawberry R2R3-MYB Transcription Factor, Improved Salt Stress Tolerance in Transgenic Arabidopsis. Biochemical and Biophysical Research Communications 2021, 569, 93–99. [Google Scholar] [CrossRef]
- Li, W.; Zhong, J.; Zhang, L.; Wang, Y.; Song, P.; Liu, W.; Li, X.; Han, D. Overexpression of a Fragaria Vesca MYB Transcription Factor Gene (FvMYB82) Increases Salt and Cold Tolerance in Arabidopsis Thaliana. International Journal of Molecular Sciences 2022, 23, 10538. [Google Scholar] [CrossRef]
- Kula, N.; Öztürk Erdem, S.; Ünal, D. Salt Tolerance Mechanisms in Strawberry Cultivars: Comparing the Physiological and Molecular Responses of Petaluma and Cabrillo. Applied Fruit Science 2025, 67, 379. [Google Scholar] [CrossRef]
- López, M.-E.; Roquis, D.; Becker, C.; Denoyes, B.; Bucher, E. DNA Methylation Dynamics during Stress Response in Woodland Strawberry (Fragaria Vesca). Horticulture research 2022, 9, uhac174. [Google Scholar] [CrossRef]
- Chu, C.-Y.; Lin, L.-F.; Lai, S.-C.; Yang, J.-H.; Chou, M.-L. FaTEDT1L of Octoploid Cultivated Strawberry Functions as a Transcriptional Activator and Enhances Abiotic Stress Tolerance in Transgenic Arabidopsis. International Journal of Molecular Sciences 2024, 25, 10091. [Google Scholar] [CrossRef] [PubMed]
- Tibbs Cortes, L.; Zhang, Z.; Yu, J. Status and Prospects of Genome-wide Association Studies in Plants. The Plant Genome 2021, 14, e20077. [Google Scholar] [CrossRef] [PubMed]
- Lei, D.; Lin, Y.; Luo, M.; Zhao, B.; Tang, H.; Zhou, X.; Yao, W.; Zhang, Y.; Wang, Y.; Li, M. Genome-Wide Investigation of G6PDH Gene in Strawberry: Evolution and Expression Analysis during Development and Stress. International Journal of Molecular Sciences 2022, 23, 4728. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-H.; Tjong, W.-Y.; Yang, H.-C.; Liu, H.-Y.; Stern, A.; Chiu, D.T.-Y. Glucose-6-Phosphate Dehydrogenase, Redox Homeostasis and Embryogenesis. International Journal of Molecular Sciences 2022, 23. [Google Scholar] [CrossRef]
- Crizel, R.L.; Perin, E.C.; Vighi, I.L.; Woloski, R.; Seixas, A.; da Silva Pinto, L.; Rombaldi, C.V.; Galli, V. Genome-Wide Identification, and Characterization of the CDPK Gene Family Reveal Their Involvement in Abiotic Stress Response in Fragaria x Ananassa. Scientific reports 2020, 10, 11040. [Google Scholar] [CrossRef]
- Li, H.; Yue, M.; Jiang, L.; Liu, Y.; Zhang, N.; Liu, X.; Ye, Y.; Lin, X.; Zhang, Y.; Lin, Y. Genome-Wide Identification of Strawberry C2H2-ZFP C1-2i Subclass and the Potential Function of FaZAT10 in Abiotic Stress. International Journal of Molecular Sciences 2022, 23, 13079. [Google Scholar] [CrossRef]
- Jiang, L.; Lin, Y.; Wang, L.; Peng, Y.; Yang, M.; Jiang, Y.; Hou, G.; Liu, X.; Li, M.; Zhang, Y. Genome-Wide Identification and Expression Profiling Reveal the Regulatory Role of U-Box E3 Ubiquitin Ligase Genes in Strawberry Fruit Ripening and Abiotic Stresses Resistance. Frontiers in Plant Science 2023, 14, 1171056. [Google Scholar] [CrossRef]
- Zhang, J.-L.; Shi, H. Physiological and Molecular Mechanisms of Plant Salt Tolerance. Photosynthesis research 2013, 115, 1–22. [Google Scholar] [CrossRef]
- Singh, N.K.; Nelson, D.E.; Kuhn, D.; Hasegawa, P.M.; Bressan, R.A. Molecular Cloning of Osmotin and Regulation of Its Expression by ABA and Adaptation to Low Water Potential. Plant physiology 1989, 90, 1096–1101. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





