Submitted:
16 December 2025
Posted:
17 December 2025
You are already at the latest version
Abstract
Keywords:
Introduction
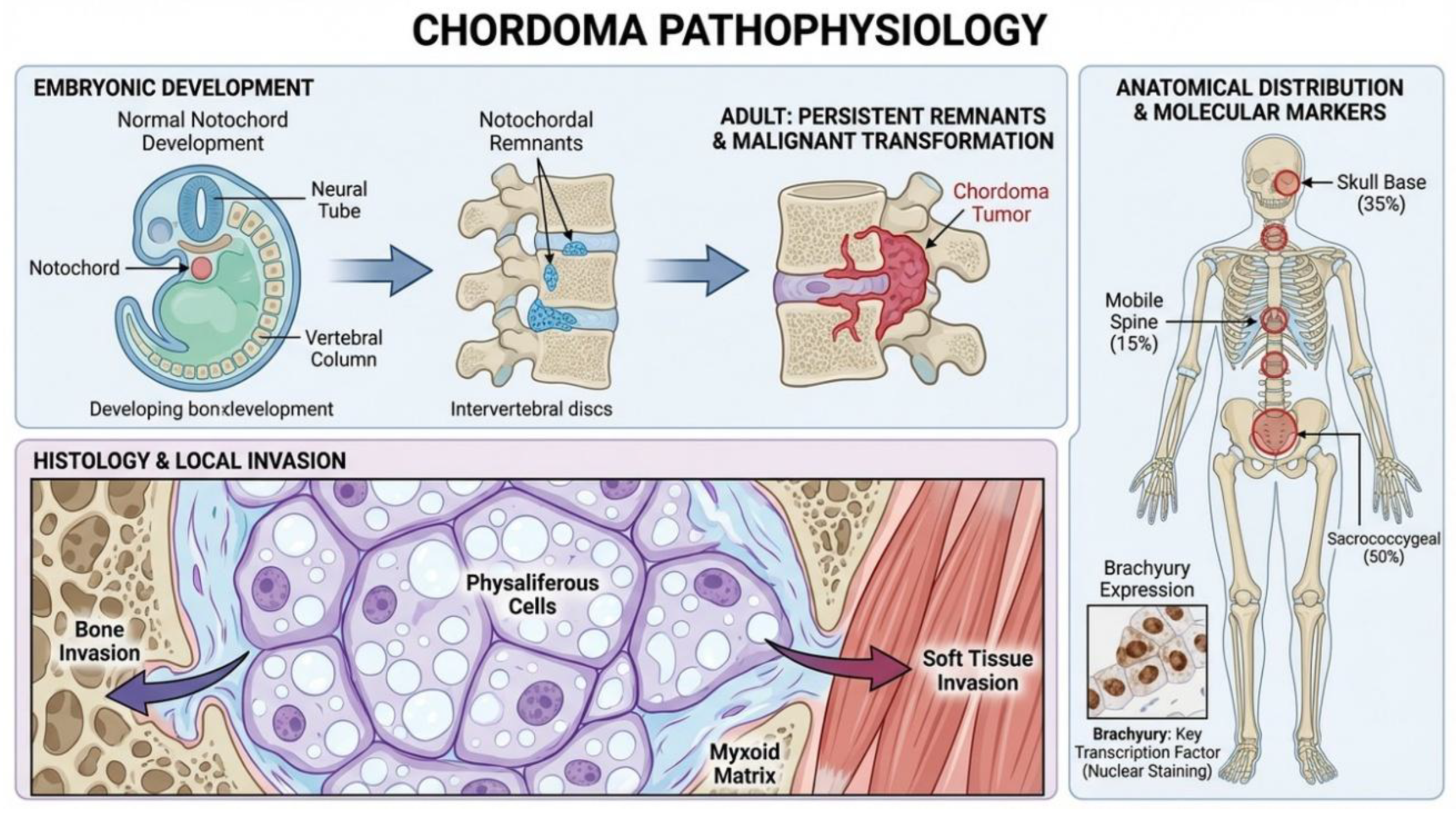
Chordoma: Characteristics and Pathophysiology
Pathophysiology
Circular RNAs in Cancer Immunometabolism
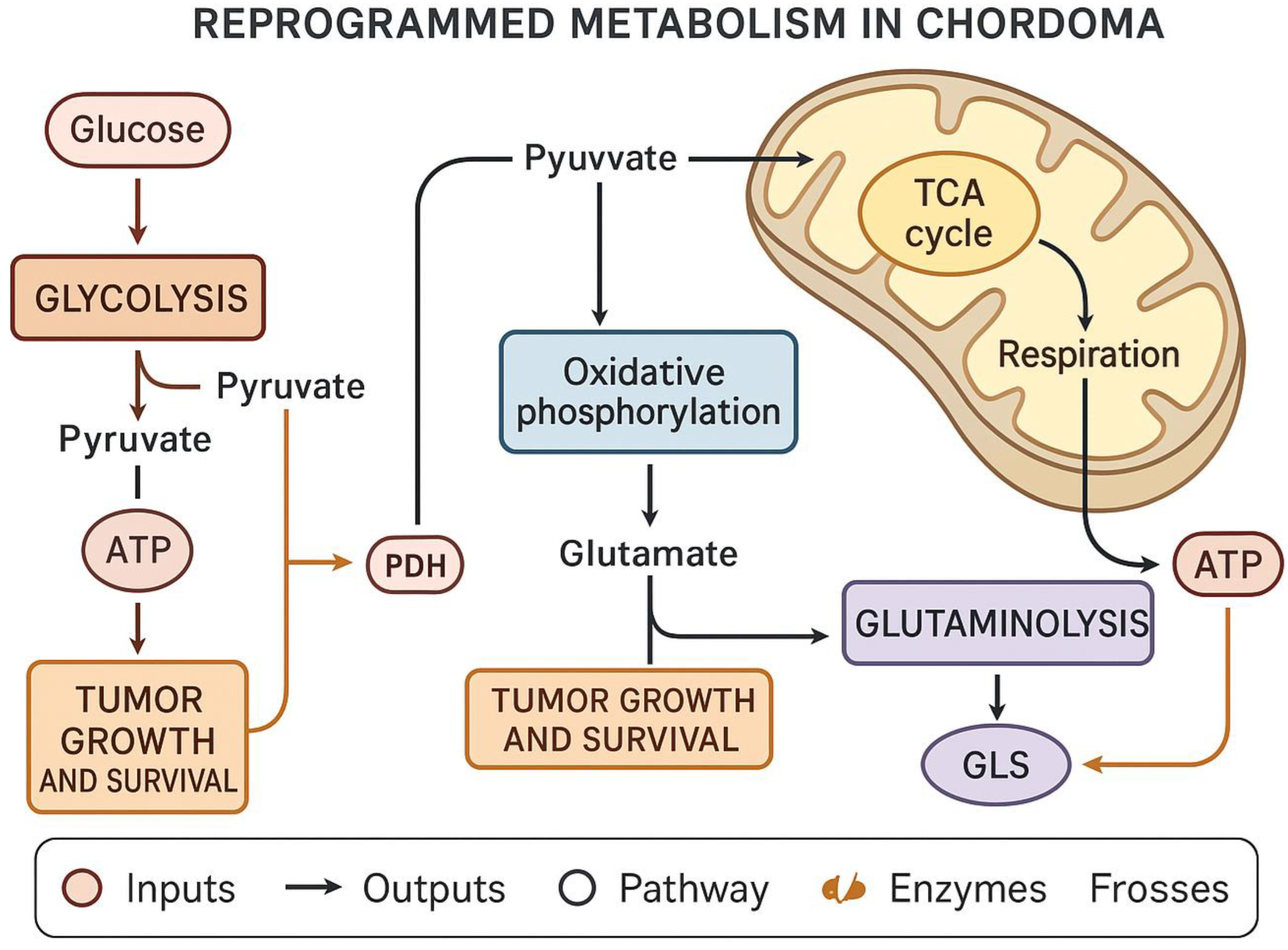
Role in Metabolic Reprogramming
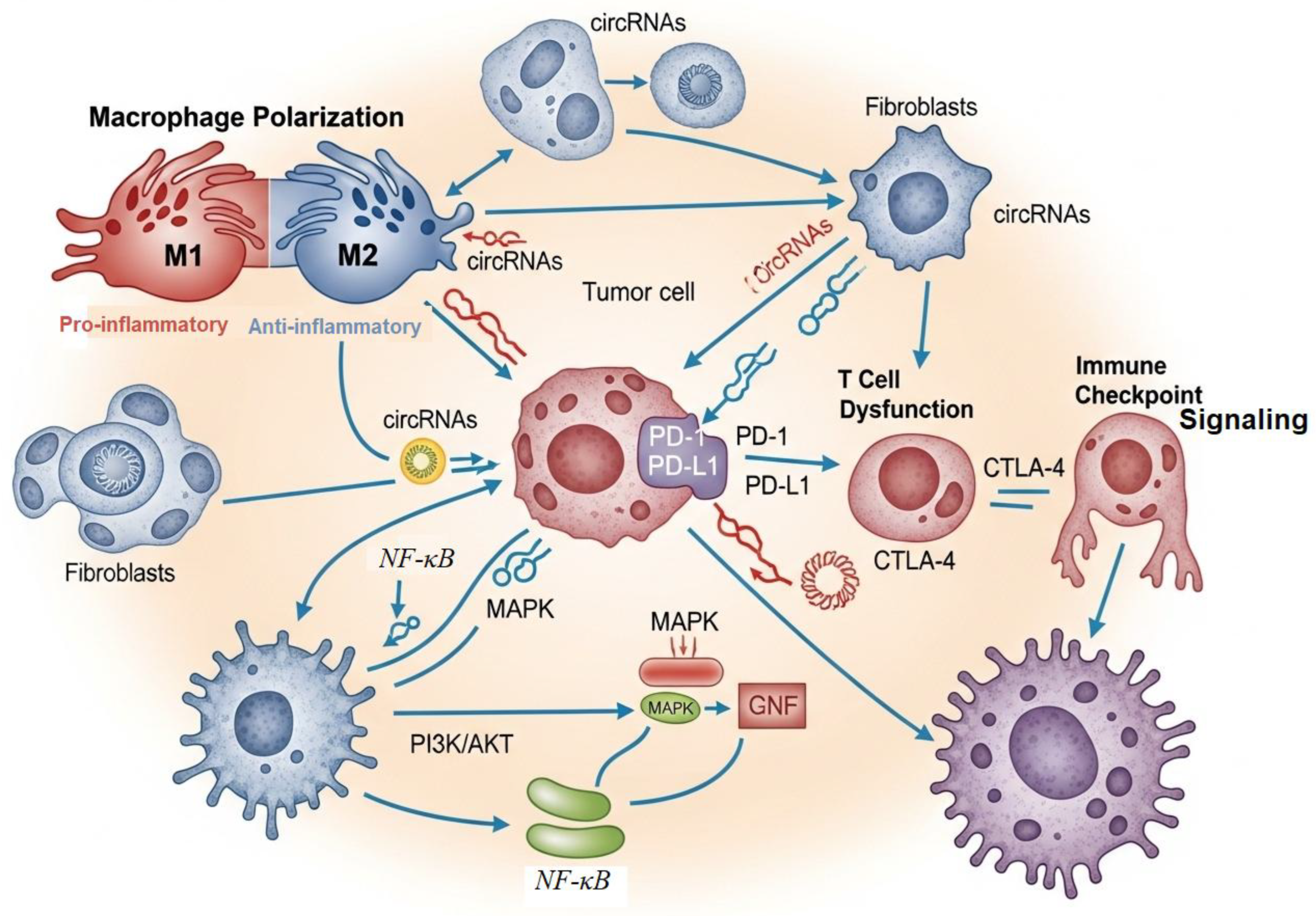
Role in Immune Regulation
Molecular Crosstalk in Immunometabolism
Specific circRNA Mechanisms in Chordoma: Bridging to Immunometabolism
Potential Immunometabolic Roles
Therapeutic Strategies Targeting circRNAs in Chordoma
Direct Targeting of circRNAs
Indirect Targeting of circRNA-Regulated Pathways
Molecular Crosstalk and Combination Therapies
Comparative Analysis of Chordoma Molecular and Immunological Studies
Synthesis and Comparative Insights
circRNA-Based Vaccines
Challenges and Future Perspectives
Conclusions
Conflicts of Interest
References
- Walcott BP, Nahed B V., Mohyeldin A, Coumans JV, Kahle KT, Ferreira MJ. Chordoma: Current concepts, management, and future directions. Lancet Oncol 2012;13. [CrossRef]
- Pennington Z, Ehresman J, McCarthy EF, Ahmed AK, Pittman PD, Lubelski D, et al. Chordoma of the sacrum and mobile spine: a narrative review. Spine Journal 2021;21:500–17. [CrossRef]
- Salisbury JR. The pathology of the human notochord. J Pathol 1993;171:253–5. [CrossRef]
- Niu HQ, Zheng BY, Zou MX, Zheng BW. Complex immune microenvironment of chordoma: a road map for future treatment. J Immunother Cancer 2024;12. [CrossRef]
- Chen Y, Zhang H. Immune microenvironment and immunotherapy for chordoma. Front Oncol 2024;14. [CrossRef]
- Yang XR, Ng D, Alcorta DA, Liebsch NJ, Sheridan E, Li S, et al. T (brachyury) gene duplication confers major susceptibility to familial chordoma. Nat Genet 2009;41:1176–8. [CrossRef]
- Sun X, Hornicek F, Schwab JH. Chordoma: an update on the pathophysiology and molecular mechanisms. Curr Rev Musculoskelet Med 2015;8:344. [CrossRef]
- Bai J, Shi J, Zhang Y, Li C, Xiong Y, Koka H, et al. Gene Expression Profiling Identifies Two Chordoma Subtypes Associated with Distinct Molecular Mechanisms and Clinical Outcomes. Clin Cancer Res 2023;29:261–70. [CrossRef]
- Presneau, Shalaby A, Idowu B, Gikas P, Cannon SR, Gout I, et al. Potential therapeutic targets for chordoma: PI3K/AKT/TSC1/TSC2/mTOR pathway. Br J Cancer 2009;100:1406–14. [CrossRef]
- Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C, et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet 2009;41:535–43. [CrossRef]
- Yu X, Li Z. Epigenetic deregulations in chordoma. Cell Prolif 2015;48:497–502. [CrossRef]
- Conn VM, Chinnaiyan AM, Conn SJ. Circular RNA in cancer. Nat Rev Cancer 2024;24:597–613. [CrossRef]
- Zheng Y, Ren S, Zhang Y, Liu S, Meng L, Liu F, et al. Circular RNA circWWC3 augments breast cancer progression through promoting M2 macrophage polarization and tumor immune escape via regulating the expression and secretion of IL-4. Cancer Cell Int 2022;22. [CrossRef]
- Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675–91. [CrossRef]
- Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol 2022;19:188–206. [CrossRef]
- Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol 2019;12. [CrossRef]
- Ji X, Sun W, Lv C, Huang J, Zhang H. Circular RNAs Regulate Glucose Metabolism in Cancer Cells. Onco Targets Ther 2021;14:4005. [CrossRef]
- Li T, Xian HC, Dai L, Tang YL, Liang XH. Tip of the Iceberg: Roles of CircRNAs in Cancer Glycolysis. Onco Targets Ther 2021;14:2379. [CrossRef]
- Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer 2020;19:1–19. [CrossRef]
- Liu T, Long K, Zhu Z, Song Y, Chen C, Xu G, et al. Roles of circRNAs in regulating the tumor microenvironment. Med Oncol 2023;40:329. [CrossRef]
- Gonzalez C, Cimini M, Cheng Z, Benedict C, Wang C, Trungcao M, et al. Role of circular RNA cdr1as in modulation of macrophage phenotype. Life Sci 2022;309:121003. [CrossRef]
- He L, Tam PKH, Deng CX. Orchestration of Tumor-Associated Macrophages in the Tumor Cell-Macrophage-CD8+ T Cell Loop for Cancer Immunotherapy. Int J Biol Sci 2025;21:4098–116. [CrossRef]
- Carlos-Reyes Á, Romero-Garcia S, Contreras-Sanzón E, Ruiz V, Prado-Garcia H. Role of Circular RNAs in the Regulation of Immune Cells in Response to Cancer Therapies. Front Genet 2022;13:823238. [CrossRef]
- Jiang W, Pan S, Chen X, Wang Z wei, Zhu X. The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in cancer immunotherapy. Mol Cancer 2021;20:1–17. [CrossRef]
- Zhang W, Xu C, Yang Z, Zhou J, Peng W, Zhang X, et al. Circular RNAs in tumor immunity and immunotherapy. Mol Cancer 2024;23:1–15. [CrossRef]
- Geng Y, Wang M, Wu Z, Jia J, Yang T, Yu L. Research progress of circRNA in malignant tumour metabolic reprogramming. RNA Biol 2023;20:641. [CrossRef]
- Wang Y, Cui Y, Li X, Jin SH, Wang H, Gaipl US, et al. CircRNAs: functions and emerging roles in cancer and immunotherapy. BMC Med 2025;23:1–18. [CrossRef]
- Ghosh PK, Ghosh A. Dysregulation of noncoding RNA in chordoma; implications in identifying potential targets for novel therapeutic approaches. Mol Biol Rep 2024;51. [CrossRef]
- Li H, Tang Y, Ruan X, Zhang J, Liu H, Yu S, et al. N6-methyladenosine-modified circTEAD1 stabilizes Yap1 mRNA to promote chordoma tumorigenesis. Clin Transl Med 2024;14. [CrossRef]
- Niu HQ, Zheng BY, Zou MX, Zheng BW. Complex immune microenvironment of chordoma: a road map for future treatment. J Immunother Cancer 2024;12. [CrossRef]
- Mammar H, Polivka M, Belkacemi Y, Lot G, Froelich S, Carpentier A, et al. Hypoxia and Metabolism Regulation in Chordomas: Correlation Between Biology and Clinical Features for Potential Targeted Therapy. International Journal of Radiation Oncology*Biology*Physics 2015;93:E89–90. [CrossRef]
- Xu J, Shi Q, Wang B, Ji T, Guo W, Ren T, et al. The role of tumor immune microenvironment in chordoma: promising immunotherapy strategies. Front Immunol 2023;14:1257254. [CrossRef]
- Barber SM, Sadrameli SS, Lee JJ, Fridley JS, Teh BS, Oyelese AA, et al. Chordoma-Current Understanding and Modern Treatment Paradigms. J Clin Med 2021;10:1–18. [CrossRef]
- Jeoung J, Kim W, Jo H, Jeoung D. Circular RNAs as Targets for Developing Anticancer Therapeutics. Cells 2025, Vol 14, Page 1106 2025;14:1106. [CrossRef]
- Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, et al. CircRNAs in cancer metabolism: A review. J Hematol Oncol 2019;12:1–10. [CrossRef]
- He AT, Liu J, Li F, Yang BB. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther 2021;6:1–14. [CrossRef]
- Han C, Seebacher NA, Hornicek FJ, Kan Q, Duan Z, Han C, et al. Regulation of microRNAs function by circular RNAs in human cancer. Oncotarget 2017;8:64622–37. [CrossRef]
- Koch L. CRISPR–Cas13 targets circRNAs. Nat Rev Genet 2021;22:68. [CrossRef]
- He AT, Liu J, Li F, Yang BB. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduction and Targeted Therapy 2021 6:1 2021;6:185-. [CrossRef]
- Dawoud A, Elmasri RA, Mohamed AH, Mahmoud A, Rostom MM, Youness RA. Involvement of CircRNAs in regulating The “New Generation of Cancer Hallmarks”: A Special Depiction on Hepatocellular Carcinoma. Crit Rev Oncol Hematol 2024;196:104312. [CrossRef]
- Gong Z, Hu W, Zhou C, Guo J, Yang L, Wang B. Recent advances and perspectives on the development of circular RNA cancer vaccines. NPJ Vaccines 2025;10. [CrossRef]
- Zhao Y, Wang H. Artificial intelligence-driven circRNA vaccine development: multimodal collaborative optimization and a new paradigm for biomedical applications. Brief Bioinform 2025;26. [CrossRef]
- Jiang W, Pan S, Chen X, Wang Z wei, Zhu X. The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in cancer immunotherapy. Molecular Cancer 2021 20:1 2021;20:116-. [CrossRef]
- Allegra A, Cicero N, Tonacci A, Musolino C, Gangemi S. Circular RNA as a Novel Biomarker for Diagnosis and Prognosis and Potential Therapeutic Targets in Multiple Myeloma. Cancers (Basel) 2022;14. [CrossRef]
- Piras R, Ko EY, Barrett C, De Simone M, Lin X, Broz MT, et al. circCsnk1g3- and circAnkib1-regulated interferon responses in sarcoma promote tumorigenesis by shaping the immune microenvironment. Nature Communications 2022 13:1 2022;13:7243-. [CrossRef]
- Wei CY, Zhu MX, Lu NH, Liu JQ, Yang YW, Zhang Y, et al. Circular RNA circ_0020710 drives tumor progression and immune evasion by regulating the miR-370-3p/CXCL12 axis in melanoma. Mol Cancer 2020;19. [CrossRef]
- Dang Q, Li B, Jin B, Ye Z, Lou X, Wang T, et al. Cancer immunometabolism: advent, challenges, and perspective. Mol Cancer 2024;23:1–22. [CrossRef]
- Das U, Banerjee S, Sarkar M, Muhammad L F, Soni TK, Saha M, et al. Circular RNA vaccines: Pioneering the next-gen cancer immunotherapy. Cancer Pathogenesis and Therapy 2025;3:309–21. [CrossRef]
| Pathway | Role in Chordoma | Associated CircRNAs | Therapeutic Implications |
|---|---|---|---|
| PI3K/AKT/mTOR | Promotes cell growth, proliferation, and survival; involved in metabolic reprogramming | circRNAs modulating PI3K/AKT/mTOR pathway | Targeting circRNAs to inhibit pathway activity and reduce tumor growth |
| NRF2 | Regulates antioxidant response and metabolic adaptation | circRNAs modulating NRF2 pathway | Targeting circRNAs to enhance oxidative stress and improve therapeutic response |
| Glycolysis (Warburg Effect) | Provides rapid ATP production and metabolic intermediates for biosynthesis | circRNAs regulating glycolytic enzymes | Targeting circRNAs to reverse metabolic reprogramming and sensitize to therapy |
| Immune Checkpoint Signaling | Contributes to immune evasion by suppressing anti-tumor immune responses | circRNAs modulating immune checkpoint molecules (e.g., PD-1, CTLA-4) | Targeting circRNAs to enhance anti-tumor immunity and improve immunotherapy efficacy |
| Strategy | Mechanism | Examples/Approach | Potential Benefits | Kaynak |
|---|---|---|---|---|
| Inhibition of oncogenic circRNAs | Degradation or blocking of circRNA function | Antisense oligonucleotides (ASOs), siRNAs, CRISPR-based technologies | Suppress tumor growth, reduce proliferation, overcome drug resistance | [34,38,39,40] |
| Restoration of tumor-suppressive circRNAs | Re-expression or upregulation of circRNA | Gene therapy, small molecules | Inhibit tumor growth, promote apoptosis, enhance chemosensitivity | [41,42] |
| Indirect targeting via immunometabolism | Modulating circRNAs that influence metabolic pathways or immune checkpoints | Targeting circRNAs regulating glycolytic enzymes, macrophage polarization, immune checkpoint molecules | Reverse metabolic reprogramming, enhance anti-tumor immunity, improve immunotherapy efficacy | [43,44,45,46] |
| Combination therapies | Combining circRNA-targeted therapies with other treatments | CircRNA inhibition + metabolic inhibitors, CircRNA inhibition + immune checkpoint blockade | Synergistic anti-tumor effects, overcome resistance mechanisms | [47,48] |
| Paper Title | Year | Key Focus | Main Findings | Relevance to circRNA Review |
|---|---|---|---|---|
| Chordoma: an update on pathophysiology and molecular mechanisms | 2015 | Pathophysiology, molecular mechanisms | - Brachyury as a key driver- Role of chromosomal alterations, DNA methylation, and microRNAs | Foundational understanding of chordoma molecular biology, including non-coding RNAs. |
| Gene Expression Profiling Identifies Two Chordoma Subtypes Associated with Distinct Molecular Mechanisms and Clinical Outcomes | 2023 | Molecular subtyping, clinical outcomes | - Identified two subtypes with distinct molecular mechanisms (chromatin remodeling vs. EMT/Hedgehog pathways)- Subtypes correlate with clinical outcomes | Highlights molecular heterogeneity and the importance of RNA-based classification. |
| N6-methyladenosine-modified circTEAD1 stabilizes Yap1 mRNA to promote chordoma tumorigenesis | 2024 | circRNA function in chordoma | - First study on circRNAs in chordoma- circTEAD1 is upregulated and promotes tumorigenesis- m6A modification is crucial for circTEAD1 function- circTEAD1 stabilizes Yap1 mRNA via an RNA-protein complex | Core paper for the review, providing direct evidence of circRNA involvement in chordoma. |
| Immune microenvironment and immunotherapy for chordoma | 2024 | Immune microenvironment, immunotherapy | - Chordoma TME is characterized by high infiltration of M2 macrophages and regulatory T cells- Reviews current immunotherapy trials and their limitations | Provides the immunological context for the review, highlighting the immunosuppressive microenvironment that circRNAs may influence. |
| Proteogenomic characterization of skull-base chordoma | 2024 | Proteogenomics, chromosome instability | - Chromosome instability is a key prognostic factor- Identified immune cold subtype linked to chromosome 9p/10q loss- Proteomics-based classification reveals subtypes with high CIN and immune cold features | Reinforces the concept of molecular subtypes and links genomic instability to the immune landscape. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




