Submitted:
26 June 2023
Posted:
27 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and methods
2.1. Data and clinical sample collection
2.2. Identification of OS cells and cell types in the TME
2.3. Pseudotime trajectory analysis of disulfidptosis-related genes in TME cells
2.4. Non-negative matrix factorization of disulfidptosis-related genes in TME Cells
2.5. Identification of marker genes of disulfidptosis-related cell subtypes in TME cells
2.6. Cell-cell communication analysis of disulfidptosis-related cell subtypes
2.7. SCENIC analysis for disulfidptosis-related subtypes
2.8. Survival analysis of disulfidptosis-related signatures in public bulk RNA sequence database
2.9. Analysis of immunotherapy in disulfidptosis-related clusters
2.10. Immunofluorescent staining
2.11. Statistical analysis
3. Results
3.1. Landscape of disulfidptosis-related genes in the tumor microenvironment (TME) of osteosarcoma
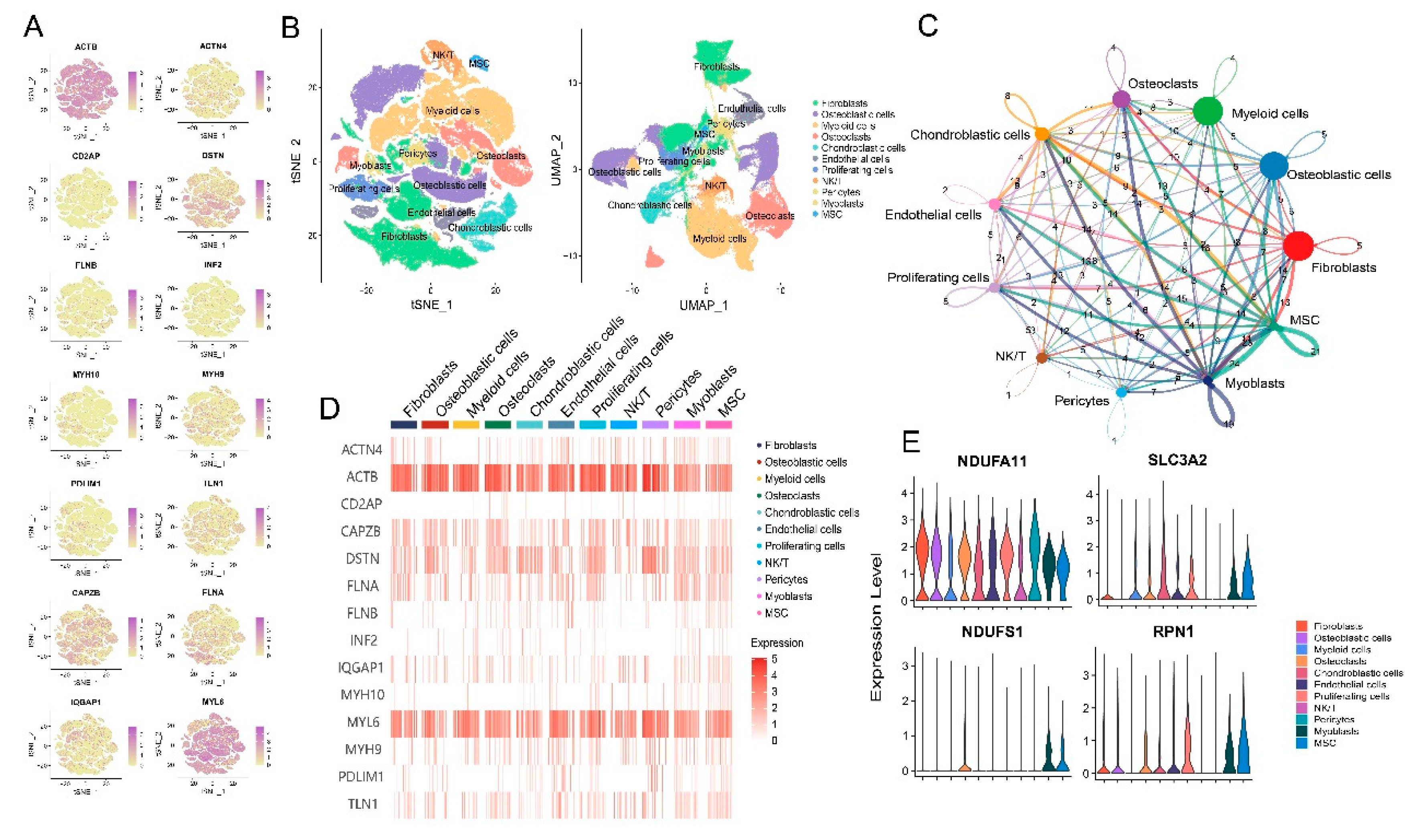
3.2. Disulfidptosis-driven fibroblasts shape the TME of osteosarcoma
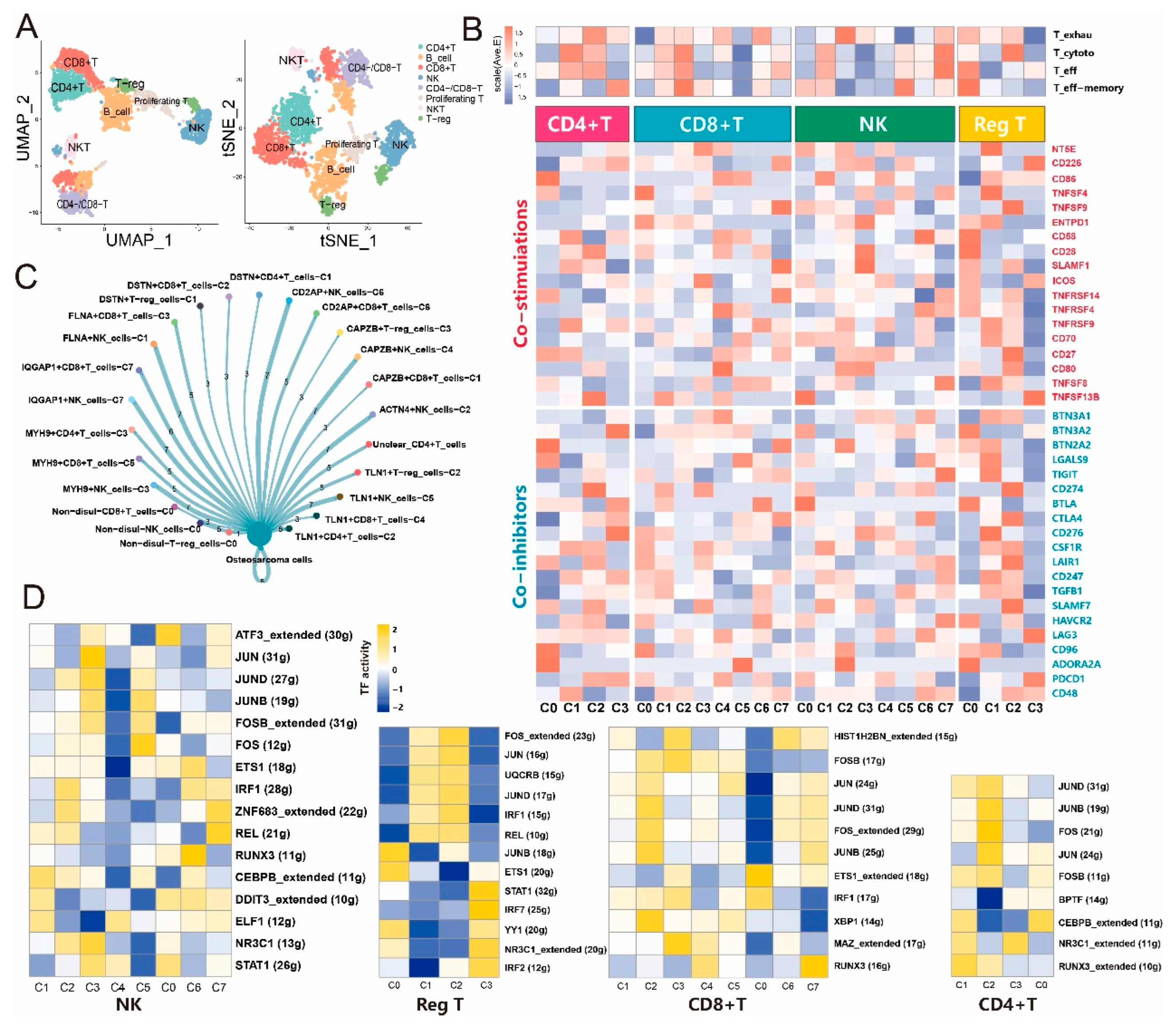
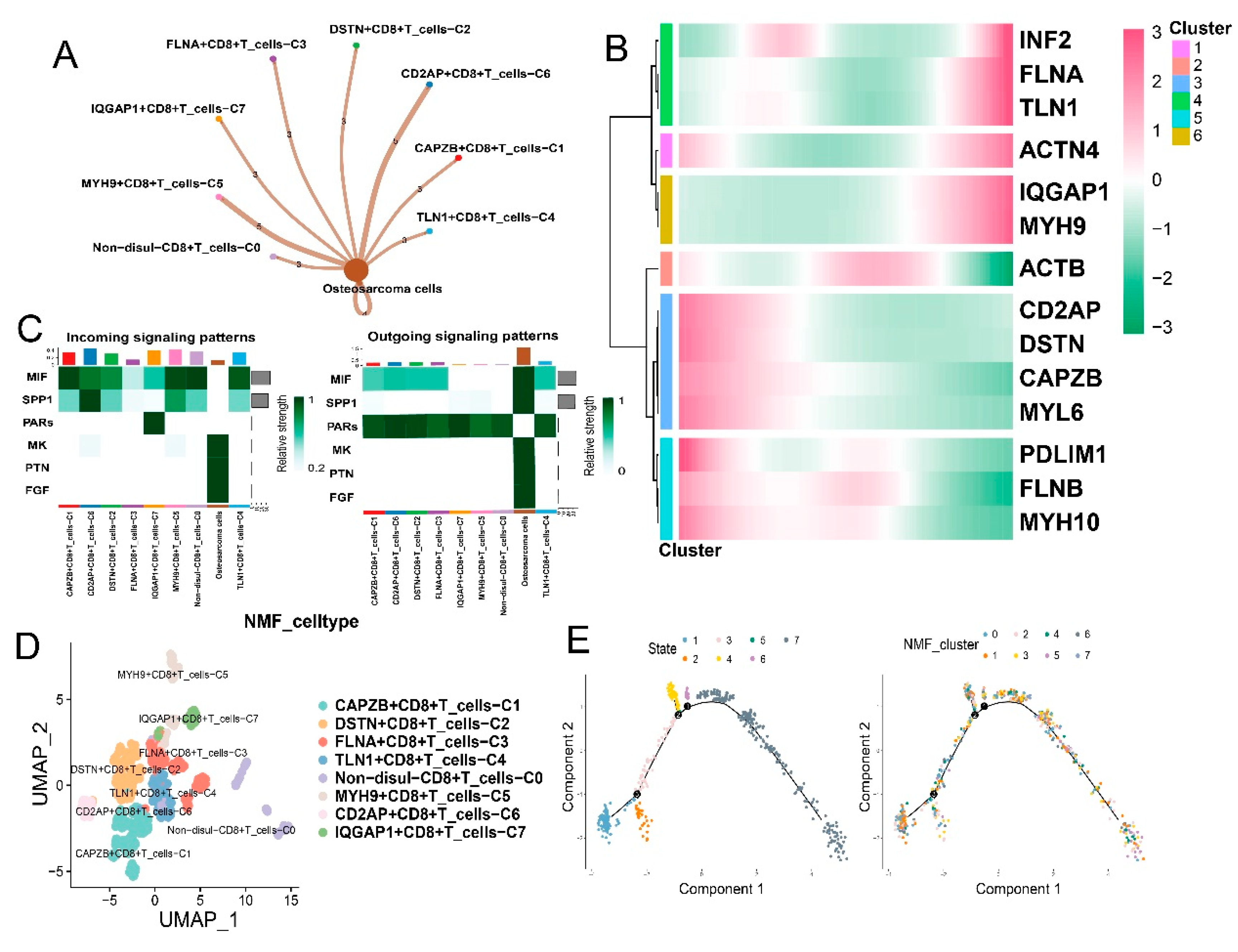
3.3. Disulfidptosis-driven subtypes of tumor-infiltrating lymphocytes (TILs) impact the immune response in osteosarcoma
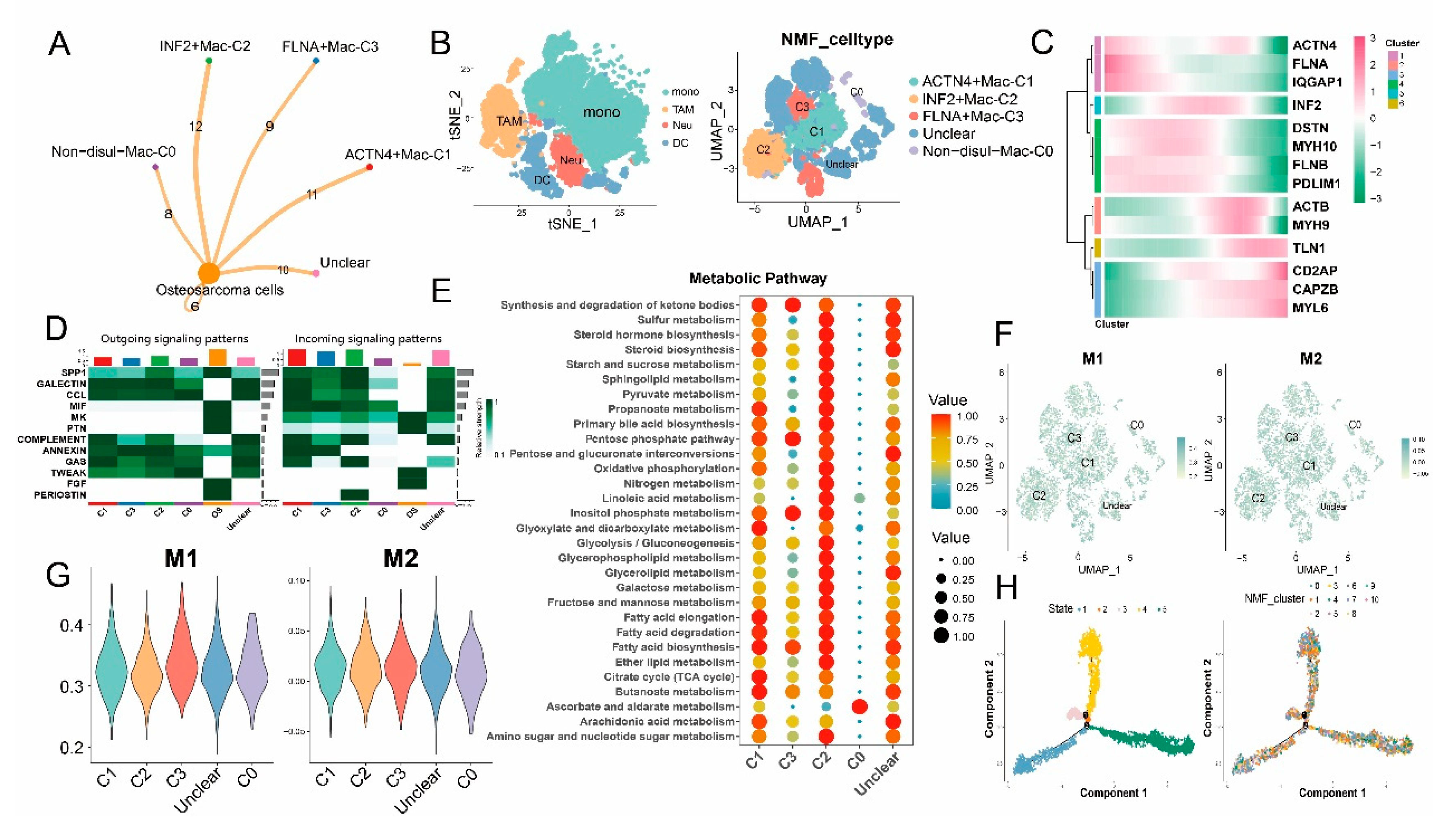
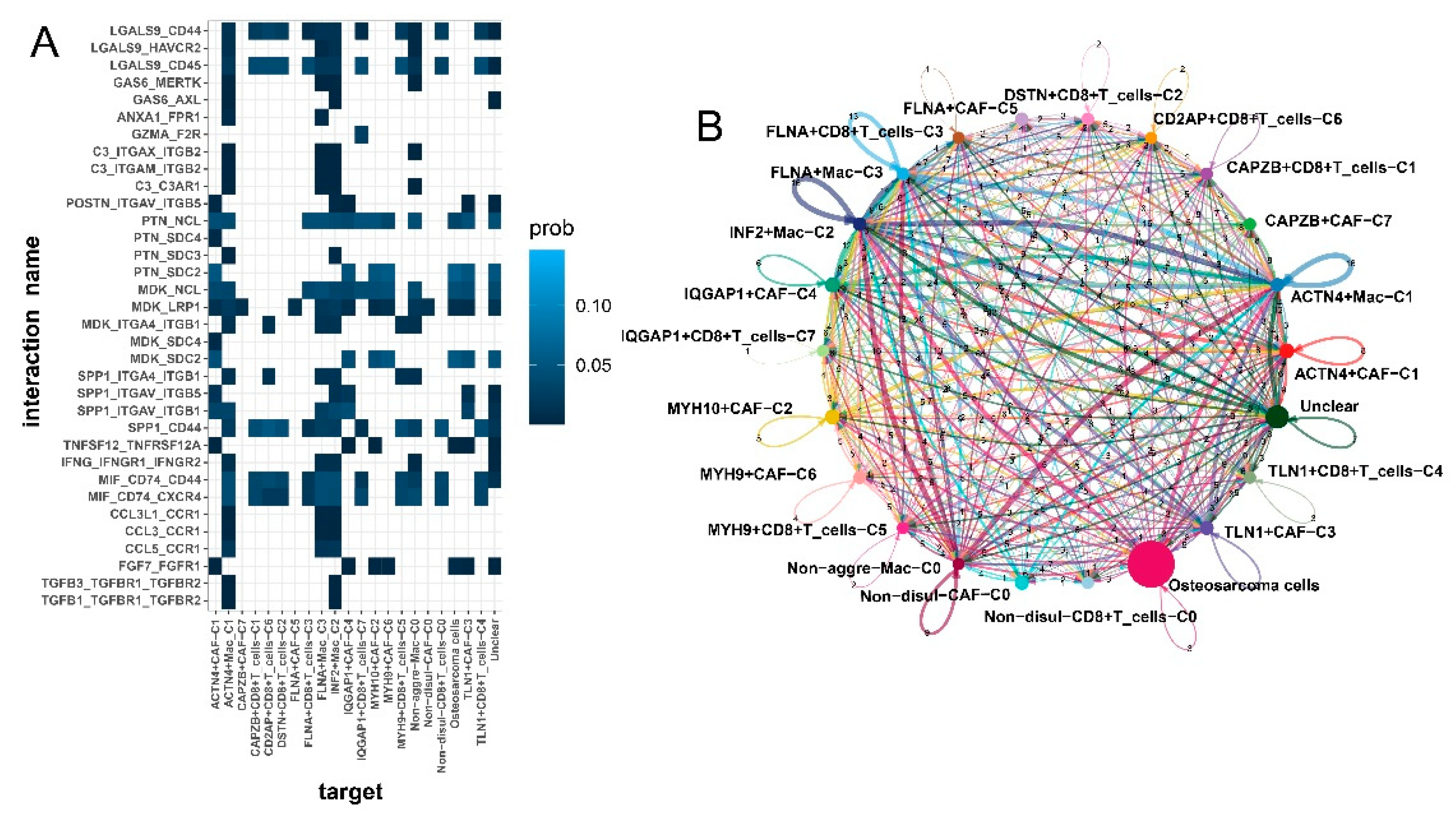
3.4. Disulfidptosis-driven macrophages: Impact on cellular metabolism and intercellular communication
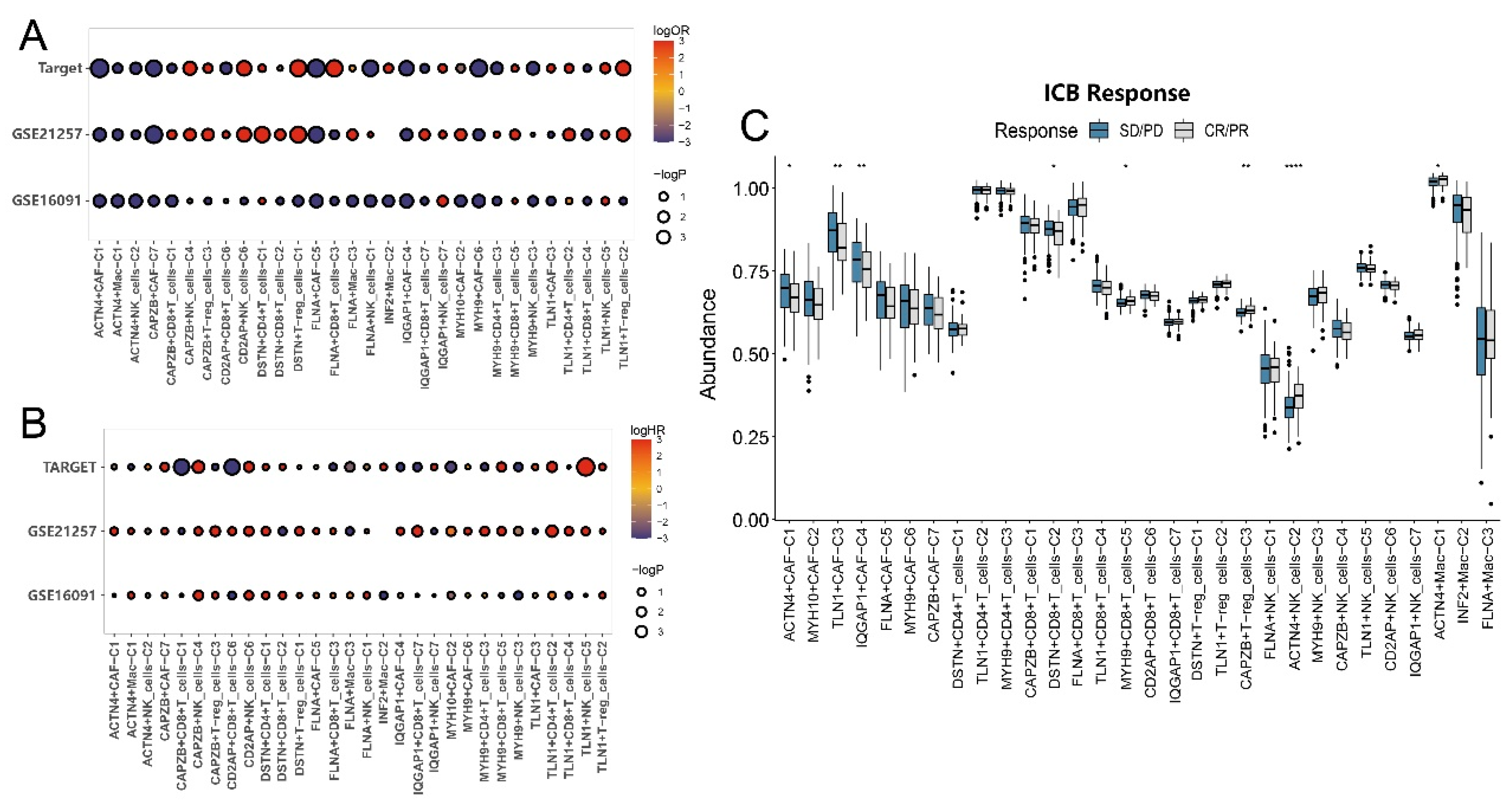
3.5. Impact of disulfidptosis-driven TME on osteosarcoma prognosis
3.6. Impact of disulfidptosis-driven TME on immunotherapy in osteosarcoma
3.7. Enhanced intercellular communication in disulfidptosis-mediated TME patterns
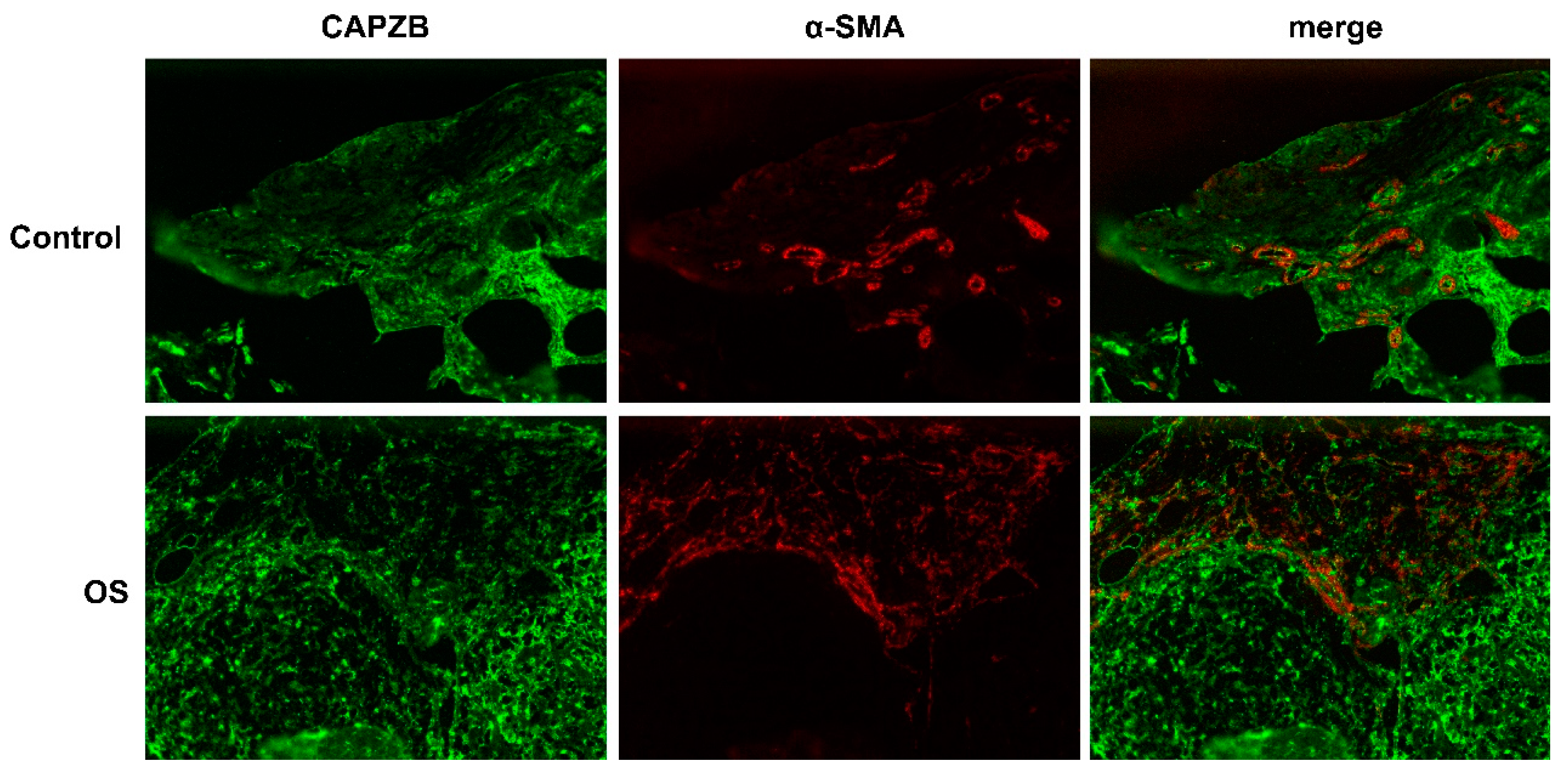
3.8. Histological localization of CAPZB-CAF-C7 in osteosarcoma and osteoblastoma
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Competing interests
Ethics statement
References
- Mirabello, L.; Troisi, R.J.; Savage, S.A. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer 2009, 115, 1531–43. [Google Scholar] [CrossRef] [PubMed]
- Beird, H.C.; et al. Osteosarcoma. Nat Rev Dis Primers 2022, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Peng, F.; et al. Regulated cell death (RCD) in cancer: key pathways and targeted therapies. Signal Transduct Target Ther 2022, 7, 286. [Google Scholar] [CrossRef]
- Tang, D.; et al. Ferroptosis: molecular mechanisms and health implications. Cell Res 2021, 31, 107–125. [Google Scholar] [CrossRef]
- Shin, C.S.; et al. The glutamate/cystine xCT antiporter antagonizes glutamine metabolism and reduces nutrient flexibility. Nat Commun 2017, 8, 15074. [Google Scholar] [CrossRef] [PubMed]
- Koppula, P.; et al. The glutamate/cystine antiporter SLC7A11/xCT enhances cancer cell dependency on glucose by exporting glutamate. J Biol Chem 2017, 292, 14240–14249. [Google Scholar] [CrossRef]
- Goji, T.; et al. Cystine uptake through the cystine/glutamate antiporter xCT triggers glioblastoma cell death under glucose deprivation. J Biol Chem 2017, 292, 19721–19732. [Google Scholar] [CrossRef]
- Liu, X.; et al. Cystine transporter regulation of pentose phosphate pathway dependency and disulfide stress exposes a targetable metabolic vulnerability in cancer. Nat Cell Biol 2020, 22, 476–486. [Google Scholar] [CrossRef]
- Liu, X.; et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat Cell Biol 2023, 25, 404–414. [Google Scholar] [CrossRef]
- Xiao, Y.; Yu, D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol Ther 2021, 221, 107753. [Google Scholar] [CrossRef] [PubMed]
- AlMusawi, S.; Ahmed, M.; Nateri, A.S. Understanding cell-cell communication and signaling in the colorectal cancer microenvironment. Clin Transl Med 2021, 11, e308. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; et al. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat Commun 2020, 11, 6322. [Google Scholar] [CrossRef]
- Stine, Z.E.; et al. Targeting cancer metabolism in the era of precision oncology. Nat Rev Drug Discov 2022, 21, 141–162. [Google Scholar] [CrossRef]
- Machesky, L.M. Deadly actin collapse by disulfidptosis. Nat Cell Biol 2023, 25, 375–376. [Google Scholar] [CrossRef] [PubMed]
- Satija, R.; et al. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 2015, 33, 495–502. [Google Scholar] [CrossRef]
- Cao, J.; et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019, 566, 496–502. [Google Scholar] [CrossRef]
- Wu, P.; et al. A robust semi-supervised NMF model for single cell RNA-seq data. PeerJ 2020, 8, e10091. [Google Scholar] [CrossRef]
- Jin, S.; et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun 2021, 12, 1088. [Google Scholar] [CrossRef]
- Aibar, S.; et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017, 14, 1083–1086. [Google Scholar] [CrossRef]
- Van de Sande, B.; et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc 2020, 15, 2247–2276. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, S., R. Castelo, and J. Guinney, GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013, 14, 7. [Google Scholar] [CrossRef]
- Jiang, P.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Necchi, A.; et al. Atezolizumab in platinum-treated locally advanced or metastatic urothelial carcinoma: post-progression outcomes from the phase II IMvigor210 study. Ann Oncol 2017, 28, 3044–3050. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; et al. Single-cell characteristics and malignancy regulation of alpha-fetoprotein-producing gastric cancer. Cancer Med 2023, 12, 12018–12033. [Google Scholar] [CrossRef]
- Galbo, P.M., Jr.; Zang, X.; Zheng, D. Molecular Features of Cancer-associated Fibroblast Subtypes and their Implication on Cancer Pathogenesis, Prognosis, and Immunotherapy Resistance. Clin Cancer Res 2021, 27, 2636–2647. [Google Scholar] [CrossRef] [PubMed]
- Koren, E.; Fuchs, Y. Modes of Regulated Cell Death in Cancer. Cancer Discov 2021, 11, 245–265. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; et al. The molecular machinery of regulated cell death. Cell Res 2019, 29, 347–364. [Google Scholar] [CrossRef]
- Gao, W.; et al. Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor immunotherapy. Signal Transduct Target Ther 2022, 7, 196. [Google Scholar] [CrossRef]
- Tong, X.; et al. Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research. J Hematol Oncol 2022, 15, 174. [Google Scholar] [CrossRef]
- Dahlin, D.C.; Unni, K.K. Osteosarcoma of bone and its important recognizable varieties. Am J Surg Pathol 1977, 1, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; et al. Subtype Classification and Prognosis Signature Construction of Osteosarcoma Based on Cellular Senescence-Related Genes. J Oncol 2022, 2022, 4421952. [Google Scholar] [CrossRef] [PubMed]
- Ligon, J.A.; et al. Pathways of immune exclusion in metastatic osteosarcoma are associated with inferior patient outcomes. J Immunother Cancer 2021, 9. [Google Scholar] [CrossRef]
- Sundara, Y.T.; et al. Increased PD-L1 and T-cell infiltration in the presence of HLA class I expression in metastatic high-grade osteosarcoma: a rationale for T-cell-based immunotherapy. Cancer Immunol Immunother 2017, 66, 119–128. [Google Scholar] [CrossRef]
- Han, Q.; Shi, H.; Liu, F. CD163(+) M2-type tumor-associated macrophage support the suppression of tumor-infiltrating T cells in osteosarcoma. Int Immunopharmacol 2016, 34, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; et al. Managing the immune microenvironment of osteosarcoma: the outlook for osteosarcoma treatment. Bone Res 2023, 11, 11. [Google Scholar] [CrossRef]
- Chen, Y.; McAndrews, K.M.; Kalluri, R. Clinical and therapeutic relevance of cancer-associated fibroblasts. Nat Rev Clin Oncol 2021, 18, 792–804. [Google Scholar] [CrossRef]
- Zhou, B.B.; et al. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov 2009, 8, 806–23. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat Rev Cancer 2016, 16, 582–98. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013, 19, 1423–37. [Google Scholar] [CrossRef]
- Su, S.; et al. CD10(+)GPR77(+) Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell 2018, 172, 841–856.e16. [Google Scholar] [CrossRef] [PubMed]
- Sjöberg, E.; et al. A Novel ACKR2-Dependent Role of Fibroblast-Derived CXCL14 in Epithelial-to-Mesenchymal Transition and Metastasis of Breast Cancer. Clin Cancer Res 2019, 25, 3702–3717. [Google Scholar] [CrossRef]
- Hornburg, M.; et al. Single-cell dissection of cellular components and interactions shaping the tumor immune phenotypes in ovarian cancer. Cancer Cell 2021, 39, 928–944.e6. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.; et al. Manipulation of Transgene Expression in Fibroblast Cells by a Multifunctional Linear-Branched Hybrid Poly(β-Amino Ester) Synthesized through an Oligomer Combination Approach. Nano Lett 2019, 19, 381–391. [Google Scholar] [CrossRef]
- Wu, H.W.; et al. The Effects of Programmed Cell Death of Mesenchymal Stem Cells on the Development of Liver Fibrosis. Stem Cells Int 2023, 2023, 4586398. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R.; et al. Immunogenic and tolerogenic cell death. Nat Rev Immunol 2009, 9, 353–63. [Google Scholar] [CrossRef]
- Horton, B.L.; et al. Intratumoral CD8(+) T-cell Apoptosis Is a Major Component of T-cell Dysfunction and Impedes Antitumor Immunity. Cancer Immunol Res 2018, 6, 14–24. [Google Scholar] [CrossRef]
- Jarosz-Biej, M.; et al. Tumor Microenvironment as A "Game Changer" in Cancer Radiotherapy. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef]
- Jung, J.; Zeng, H.; Horng, T. , Metabolism as a guiding force for immunity. Nat Cell Biol 2019, 21, 85–93. [Google Scholar] [CrossRef]
- Lei, S.; et al. JUND/linc00976 promotes cholangiocarcinoma progression and metastasis, inhibits ferroptosis by regulating the miR-3202/GPX4 axis. Cell Death Dis 2022, 13, 967. [Google Scholar] [CrossRef]
- Liu, Z.; et al. PCDH7 knockdown potentiates colon cancer cells to chemotherapy via inducing ferroptosis and changes in autophagy through restraining MEK1/2/ERK/c-Fos axis. Biochem Cell Biol 2022, 100, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; et al. IRF1/ZNF350/GPX4-mediated ferroptosis of renal tubular epithelial cells promote chronic renal allograft interstitial fibrosis. Free Radic Biol Med 2022, 193 Pt 2, 579–594. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



