Submitted:
10 December 2025
Posted:
12 December 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
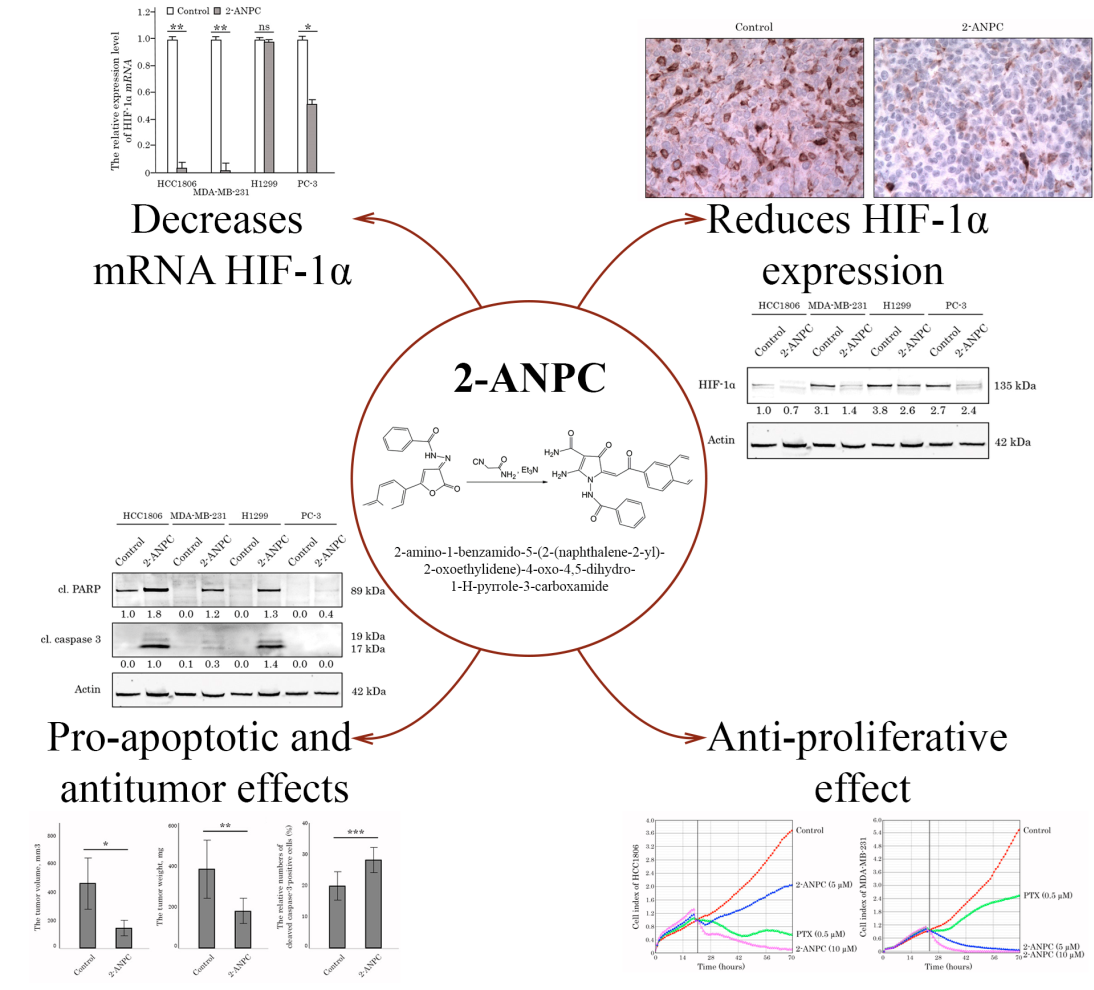
2.1. Chemistry
2.2. Cell Lines and Culture Conditions
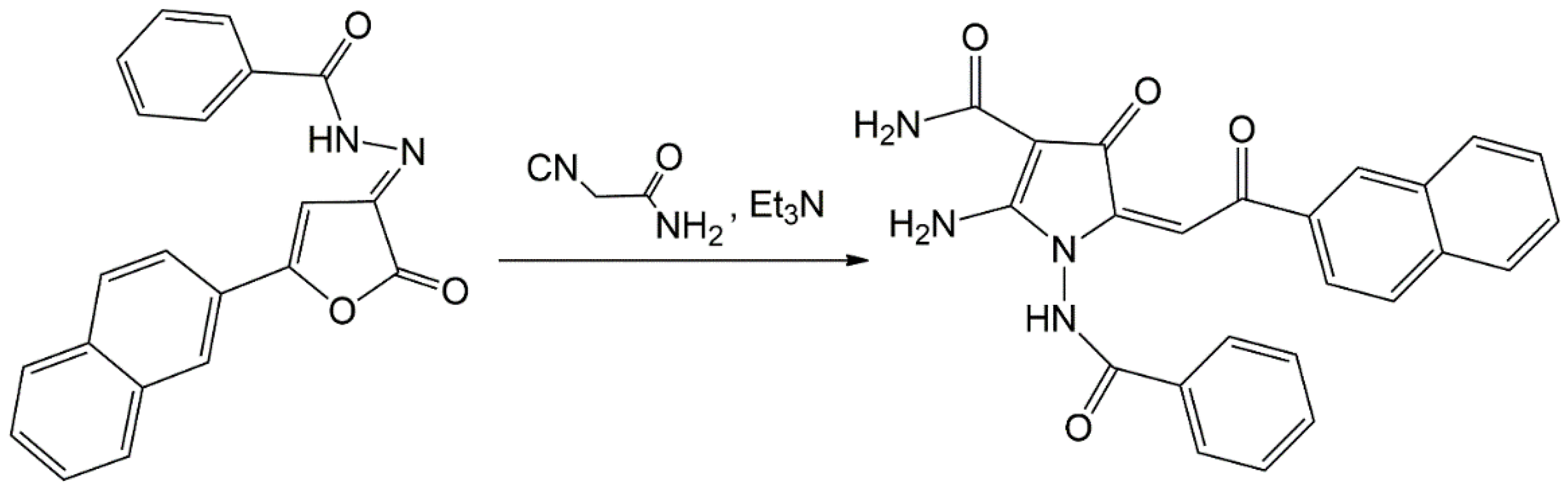
2.3. Real-Time Monitoring of Cell Proliferation
2.4. Antibodies
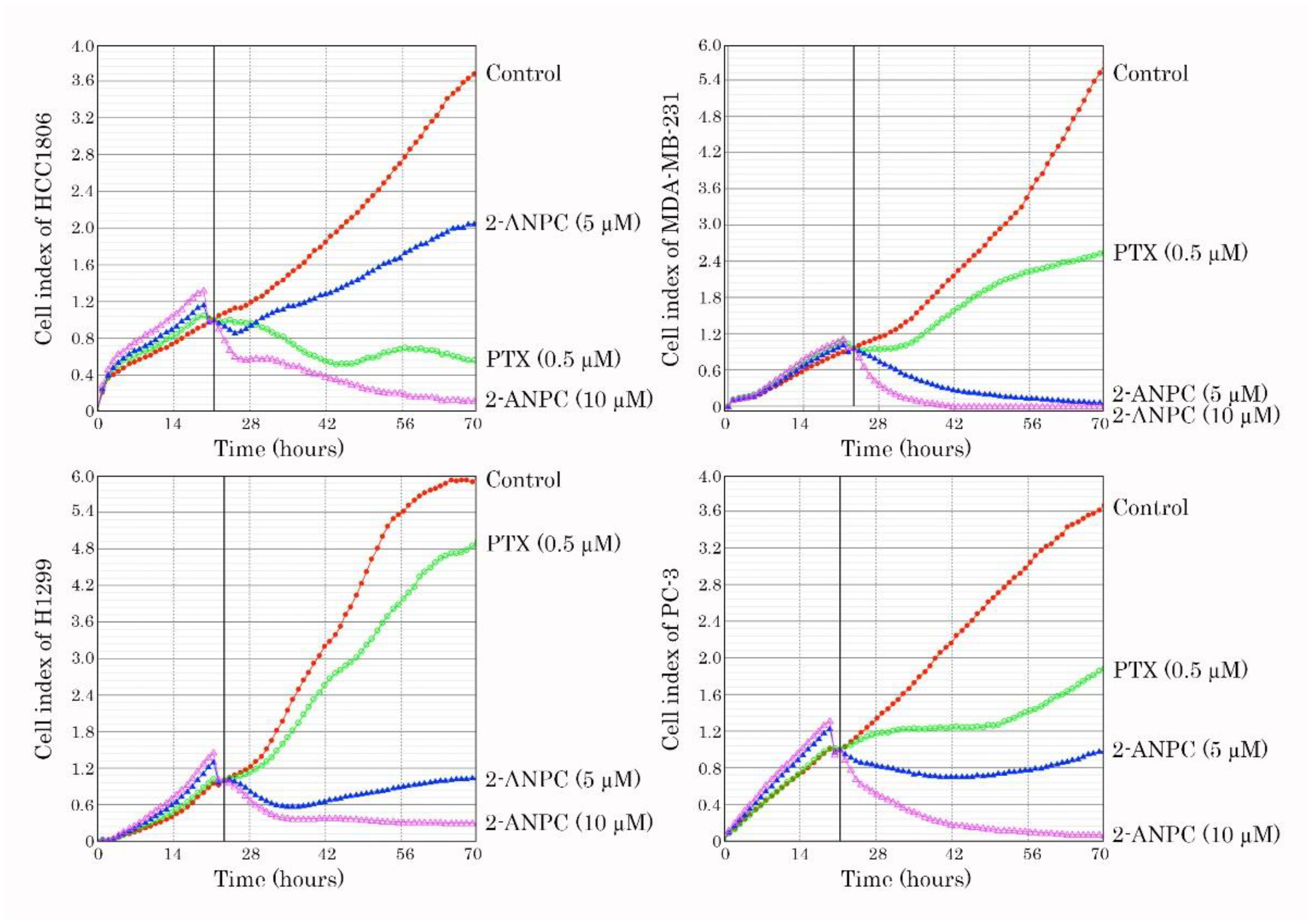
2.5. Western Blotting
2.6. RNA Extraction and RT-PCR
2.7. Study of Antitumor Activity
2.8. Statistics
2.9. HIF-1α Modelling
2.10. HIF-1 Protein Complex Modelling
2.11. Site Mapping and Docking
3. Results
3.1. 2-ANPC Exhibits Potent Anti-Proliferative and Pro-Apoptotic Activities Against Breast, Lung, and Prostate Cancer Cell Lines
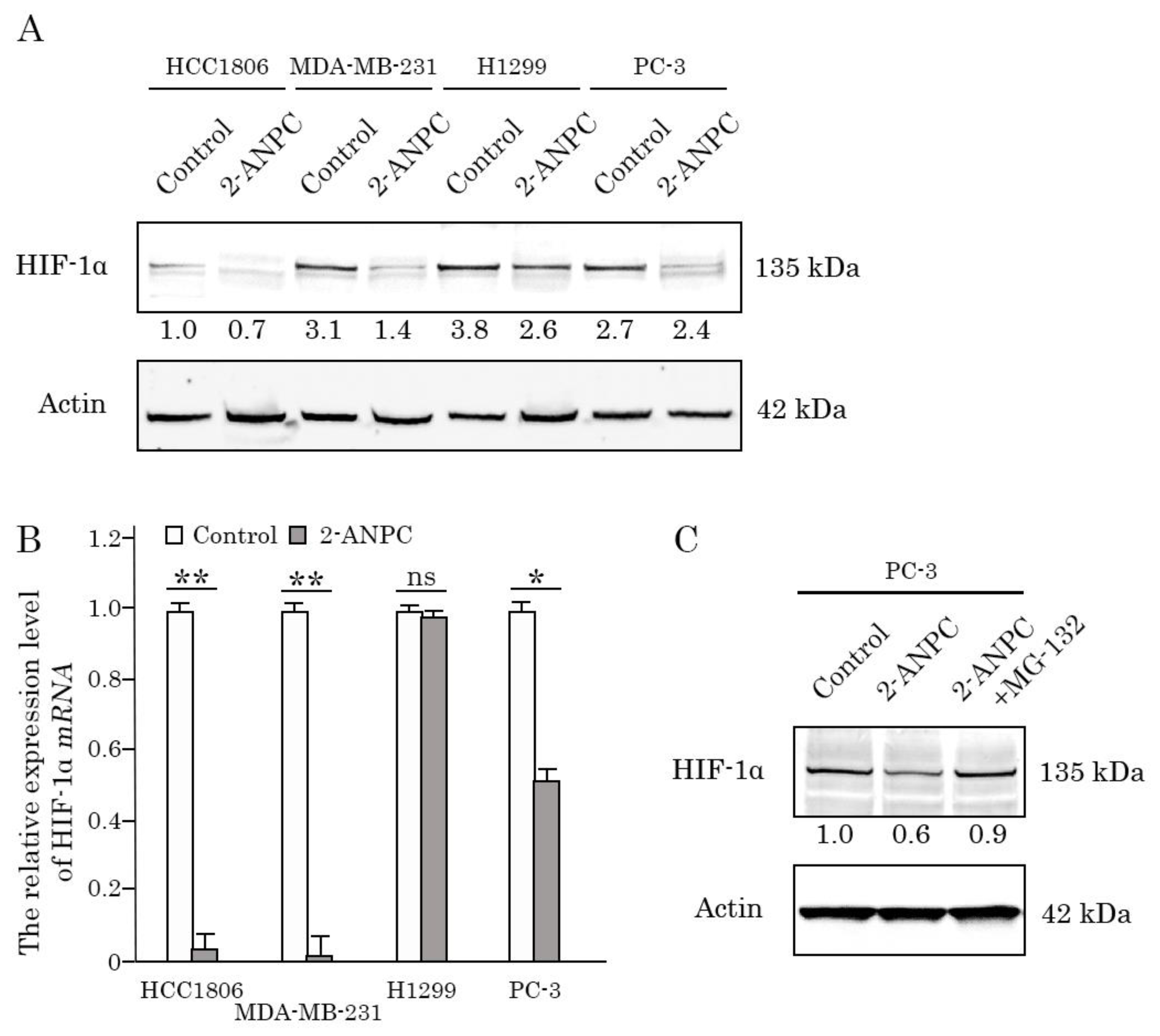
3.2. 2-ANPC Effectively Decreases HIF-1α Expression In Vitro in Epithelial Cancer Cells by Promoting Its Proteasome-Dependent Degradation
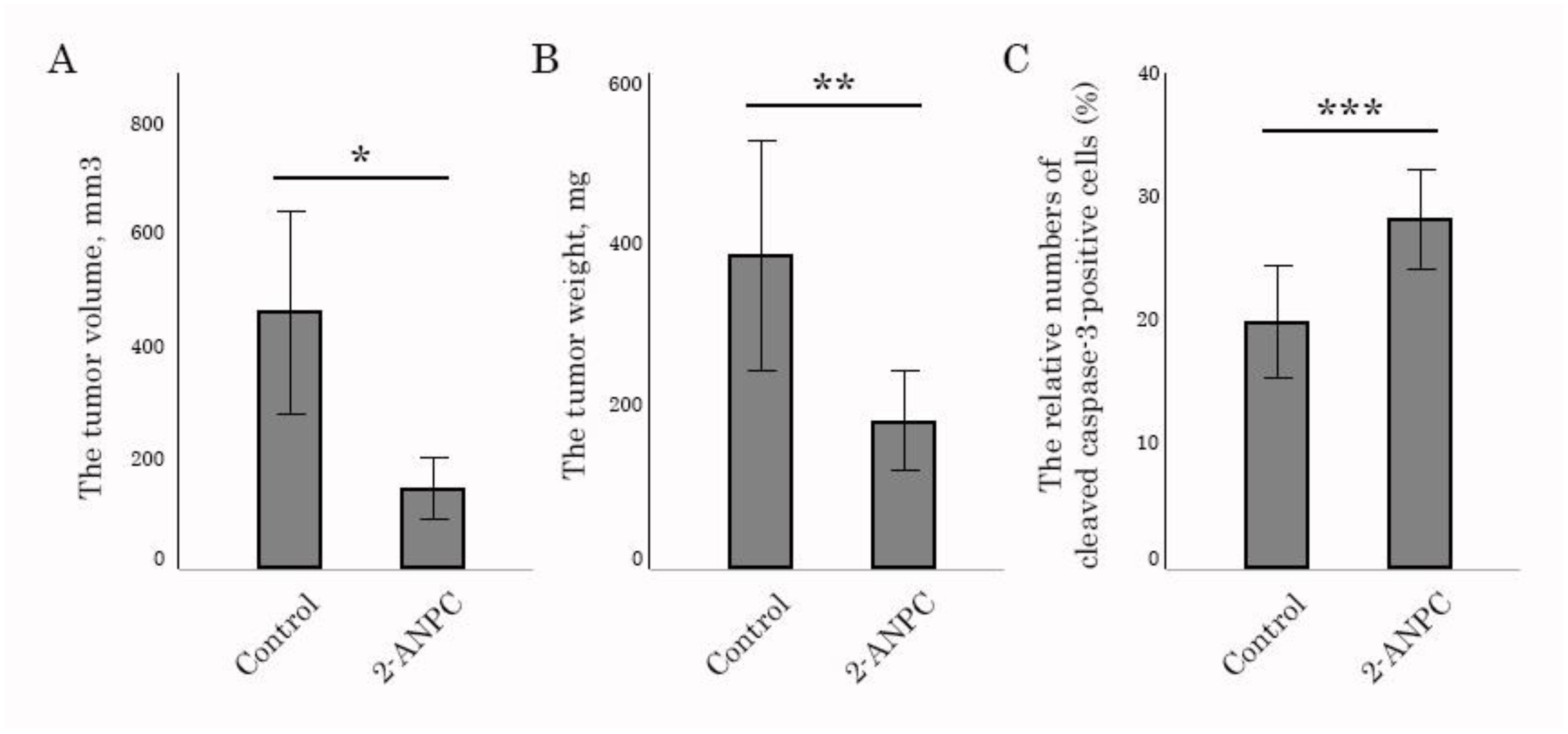
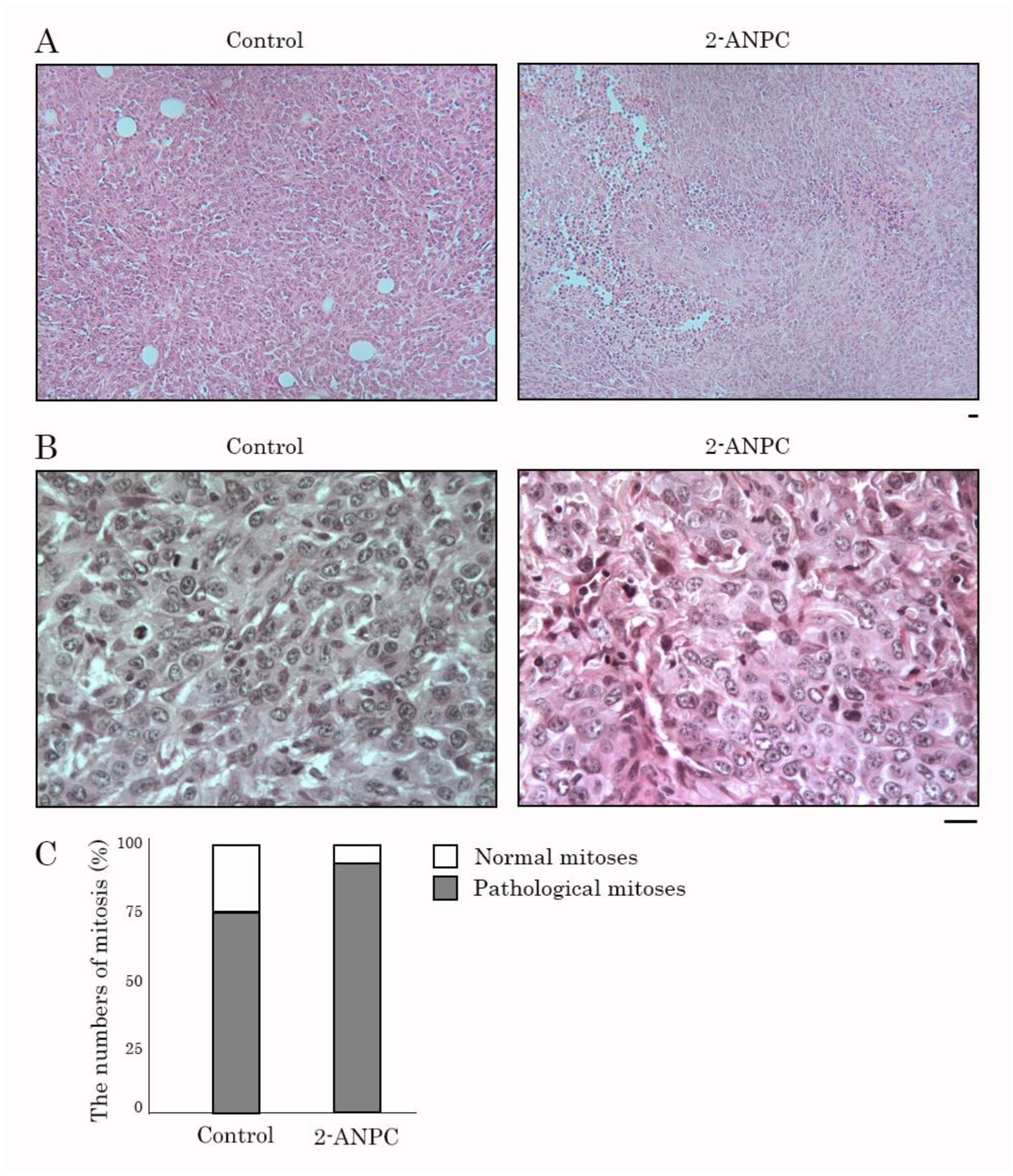
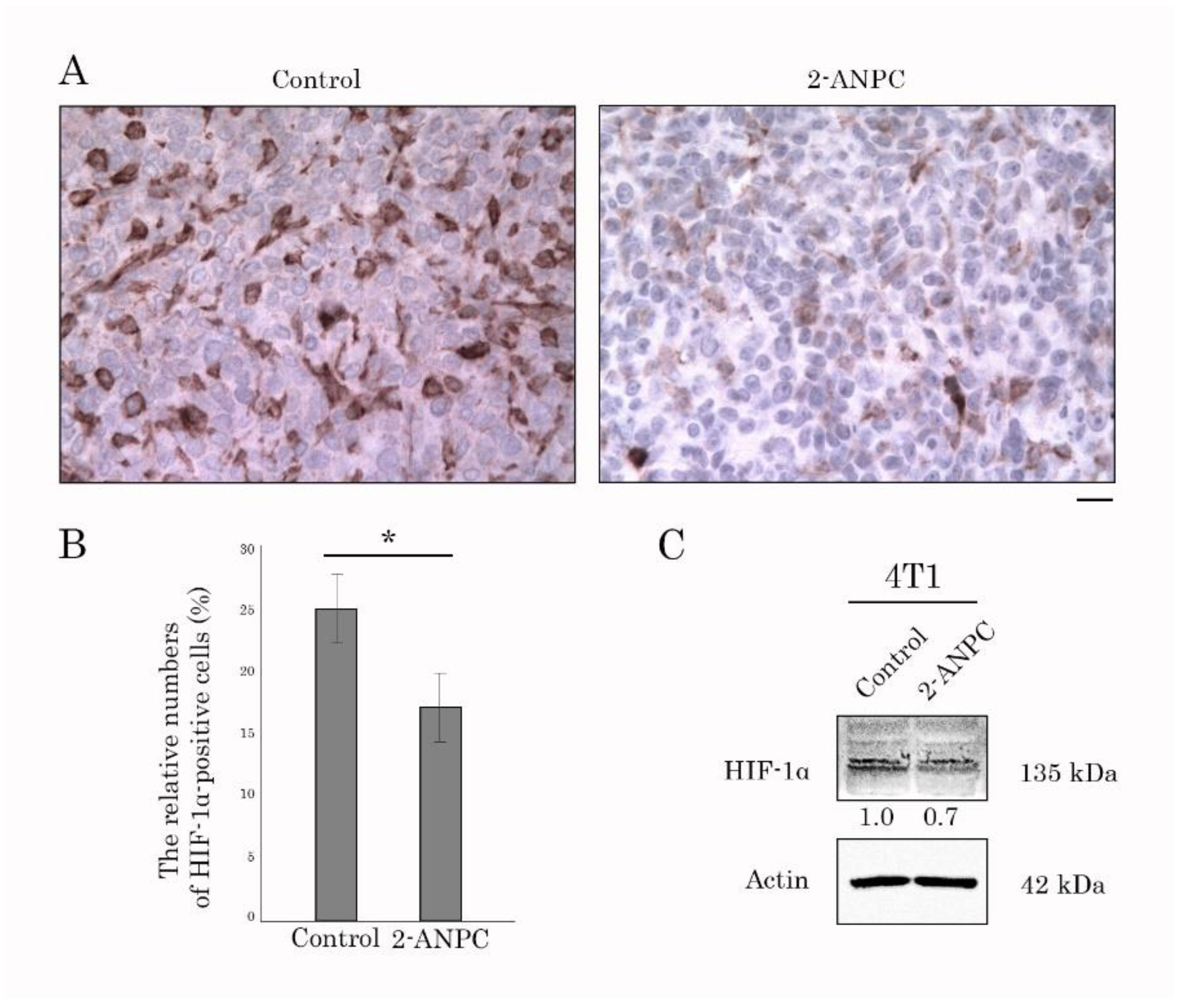
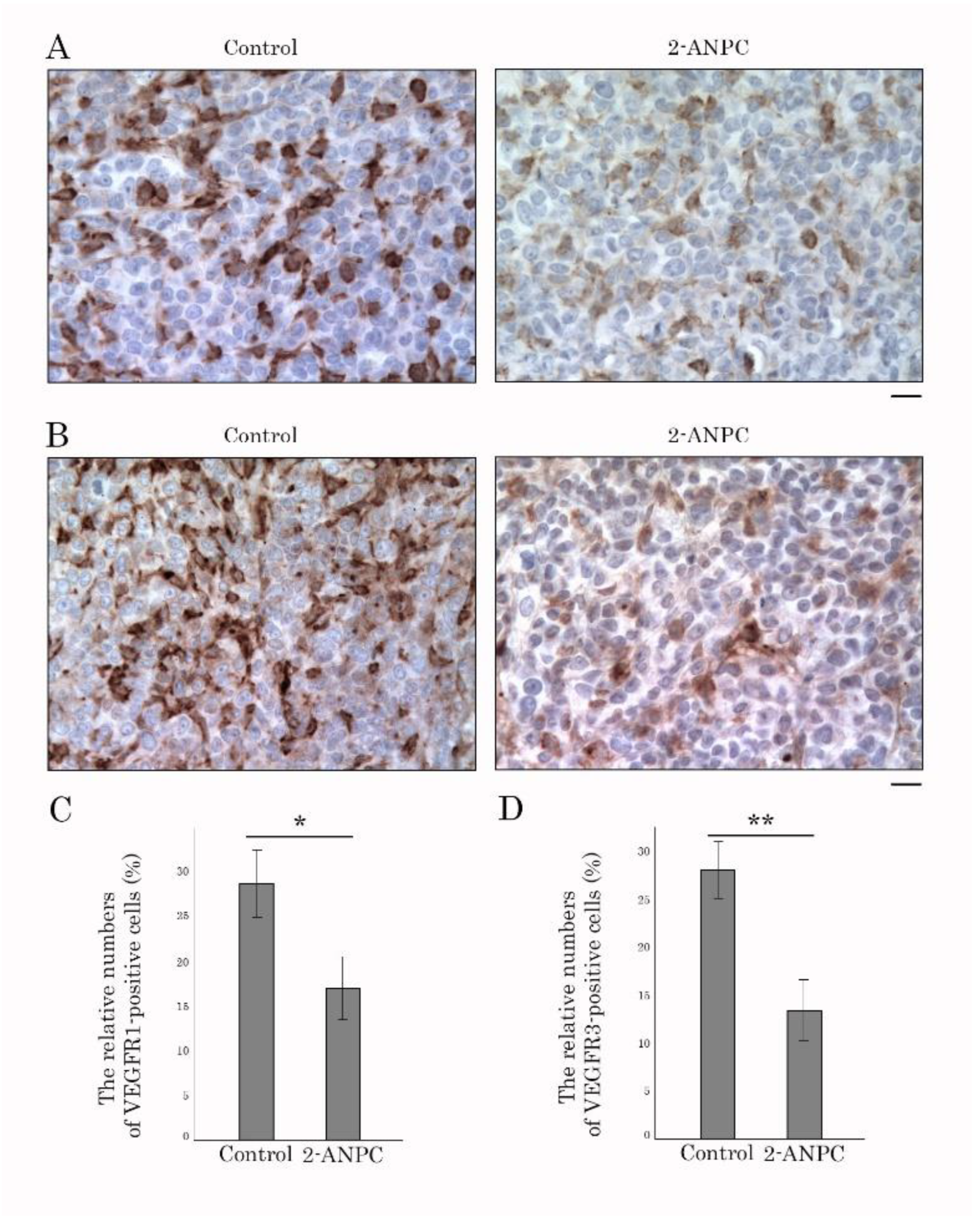
3.3. 2-ANPC Inhibits Tumor Growth and Decreases HIF-1α, VEGFR1 and VEGFR3 Expression In Vivo
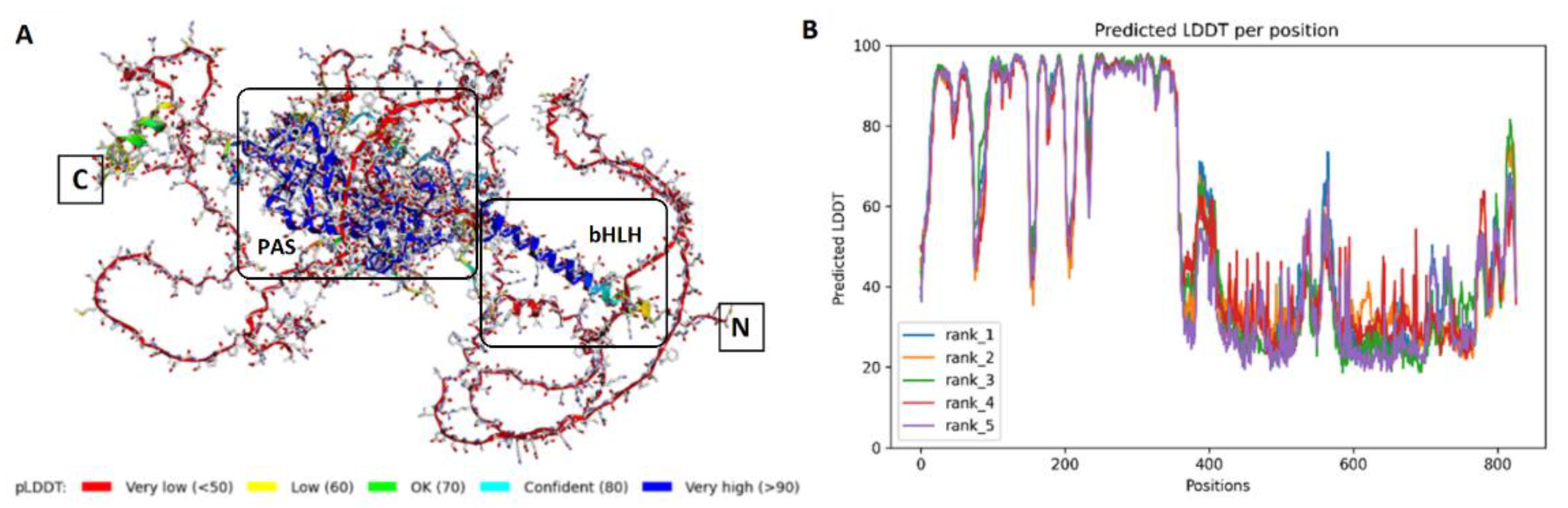
3.4. Molecular Modeling Studies
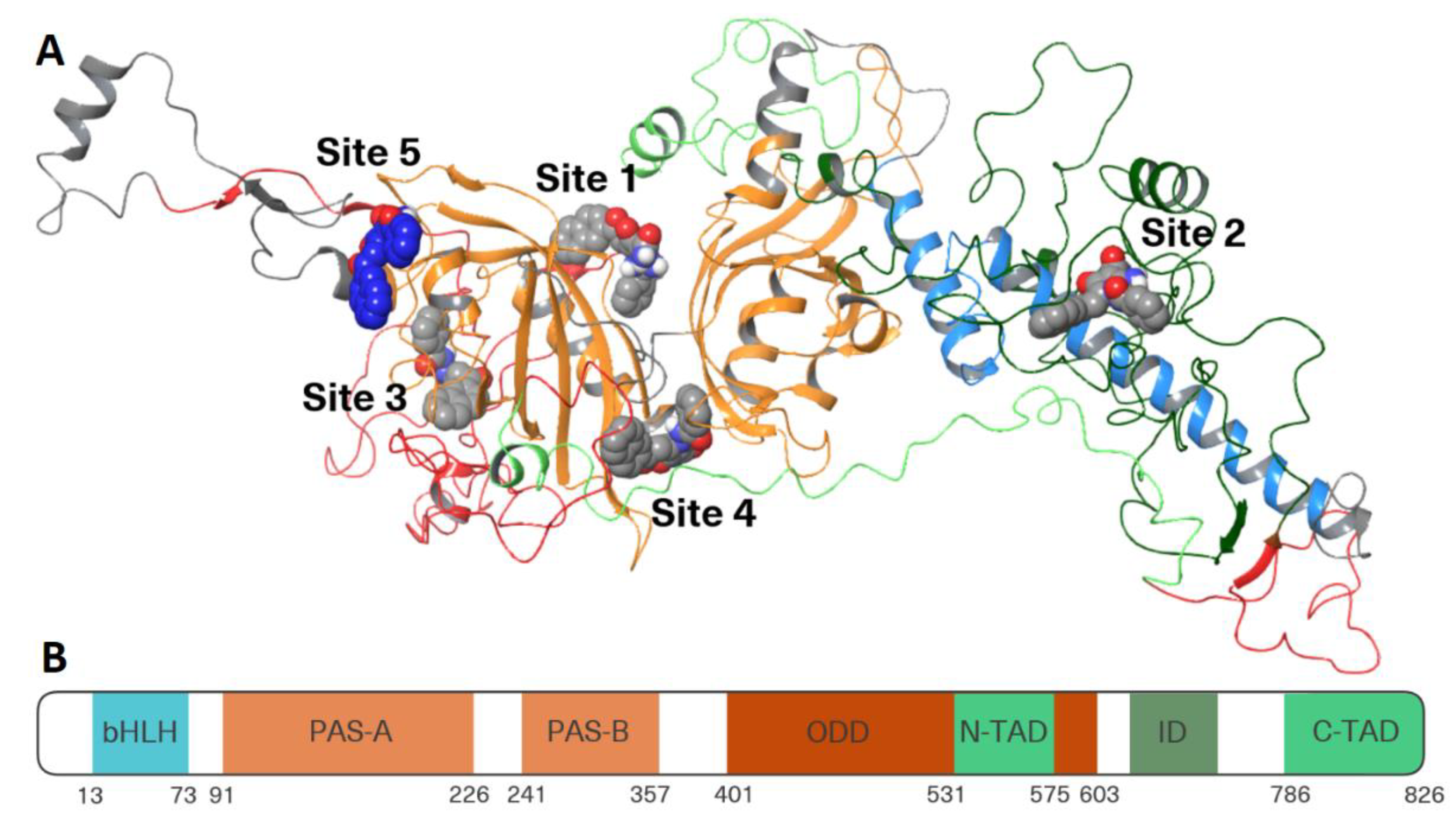
3.4.1. Folding and Binding Sites Search
3.4.2. Site Mapping and Multi-Ligand Dynamics
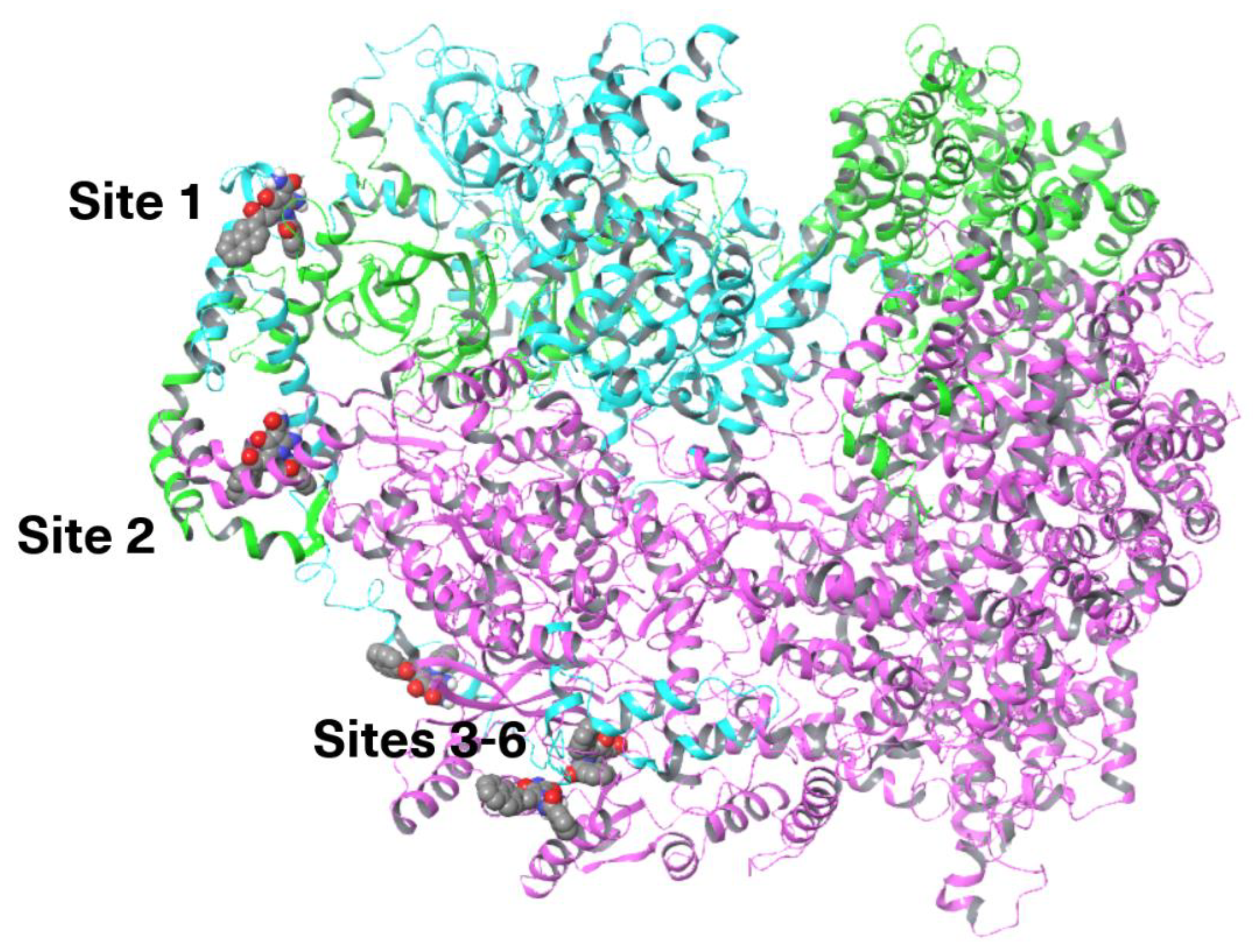
3.5. HIF Active Complex Binding Hypothesis
3.5.1. Folding and Molecular Dynamics
3.5.2. Site Mapping and Docking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2-ANPC | 2-amino-1-benzamido-5-(2-(naphthalene-2-yl)-2-oxoethylidene)-4-oxo-4,5-dihydro-1-H-pyrrole-3-carboxamide |
| 2-ME2 | 2-Methoxyestradiol |
| ARNT/HIF-1 β | Aryl hydrocarbon receptor nuclear translocator/Hypoxia-inducible factor-1β |
| bHLH | basic helix–loop–helix |
| CBP | CREB-binding protein |
| CRM1 | Chromosome region maintenance 1 |
| CTAD | C-terminal transactivation domain |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| H&E | Hematoxylin and eosin |
| HDAC | Class II histone deacetylase |
| HIF-1α | Hypoxia-inducible factor-1α |
| HSP90 | Heat shock protein 90 |
| ID | Inhibitory Domain |
| IDRs | Intrinsically Disordered Regions |
| IFD score | Energy spent on formation of the laying of the compound in the binding site and binding energy of ligand and protein |
| IHC | Immunohistochemical |
| MTA | Microtubule-targeting agents |
| NAMD | Nanoscale Molecular Dynamics |
| NMR | Nuclear Magnetic Resonance |
| NSCLC | Non-small cell lung cancer |
| NTAD | N-terminal transcription activation domain |
| ODD | Oxygen-dependent degradation |
| PARP | Poly(ADP)-ribose polymerase |
| PAS | Per-ARNT-Sim domain |
| PDB | Protein data bank |
| PPI | Protein-protein interaction |
| PTX | Paclitaxel |
| pVHL | von Hippel–Lindau tumor suppressor protein |
| RT-PCR | Real-time polymerase chain reaction |
| SD | Standard deviation |
| TMA | Tissue microarrays |
| TME | Tumor microenvironment |
| TNBC | Triple-negative breast cancer |
| VEGF | Vascular endothelial growth factor |
| VEGFR | Vascular endothelial growth factor receptor |
| VMD | Visual Molecular Dynamics |
| YC-1 | Lificiguat |
References
- Semenza, G.L. HIF-1 and Mechanisms of Hypoxia Sensing. Curr Opin Cell Biol 2001, 13, 167–171. [CrossRef]
- Sebestyén, A.; Kopper, L.; Dankó, T.; Tímár, J. Hypoxia Signaling in Cancer: From Basics to Clinical Practice. Pathology and Oncology Research 2021, 27, 1609802. [CrossRef]
- Rashid, M.; Zadeh, L.R.; Baradaran, B.; Molavi, O.; Ghesmati, Z.; Sabzichi, M.; Ramezani, F. Up-down Regulation of HIF-1α in Cancer Progression. Gene 2021, 798, 145796. [CrossRef]
- Zhao, Y.; Xing, C.; Deng, Y.; Ye, C.; Peng, H. HIF-1α Signaling: Essential Roles in Tumorigenesis and Implications in Targeted Therapies. Genes Dis 2024, 11, 234–251. [CrossRef]
- Zhang, C.; Liu, J.; Wang, J.; Zhang, T.; Xu, D.; Hu, W.; Feng, Z. The Interplay Between Tumor Suppressor p53 and Hypoxia Signaling Pathways in Cancer. Front. Cell Dev. Biol. 2021, 9, 648808. [CrossRef]
- Zhang, C.; Liu, J.; Xu, D.; Zhang, T.; Hu, W.; Feng, Z. Gain-of-Function Mutant P53 in Cancer Progression and Therapy. J Mol Cell Biol 2020, 12, 674–687. [CrossRef]
- Alam, M.M.; Fermin, J.M.; Knackstedt, M.; Noonan, M.J.; Powell, T.; Goodreau, L.; Daniel, E.K.; Rong, X.; Moore-Medlin, T.; Khandelwal, A.R.; et al. Everolimus downregulates STAT3/HIF-1α/VEGF pathway to inhibit angiogenesis and lymphangiogenesis in TP53 mutant head and neck squamous cell carcinoma (HNSCC). Oncotarget 2023, 14, 85–95. [CrossRef]
- Kilic, M.; Kasperczyk, H.; Fulda, S.; Debatin, K.M. Role of Hypoxia Inducible Factor-1 Alpha in Modulation of Apoptosis Resistance. Oncogene 2007, 26, 2027–2038. [CrossRef]
- Qannita, R.A.; Alalami, A.I.; Harb, A.A.; Aleidi, S.M.; Taneera, J.; Abu-Gharbieh, E.; El-Huneidi, W.; Saleh, M.A.; Alzoubi, K.H.; Semreen, M.H.; et al. Targeting Hypoxia-Inducible Factor-1 (HIF-1) in Cancer: Emerging Therapeutic Strategies and Pathway Regulation. Pharmaceuticals 2024, 17, 195. [CrossRef]
- Shirai, Y.; Chow, C.C.T.; Kambe, G.; Suwa, T.; Kobayashi, M.; Takahashi, I.; Harada, H.; Nam, J.M. An overview of the recent development of anticancer agents targeting the hif-1 transcription factor. Cancers 2021, 13, 2813. [CrossRef]
- Seredinski, S.; Boos, F.; Günther, S.; Oo, J.A.; Warwick, T.; Izquierdo Ponce, J.; Lillich, F.F.; Proschak, E.; Knapp, S.; Gilsbach, R.; et al. DNA topoisomerase inhibition with the HIF inhibitor acriflavine promotes transcription of lncRNAs in endothelial cells. Mol. Ther. Nucl. Acids 2022, 27, 1023–1035. [CrossRef]
- Xiang, L.; Gilkes, D.M.; Chaturvedi, P.; Luo, W.; Hu, H.; Takano, N.; Liang, H.; Semenza, G.L. Ganetespib blocks HIF-1 activity and inhibits tumor growth, vascularization, stem cell maintenance, invasion, and metastasis in orthotopic mouse models of triple-negative breast cancer. J. Mol. Med. 2014, 92, 151–164. [CrossRef]
- Takamori, H.; Yamasaki, T.; Kitadai, R.; Minamishima, Y.A.; Nakamura, E. Development of drugs targeting hypoxia-inducible factor against tumor cells with VHL mutation: Story of 127 years. Cancer Sci. 2023, 114, 1208–1217. [CrossRef]
- Kim, S.H.; Jeong, J.W.; Park, J.A.; Lee, J.W.; Seo, J.H.; Jung, B.K.; Bae, M.K.; Kim, K.W. Regulation of the HIF-1alpha stability by histone deacetylases. Oncol. Rep. 2007, 17, 647–651.
- Schoepflin, Z.R.; Shapiro, I.M.; Risbud, M.V. Class I and IIa HDACs Mediate HIF-1alpha Stability Through PHD2-Dependent Mechanism, While HDAC6, a Class IIb Member, Promotes HIF-1alpha Transcriptional Activity in Nucleus Pulposus Cells of the Intervertebral Disc. J. Bone Miner. Res. 2016, 31, 1287–1299. [CrossRef]
- Zhang, C.; Yang, C.; Feldman, M.J.; Wang, H.; Pang, Y.; Maggio, D.M.; Zhu, D.; Nesvick, C.L.; Dmitriev, P.; Bullova, P.; et al. Vorinostat suppresses hypoxia signaling by modulating nuclear translocation of hypoxia inducible factor 1 alpha. Oncotarget 2017, 8, 56110–56125. [CrossRef]
- Tanaka, T.; Yamaguchi, J.; Shoji, K.; Nangaku, M. Anthracycline inhibits recruitment of hypoxia-inducible transcription factors and suppresses tumor cell migration and cardiac angiogenic response in the host. J. Biol. Chem. 2012, 287, 34866–34882. [CrossRef]
- Galembikova A.R., Dunaev P.D., Bikinieva F.F., Mustafin I.G., Kopnin P.B., Zykova S.S., Mukhutdinova F.I., Sarbazyan E.A., Boichuk S.V. Mechanisms of cytotoxic activity of pyrrole-carboxamides against multidrug-resistant tumor cell sublines. Usp. mol. onkol. 2023, 10, 59-71. [CrossRef]
- Boichuk, S.; Galembikova, A.; Syuzov, K.; Dunaev, P.; Bikinieva, F.; Aukhadieva, A.; Zykova, S.; Igidov, N.; Gankova, K.; Novikova, M.; et al. The Design, Synthesis, and Biological Activities of Pyrrole-Based Carboxamides: The Novel Tubulin Inhibitors Targeting the Colchicine-Binding Site. Molecules 2021, 26, 5780. [CrossRef]
- Boichuk S.V., Zykova S.S.; et al. 2-amino-1-benzamido-5-[2-(naphthalene-2-yl)-2-oxoethylidene]-4-oxo-4,5-dihydro-1h-pyrrole-3-carboxamide and 2-amino-5-(3,3-dimethyl-2-oxobutylidene)-4-oxo-1-[2-(phenylamino)benzamido]-4,5-dihydro-1h-pyrrole-3-carboxamide inhibiting the process of polymerisation of tubulin and exhibiting cytotoxic and antitumour activity against human epithelial tumours. RU 2777209 C1, 01.08.2022.
- Kizimova, I.A.; Igidov, N.M.; Kiselev, M.A.; Dmitriev, M. V.; Chashchina, S. V.; Siutkina, A.I. Synthesis of New 2-Aminopyrrole Derivatives by Reaction of Furan-2,3-Diones 3-Acylhydrazones with CH-Nucleophiles. Russ J Gen Chem 2020, 90, 182–186. [CrossRef]
- Boichuk, S.; Galembikova, A.; Dunaev, P.; Valeeva, E.; Shagimardanova, E.; Gusev, O.; Khaiboullina, S. A Novel Receptor Tyrosine Kinase Switch Promotes Gastrointestinal Stromal Tumor Drug Resistance. Molecules 2017, 22, 2152. [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J. V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable Molecular Dynamics on CPU and GPU Architectures with NAMD. J. Chem. Phys. 2020, 153. [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ 1 and χ 2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [CrossRef]
- MacKerell, A.D.; Feig, M.; Brooks, C.L. Improved Treatment of the Protein Backbone in Empirical Force Fields. J. Am. Chem. Soc. 2004, 126, 698–699. [CrossRef]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [CrossRef]
- Bugnon, M.; Goullieux, M.; Röhrig, U.F.; Perez, M.A.S.; Daina, A.; Michielin, O.; Zoete, V. SwissParam 2023: A Modern Web-Based Tool for Efficient Small Molecule Parametrization. J. Chem. Inf. Model. 2023, 63, 6469–6475. [CrossRef]
- Allouche, A. Software News and Updates Gabedit — A Graphical User Interface for Computational Chemistry Softwares. J. Comput. Chem. 2012, 32, 174–182. [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [CrossRef]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the ACM/IEEE SC 2006 Conference (SC’06); IEEE, November 2006; pp. 43–43.
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [CrossRef]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.F.; Honig, B.; Shaw, D.E.; Friesner, R.A. A Hierarchical Approach to All-Atom Protein Loop Prediction. Proteins Struct. Funct. Genet. 2004, 55, 351–367. [CrossRef]
- Jacobson, M.P.; Friesner, R.A.; Xiang, Z.; Honig, B. On the Role of the Crystal Environment in Determining Protein Side-Chain Conformations. J. Mol. Biol. 2002, 320, 597–608. [CrossRef]
- Halgren, T. New Method for Fast and Accurate Binding-site Identification and Analysis. Chem. Biol. Drug Des. 2007, 69, 146–148. [CrossRef]
- Halgren, T.A. Identifying and Characterizing Binding Sites and Assessing Druggability. J. Chem. Inf. Model. 2009, 49, 377–389. [CrossRef]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel Procedure for Modeling Ligand/Receptor Induced Fit Effects. J. Med. Chem. 2006, 49, 534–553. [CrossRef]
- Farid, R.; Day, T.; Friesner, R.A.; Pearlstein, R.A. New Insights about HERG Blockade Obtained from Protein Modeling, Potential Energy Mapping, and Docking Studies. Bioorg. Med. Chem. 2006, 14, 3160–3173. [CrossRef]
- Sherman, W.; Beard, H.S.; Farid, R. Use of an Induced Fit Receptor Structure in Virtual Screening. Chem. Biol. Drug Des. 2006, 67, 83–84. [CrossRef]
- Sánchez-Puig, N.; Veprintsev, D.B.; Fersht, A.R. Binding of Natively Unfolded HIF-1α ODD Domain to P53. Mol. Cell 2005, 17, 11–21. [CrossRef]
- Nyqvist, I.; Dogan, J. Characterization of the Dynamics and the Conformational Entropy in the Binding between TAZ1 and CTAD-HIF-1α. Sci. Rep. 2019, 9, 20–22. [CrossRef]
- Chen, H.; Ma, D.; Yue, F.; Qi, Y.; Dou, M.; Cui, L.; Xing, Y. The Potential Role of Hypoxia-Inducible Factor-1 in the Progression and Therapy of Central Nervous System Diseases. Curr. Neuropharmacol. 2021, 20, 1651–1666. [CrossRef]
- Kim, Y.; Nam, H.J.; Lee, J.; Park, D.Y.; Kim, C.; Yu, Y.S.; Kim, D.; Park, S.W.; Bhin, J.; Hwang, D.; et al. Methylation-Dependent Regulation of HIF-1α Stability Restricts Retinal and Tumour Angiogenesis. Nat. Commun. 2016, 7. [CrossRef]
- Dewi, F.R.; Fatchiyah, F. Methylation Impact Analysis of Erythropoietin (EPO) Gene to Hypoxia Inducible Factor-1α (HIF-1α) Activity. Bioinformation 2013, 9, 782–787. [CrossRef]
- Appling, F.D.; Berlow, R.B.; Stanfield, R.L.; Dyson, H.J.; Wright, P.E. The Molecular Basis of Allostery in a Facilitated Dissociation Process. Structure 2021, 29, 1327-1338.e5. [CrossRef]
- Seoane, B.; Carbone, A. Soft Disorder Modulates the Assembly Path of Protein Complexes. PLoS Comput. Biol. 2022, 18, 1–19. [CrossRef]
- Zhang, T.; Kastrenopoulou, A.; Larrouture, Q.; Athanasou, N.A.; Knowles, H.J. Angiopoietin-like 4 promotes osteosarcoma cell proliferation and migration and stimulates osteo-clastogenesis. BMC Cancer 2018, 18, 536. [CrossRef]
- Maxwell, P.H. The HIF pathway in cancer. Semin. Cell Dev. Biol. 2005, 16, 523–53. [CrossRef]
- Wan, J.; Chai, H.; Yu, Z.; Ge, W.; Kang, N.; Xia, W.; Che, Y. HIF-1α effects on angiogenic potential in human small cell lung carcinoma. J. Exp. Clin. Cancer Res. 2011, 30, 77. [CrossRef]
- Magar, A.G.; Morya, V.K.; Kwak, M.K.; Oh, J.U.; Noh, K.C. A Molecular Perspective on HIF-1α and Angiogenic Stimulator Networks and Their Role in Solid Tumors: An Update. Int. J. Mol. Sci. 2024, 25, 3313. [CrossRef]
- Voron, T.; Marcheteau, E.; Pernot, S.; Colussi, O.; Tartour, E.; Taieb, J.; Terme, M. Control of the immune response by pro-angiogenic factors. Front. Oncol. 2014, 4, 70. [CrossRef]
- Chen, Y.; Song, Y.; Du, W.; Gong, L.; Chang, H.; Zou, Z. Tumor-associated macrophages: An accomplice in solid tumor progression. J. Biomed. Sci. 2019, 26, 78. [CrossRef]
- Rani, S.; Roy, S.; Singh, M.; Kaithwas, G. Regulation of Transactivation at C-TAD Domain of HIF-1α by Factor-Inhibiting HIF-1α (FIH-1): A Potential Target for Therapeutic Interven-tion in Cancer. Oxid. Med. Cell. Longev. 2022, 2022, 2407223. [CrossRef]
- Nagao, A.; Kobayashi, M.; Koyasu, S.; Chow, C.C.T.; Harada, H. HIF-1-Dependent Repro-gramming of Glucose Metabolic Pathway of Cancer Cells and Its Therapeutic Significance. Int. J. Mol. Sci. 2019, 20, 238. [CrossRef]
- Magar, A.G.; Morya, V.K.; Kwak, M.K.; Oh, J.U.; Noh, K.C. A Molecular Perspective on HIF-1α and Angiogenic Stimulator Networks and Their Role in Solid Tumors: An Update. Int. J. Mol. Sci. 2024, 25, 3313. [CrossRef]
- Dong, W.; Keibler, M.A.; Stephanopoulos, G. Review of metabolic pathways activated in cancer cells as determined through isotopic labeling and network analysis. Metab. Eng. 2017, 43, 113–124. [CrossRef]
- Masoud, G.N.; Li, W. HIF-1α pathway: Role, regulation and intervention for cancer thera-py. Acta Pharm. Sin. B 2015, 5, 378–389. [CrossRef]
- Roy, S.; Kumaravel, S.; Sharma, A.; Duran, C.L.; Bayless, K.J.; Chakraborty, S. Hypoxic tumor microenvironment: Implications for cancer therapy. Exp. Biol. Med. 2020, 245, 1073–1086. [CrossRef]
- Rani, S.; Roy, S.; Singh, M.; Kaithwas, G. Regulation of Transactivation at C-TAD Domain of HIF-1α by Factor-Inhibiting HIF-1α (FIH-1): A Potential Target for Therapeutic Intervention in Cancer. Oxid. Med. Cell. Longev. 2022, 2022, 2407223. [CrossRef]
- Zhao, Y.; Xing, C.; Deng, Y.; Ye, C.; Peng, H. HIF-1α signaling: Essential roles in tumorigenesis and implications in targeted therapies. Genes Dis. 2024, 11, 234–251. [CrossRef]
- Ricker, J.L.; Chen, Z.; Yang, X.P.; Pribluda, V.S.; Swartz, G.M.; Van Waes, C. 2-Methoxyestradiol Inhibits Hypoxia-Inducible Factor 1α, Tumor Growth, and Angiogenesis and Augments Paclitaxel Efficacy in Head and Neck Squamous Cell Carcinoma. Clinical Cancer Research 2004, 10, 8665–8673. [CrossRef]
- Attia, Y.M.; Mokhlis, H.A.; Ismail, A.; Doghish, A.S.; Sobhy, M.H.; Hassanein, S.S.; El-Dakroury, W.A.; Mariee, A.D.; Salama, S.A.; Sharaky, M. 2-Methoxyestradiol Sensitizes Tamoxifen-Resistant MCF-7 Breast Cancer Cells via Downregulating HIF-1α. Medical Oncology 2024, 41, 1–10. [CrossRef]
- Ouyang, C.; Zhang, J.; Lei, X.; Xie, Z.; Liu, X.; Li, Y.; Huang, S.; Wang, Z.; Tang, G. Advances in Antitumor Research of HIF-1α Inhibitor YC-1 and Its Derivatives. Bioorg Chem 2023, 133, 106400. [CrossRef]
- Yu, K.H.; Hung, H.Y. Synthetic Strategy and Structure–Activity Relationship (SAR) Studies of 3-(5′-Hydroxymethyl-2′-Furyl)-1-Benzyl Indazole (YC-1, Lificiguat): A Review. RSC Adv 2021, 12, 251–264. [CrossRef]
- Mangraviti, A.; Raghavan, T.; Volpin, F.; Skuli, N.; Gullotti, D.; Zhou, J.; Asnaghi, L.; Sankey, E.; Liu, A.; Wang, Y.; et al. HIF-1α- Targeting Acriflavine Provides Long Term Survival and Radiological Tumor Response in Brain Cancer Therapy. Scientific Reports 2017, 7, 1–13. [CrossRef]
- Mabjeesh, N.J.; Escuin, D.; LaVallee, T.M.; Pribluda, V.S.; Swartz, G.M.; Johnson, M.S.; Willard, M.T.; Zhong, H.; Simons, J.W.; Giannakakou, P. 2ME2 Inhibits Tumor Growth and Angiogenesis by Disrupting Microtubules and Dysregulating HIF. Cancer Cell 2003, 3, 363–375. [CrossRef]
- D’Amato, R.J.; Lin, C.M.; Flynn, E.; Folkman, J.; Hamel, E. 2-Methoxyestradiol, an Endogenous Mammalian Metabolite, Inhibits Tubulin Polymerization by Interacting at the Colchicine Site. Proceedings of the National Academy of Sciences 1994, 91, 3964–3968. [CrossRef]
- Poch, A.; Villanelo, F.; Henriquez, S.; Kohen, P.; Muñoz, A.; Strauss, J.F.; Devoto, L. Molecular Modelling Predicts That 2-Methoxyestradiol Disrupts HIF Function by Binding to the PAS-B Domain. Steroids 2019, 144, 21–29. [CrossRef]
- Cardoso, R.; Love, R.; Nilsson, C.L.; Bergqvist, S.; Nowlin, D.; Yan, J.; Liu, K.K.C.; Zhu, J.; Chen, P.; Deng, Y.L.; et al. Identification of Cys255 in HIF-1α as a Novel Site for Development of Covalent Inhibitors of HIF-1α/ARNT PasB Domain Protein-Protein Interaction. Protein Sci. 2012, 21, 1885–1896. [CrossRef]
- Wu, D.; Potluri, N.; Lu, J.; Kim, Y.; Rastinejad, F. Structural Integration in Hypoxia-Inducible Factors. Nature 2015, 524, 303–308. [CrossRef]
- Carbonaro, M.; O’Brate, A.; Giannakakou, P. Microtubule disruption targets HIF-1alpha mRNA to cytoplasmic P-bodies for translational repression. J. Cell Biol. 2011, 192, 83–99. [CrossRef]
- Mabjeesh, N.J.; Escuin, D.; LaVallee, T.M.; Pribluda, V.S.; Swartz, G.M.; Johnson, M.S.; Willard, M.T.; Zhong, H.; Simons, J.W.; Giannakakou, P. 2ME2 inhibits tumor growth and angiogenesis by disrupting microtubules and dysregulating HIF. Cancer Cell 2003, 3, 363–375. [CrossRef]
- LaVallee, T.M.; Burke, P.A.; Swartz, G.M.; Hamel, E.; Agoston, G.E.; Shah, J.; Suwandi, L.; Hanson, A.D.; Fogler, W.E.; Sidor, C.F.; et al. Significant antitumor activity in vivo following treatment with the microtubule agent ENMD-1198. Mol. Cancer Ther. 2008, 7, 1472–1482. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




