3.1. Differential Salivary Peptidome in T2D vs. Controls
A peptide-focused DIA-NN study revealed distinct alterations in the salivary peptidome of patients with type 2 diabetes (T2D) compared to normoglycaemic controls. A total of 199 peptides met the significance criteria (|FC| ≥ 1.5; p < 0.05), indicating a significant reconfiguration of the salivary molecular landscape associated with metabolic disease. The volcano plot demonstrates this disparity, showing that most significant features are elevated rather than reduced in T2D saliva. Twenty-one peptides showed reductions (FC 0.23–0.50), including fragments from S100A8 (P05109; ALNSIIDVYHKYSLIK), a key myeloid inflammatory protein, as well as various peptides derived from cytoskeletal or regulatory proteins such as P61247, P60660, and O95171. The limited number of downregulated features suggests targeted inhibition of specific epithelial or immune-related elements rather than a general reduction in protein expression.
Conversely, the T2D salivary profile was characterised by increased peptides associated with lipid transport, complement activation, mucosal barrier regulation, and systemic inflammation. Numerous fragments of apolipoprotein A-I and A-II (P02647, P02652) showed consistent elevation (FC ≈ 2.0–2.3), as did transthyretin (P02766), indicating an increased role for lipid-transport and plasma carrier proteins in diabetic saliva. A similar trend was observed for complement-associated proteins: α2-macroglobulin (P01023), complement C3 (P01024), C4b-binding protein (P04003), and α1-antichymotrypsin (P01011) all showed substantial elevations (FC = 2.0–2.6), indicating activation of innate immunological and protease-regulatory pathways. Notably, there was persistent enrichment of Mucin-5B (MUC5B, Q9HC84) peptides, suggesting altered salivary viscosity and epithelial barrier function in type 2 diabetes, presumably reflecting chronic metabolic stress on mucosal surfaces. Elevated levels of albumin, fibrinogen α-chain, hemopexin, and related acute-phase proteins further indicated increased plasma transudation into saliva and an enhanced systemic inflammatory response.
The most significantly altered features included haemoglobin-derived peptides (P69905, P68871, P02042), with fold-changes between 4 and nearly 8. These elevations likely reflect microvascular fragility or subclinical gingival bleeding, common complications in diabetes, providing biologically consistent justification for their marked increase in saliva. Multivariate analysis supported this global restructuring: principal component analysis showed clear separation between T2D and control participants, indicating that the combined peptide signature defines a coherent and disease-specific salivary phenotype.
Collectively, these findings indicate a dual pattern in diabetic saliva: a specific decrease in structural and regulatory peptides, alongside a widespread increase in complement-related, acute-phase, lipoprotein-derived, and haemoglobin fragments. This composite signature illustrates the interconnected effects of low-grade inflammation, epithelial barrier remodelling, and increased vascular permeability typical of metabolic dysregulation. The extent and specificity of these peptide changes highlight their potential as non-invasive biomarkers for identifying or monitoring physiological changes associated with T2D.
Figure 1.
Volcano plot showing differentially abundant salivary peptides in T2D versus normoglycemic controls. Each point represents a quantified peptide, plotted according to its log₂ fold change (x-axis) and –log₁₀(p-value) (y-axis). Vertical dashed lines indicate the fold-change threshold (|FC| > 1.5), while the horizontal dashed line marks the significance cutoff (p < 0.05). Peptides elevated in T2D appear to the right (red gradient), and those reduced in T2D are to the left (blue gradient), with colour intensity reflecting the magnitude of log₂(FC). The size of the points reflects statistical significance, with larger circles indicating lower p-values. Selected peptides showing the most significant modulation are annotated, including hemoglobin-derived fragments (e.g., P02042_VHLTPEEK), complement-related peptides (e.g., P01024_ENEGFTVTAEGK), apolipoprotein fragments (e.g., P43652_FLVNLVK), and downregulated inflammatory or structural peptides such as S100A8-derived sequences (P05109_ALNSIIDVYHKYSLIK).
Figure 1.
Volcano plot showing differentially abundant salivary peptides in T2D versus normoglycemic controls. Each point represents a quantified peptide, plotted according to its log₂ fold change (x-axis) and –log₁₀(p-value) (y-axis). Vertical dashed lines indicate the fold-change threshold (|FC| > 1.5), while the horizontal dashed line marks the significance cutoff (p < 0.05). Peptides elevated in T2D appear to the right (red gradient), and those reduced in T2D are to the left (blue gradient), with colour intensity reflecting the magnitude of log₂(FC). The size of the points reflects statistical significance, with larger circles indicating lower p-values. Selected peptides showing the most significant modulation are annotated, including hemoglobin-derived fragments (e.g., P02042_VHLTPEEK), complement-related peptides (e.g., P01024_ENEGFTVTAEGK), apolipoprotein fragments (e.g., P43652_FLVNLVK), and downregulated inflammatory or structural peptides such as S100A8-derived sequences (P05109_ALNSIIDVYHKYSLIK).
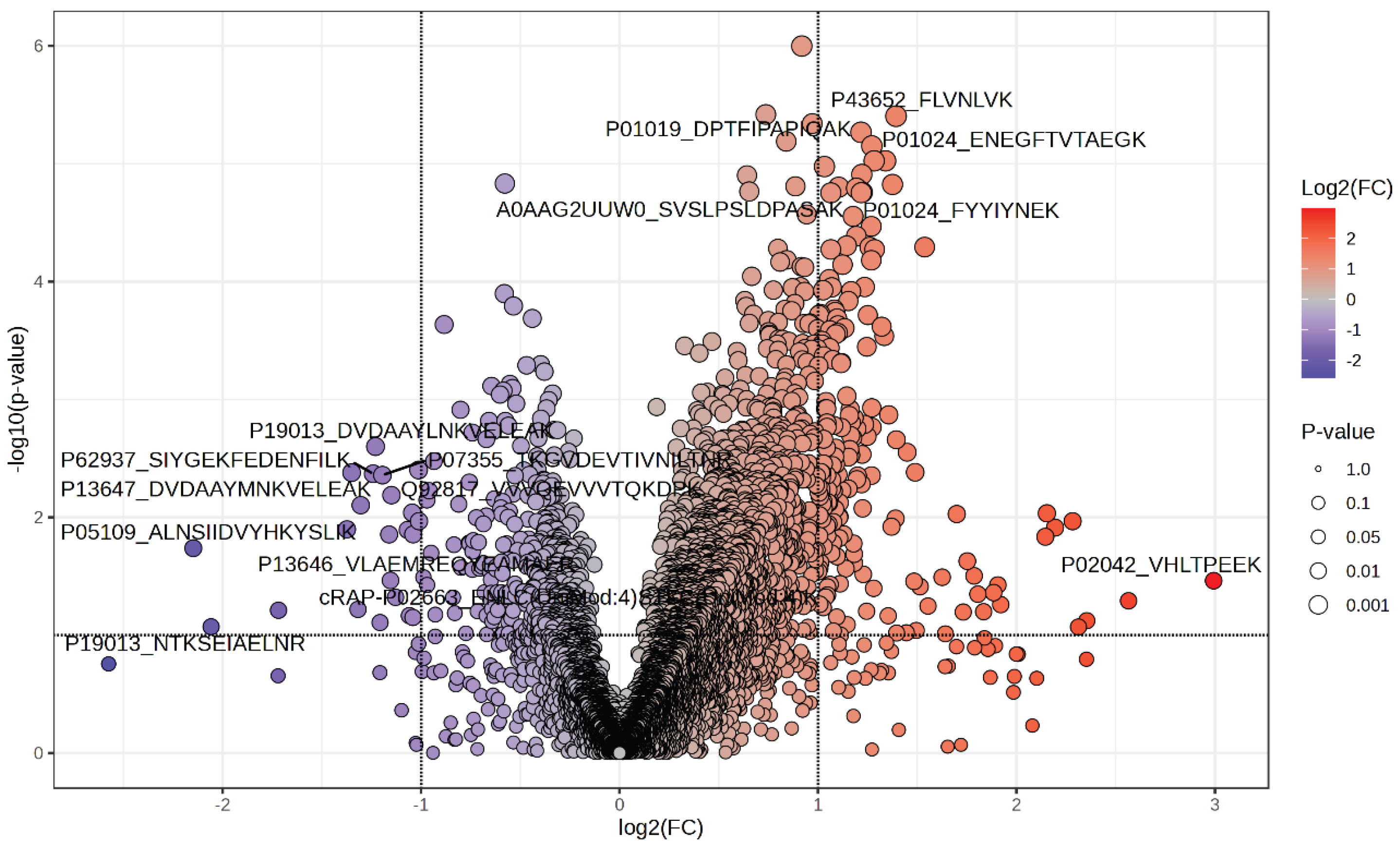
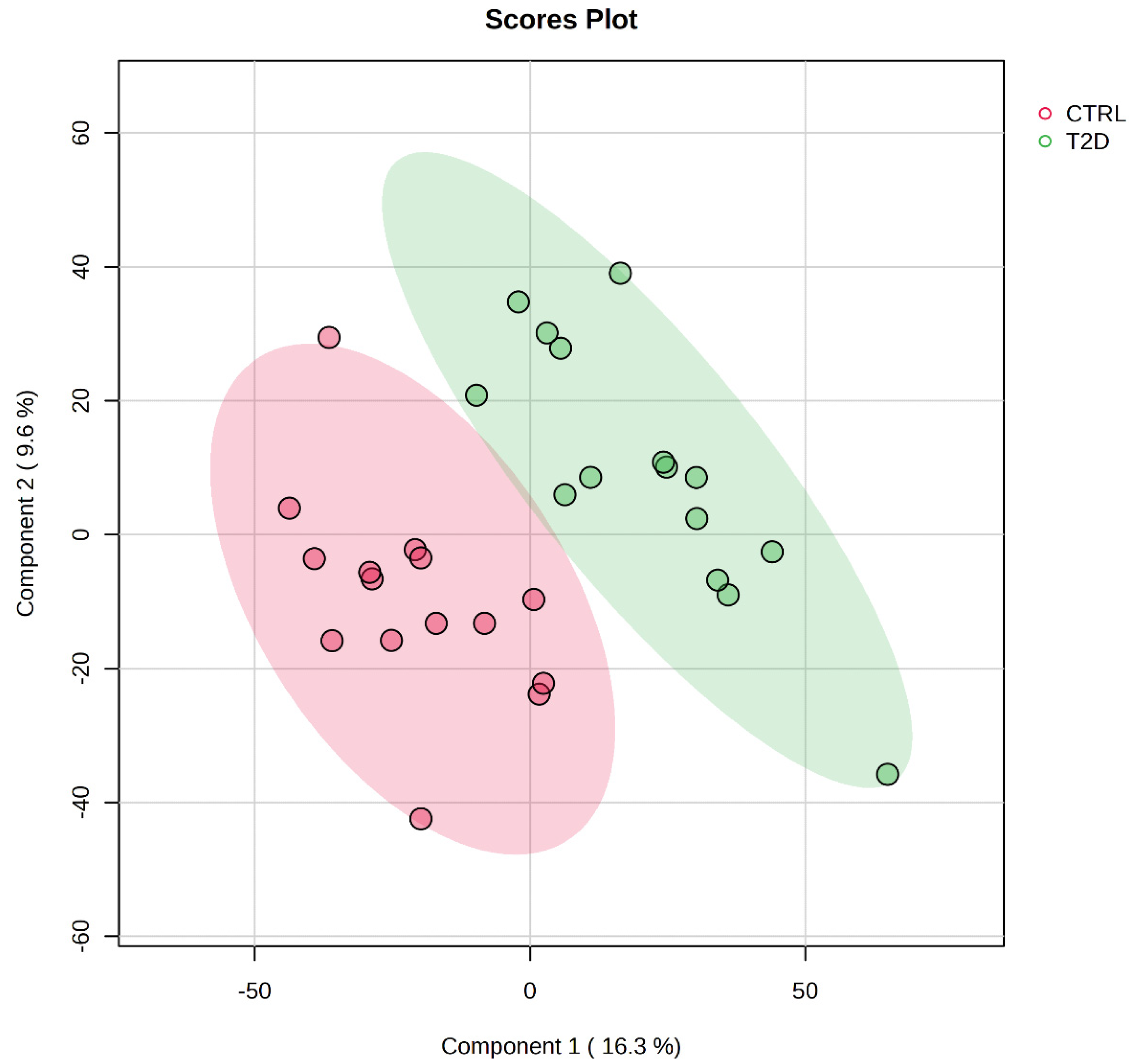
Figure 2.
Principal Component Analysis (PCA) of the salivary peptidome distinguishing T2D and control groups. Scores plot of the first two principal components (PC1: 16.3% variance; PC2: 9.6% variance) derived from peptide-level DIA-NN intensities. Each point represents a saliva sample, categorised by colour (red: controls; green: T2D). Ellipses indicate the 95% confidence intervals for each cohort. The substantial separation between groups across both components indicates consistent and disease-specific differences in the salivary peptide profile associated with type 2 diabetes.
Figure 2.
Principal Component Analysis (PCA) of the salivary peptidome distinguishing T2D and control groups. Scores plot of the first two principal components (PC1: 16.3% variance; PC2: 9.6% variance) derived from peptide-level DIA-NN intensities. Each point represents a saliva sample, categorised by colour (red: controls; green: T2D). Ellipses indicate the 95% confidence intervals for each cohort. The substantial separation between groups across both components indicates consistent and disease-specific differences in the salivary peptide profile associated with type 2 diabetes.
Scheme S1. Enhanced annotation of differentially abundant salivary peptides in T2D versus controls.
This table lists all peptides identified as significantly modulated between individuals with type 2 diabetes (T2D) and normoglycemic controls based on DIA-NN precursor quantification (|FC| ≥ 1.5; p < 0.05). For each peptide, the UniProt accession number, amino-acid sequence, and fold change (FC) are provided. Additional annotation includes peptide length, and approximate molecular weight (Approx_MW), calculated as the sum of monoisotopic masses of individual amino acids. These features support comparative structural analysis and facilitate downstream interpretation within biological pathways and bioprinting-relevant contexts. The table serves as the complete reference dataset underlying the volcano plot, PCA clustering, and structural modelling analyses presented in the manuscript.
3.2. AlphaFold Structural Modelling
AlphaFold modelling of differentially abundant peptides provided a structural perspective that enhances the quantitative analysis of salivary peptidomics. Although brief, the selected peptides displayed unique physicochemical characteristics that clarify how changes in peptide abundance associated with T2D may influence lubrication, immunological signalling, and matrix dynamics in a bioprinting context.
The two peptides derived from MUC5B, IVTENIPCGTTGTTCSK and TGLLVEQSGDYIK, showed the greatest flexibility and extension in their conformations. The extended region, containing two cysteine residues and multiple glycine/threonine repeats, was expected to adopt an open, highly dynamic conformation, consistent with the disordered segments that confer elasticity and hydration to mucin polymers. The second MUC5B peptide displayed a short turn-like structure with exposed polar groups capable of interacting with both aqueous and hydrogel environments. Together, these features highlight that the increased presence of MUC5B peptides in T2D saliva represents not only a compositional change but also a shift in structural motifs essential for mucus viscosity and epithelial barrier function.
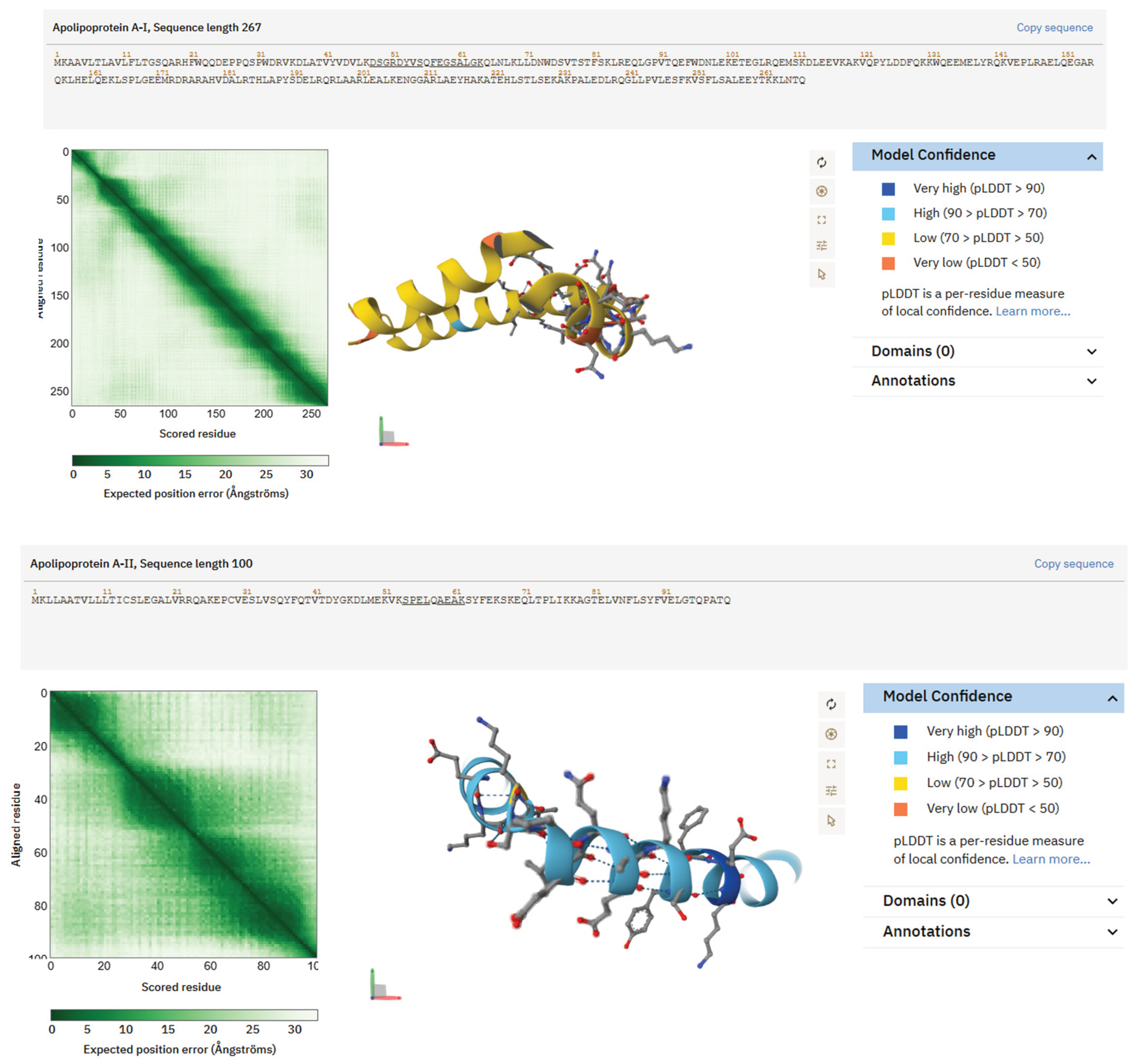
In contrast, peptides from ApoA1 and ApoA2 demonstrated distinct amphipathic properties. AlphaFold predicted a partially helical conformation for the ApoA1 fragment DSGRDYVSQFEGSALGK, with hydrophobic residues aligned along one face of the helix and acidic or polar residues grouped on the opposite side. The shorter ApoA2 peptide SPELQAEAK exhibited a compact helix-turn motif with a similar arrangement of charged and nonpolar residues. The amphipathic structures typical of lipid-binding proteins suggest that the elevated levels of apolipoprotein peptides in T2D saliva may influence interfacial tension, micelle-like behaviour, and polymer–protein interactions in mucosa-mimetic bioinks.
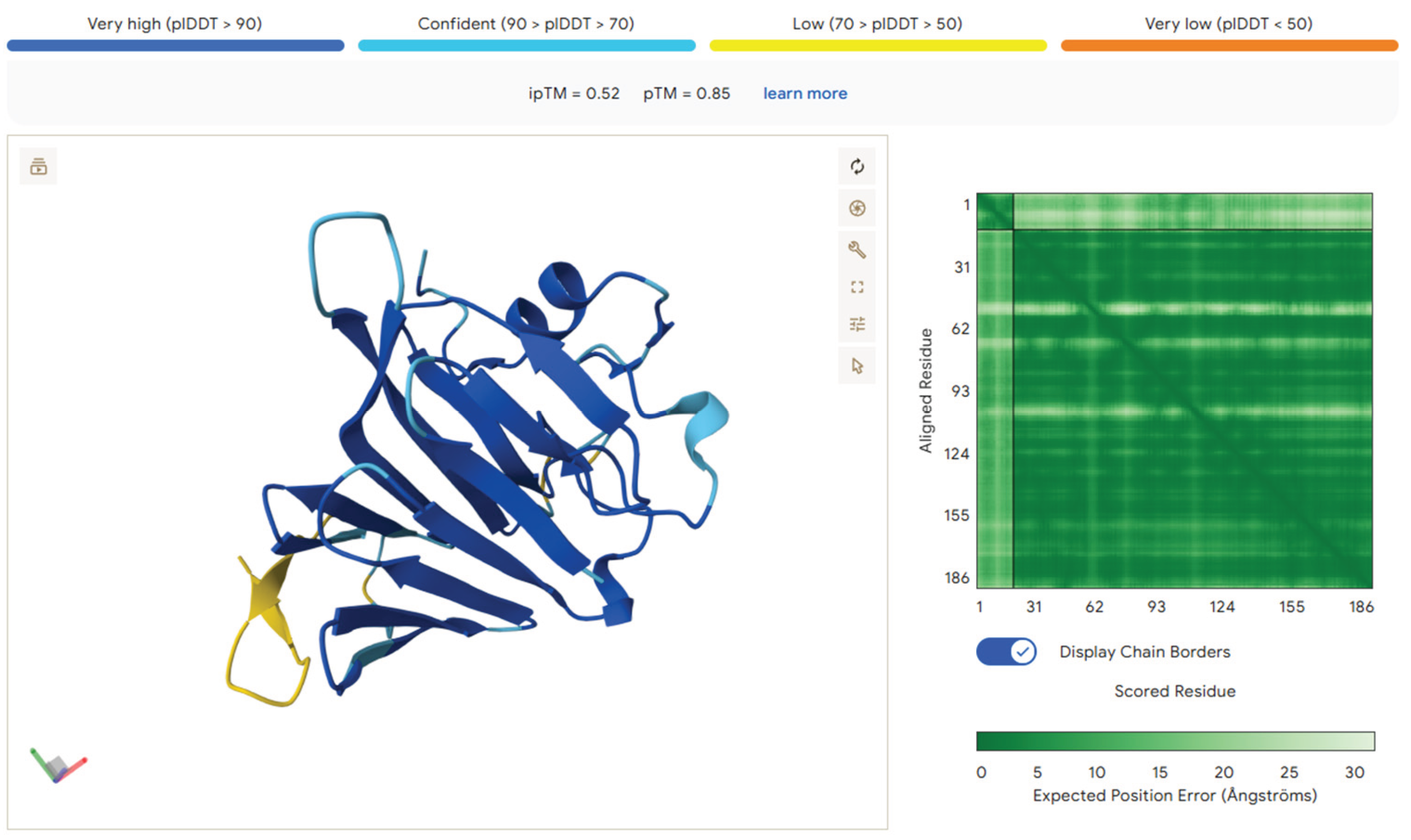
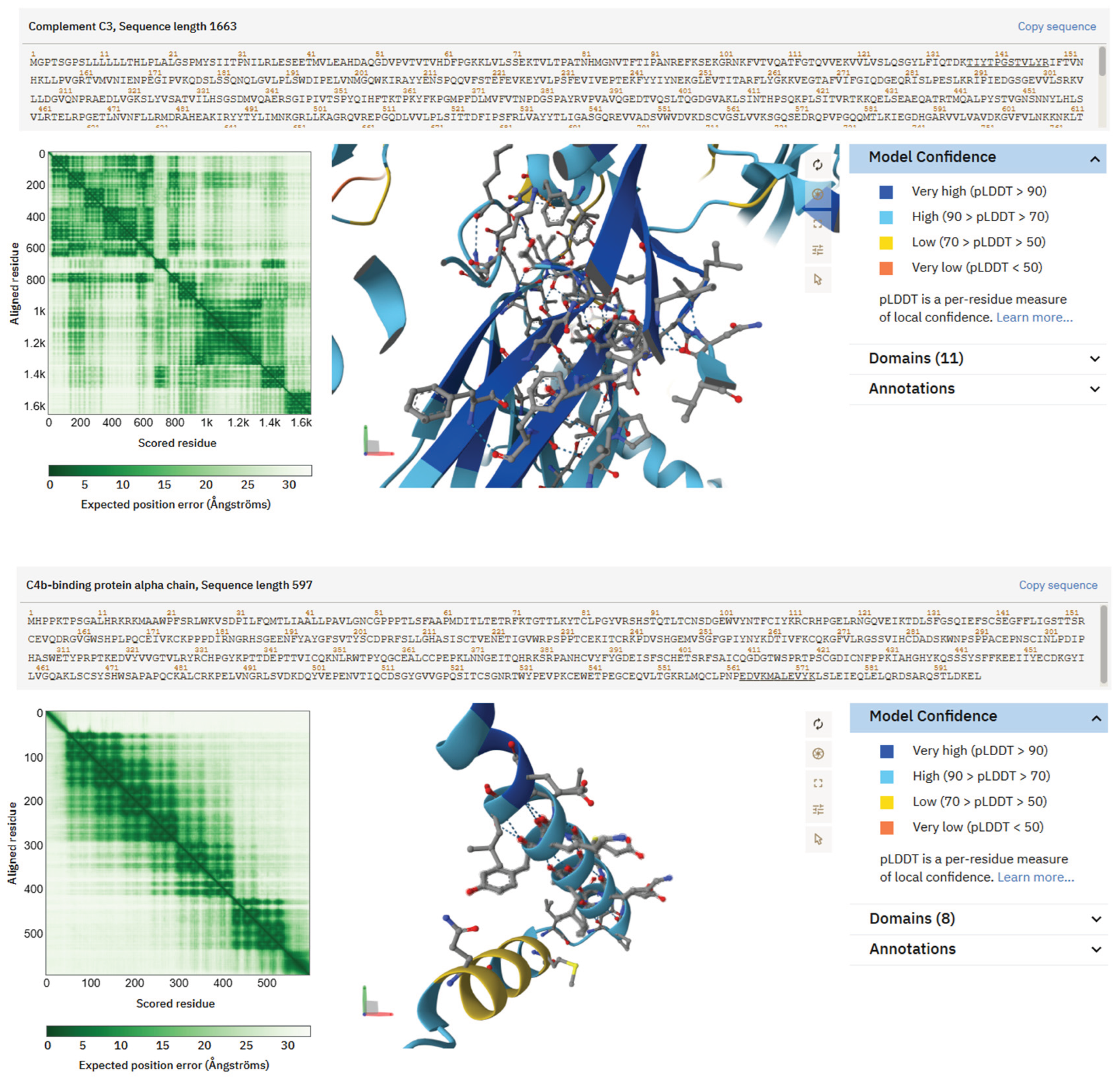
Peptides associated with complement and protease-inhibitor pathways, A2M (TEHPFTVEEFVLPK), C3 (TIYTPGSTVLYR), and C4BP (EDVYVVGTVLR)—displayed greater structural compactness and stability. AlphaFold predicted short, structured motifs with defined regions of hydrophobic and charged residues, resulting in compact interaction surfaces rather than extended flexible regions. These features support transient binding events and may regulate protease inhibition, complement signalling, or cell–matrix communication when incorporated into printed tissue. Their enrichment in T2D saliva thus introduces an immunological dimension to the biochemical environment that bioinks would encounter in vivo.
Peptides derived from haemoglobin, particularly the significantly upregulated sequences VLGAFSDGLAHLDNLK and VHLTPEEK, exhibited the most globular and helix-rich conformations. These structures resemble segments of the globin fold and present surface-exposed hydrophobic and polar groups that can participate in redox processes or interact with oxygen and reactive molecules. Their marked increase in T2D saliva, along with their consistent secondary-structure patterns, suggests they may locally affect redox balance or oxygen diffusion, factors known to influence hydrogel stability, cell viability, and tissue remodelling after bioprinting.
The AlphaFold predictions collectively show that the peptides significantly altered in T2D saliva are not random fragments but belong to specific structural categories, flexible mucin loops, amphipathic apolipoprotein helices, compact immune-regulatory motifs, and helix-rich haemoglobin segments, each with functional implications for lubrication, immune signalling, matrix interaction, and redox behaviour. Integrating these structural features with the observed fold-change patterns establishes a mechanistic link between metabolic dysregulation and the physicochemical environment relevant for developing more physiologically representative oral-tissue bioinks.
Figure 3.
Structures of upregulated MUC5B peptides in T2D saliva predicted by AlphaFold. Ribbon and surface representations of (A) IVTENIPCGTTGTTCSK and (B) TGLLVEQSGDYIK, emphasizing anticipated secondary-structure components and per-residue confidence (pLDDT). The peptide IVTENIPCGTTGTTCSK exhibits an elongated, pliable conformation, with two cysteine residues arranged to facilitate possible intrachain or interchain interactions, aligning with mucin-like polymer characteristics. TGLLVEQSGDYIK exhibits a compact turn motif characterized by flexible termini and a mixed hydrophobic/polar distribution, suggesting potential functions in hydration, lubrication, or polymer interfacing within bioink environments.
Figure 3.
Structures of upregulated MUC5B peptides in T2D saliva predicted by AlphaFold. Ribbon and surface representations of (A) IVTENIPCGTTGTTCSK and (B) TGLLVEQSGDYIK, emphasizing anticipated secondary-structure components and per-residue confidence (pLDDT). The peptide IVTENIPCGTTGTTCSK exhibits an elongated, pliable conformation, with two cysteine residues arranged to facilitate possible intrachain or interchain interactions, aligning with mucin-like polymer characteristics. TGLLVEQSGDYIK exhibits a compact turn motif characterized by flexible termini and a mixed hydrophobic/polar distribution, suggesting potential functions in hydration, lubrication, or polymer interfacing within bioink environments.
Figure 4.
Structures predicted by AlphaFold of peptides produced from ApoA1 and ApoA2, abundant in saliva of individuals with Type 2 Diabetes. Ribbon and surface representations of the ApoA1 peptide DSGRDYVSQFEGSALGK, illustrating a partly helical amphipathic structure with segregation of hydrophobic and polar residues, typical of lipid-associated domains. Structural prediction of the ApoA2 peptide SPELQAEAK, which displays a brief helical/turn motif characterized by an equitable distribution of charged and hydrophobic residues. These characteristics facilitate possible interactions with polymeric matrices, micelle-like environments, or hydrogel constituents pertinent to bioink efficacy.
Figure 4.
Structures predicted by AlphaFold of peptides produced from ApoA1 and ApoA2, abundant in saliva of individuals with Type 2 Diabetes. Ribbon and surface representations of the ApoA1 peptide DSGRDYVSQFEGSALGK, illustrating a partly helical amphipathic structure with segregation of hydrophobic and polar residues, typical of lipid-associated domains. Structural prediction of the ApoA2 peptide SPELQAEAK, which displays a brief helical/turn motif characterized by an equitable distribution of charged and hydrophobic residues. These characteristics facilitate possible interactions with polymeric matrices, micelle-like environments, or hydrogel constituents pertinent to bioink efficacy.
Figure 5.
AlphaFold structural predictions for peptides produced from complement and protease inhibitors, abundant in saliva from individuals with Type 2 Diabetes. Peptide TEHPFTVEEFVLPK from α2-Macroglobulin exhibits an acidic, partly folded motif suitable for interactions with proteases and cytokines. Complement C3 peptide TIYTPGSTVLYR assumes a compact shape, with a combination of aromatic and polar residues creating a possible regulatory interface. The peptide EDVYVVGTVLR from the C4b-binding protein α-chain exhibits hydrophobic clustering encircled by charged residues, indicating potential for scaffold anchoring or surface binding. These structural features elucidate how complement fragments may influence immunological or matrix interactions inside bioprinted tissues.
Figure 5.
AlphaFold structural predictions for peptides produced from complement and protease inhibitors, abundant in saliva from individuals with Type 2 Diabetes. Peptide TEHPFTVEEFVLPK from α2-Macroglobulin exhibits an acidic, partly folded motif suitable for interactions with proteases and cytokines. Complement C3 peptide TIYTPGSTVLYR assumes a compact shape, with a combination of aromatic and polar residues creating a possible regulatory interface. The peptide EDVYVVGTVLR from the C4b-binding protein α-chain exhibits hydrophobic clustering encircled by charged residues, indicating potential for scaffold anchoring or surface binding. These structural features elucidate how complement fragments may influence immunological or matrix interactions inside bioprinted tissues.
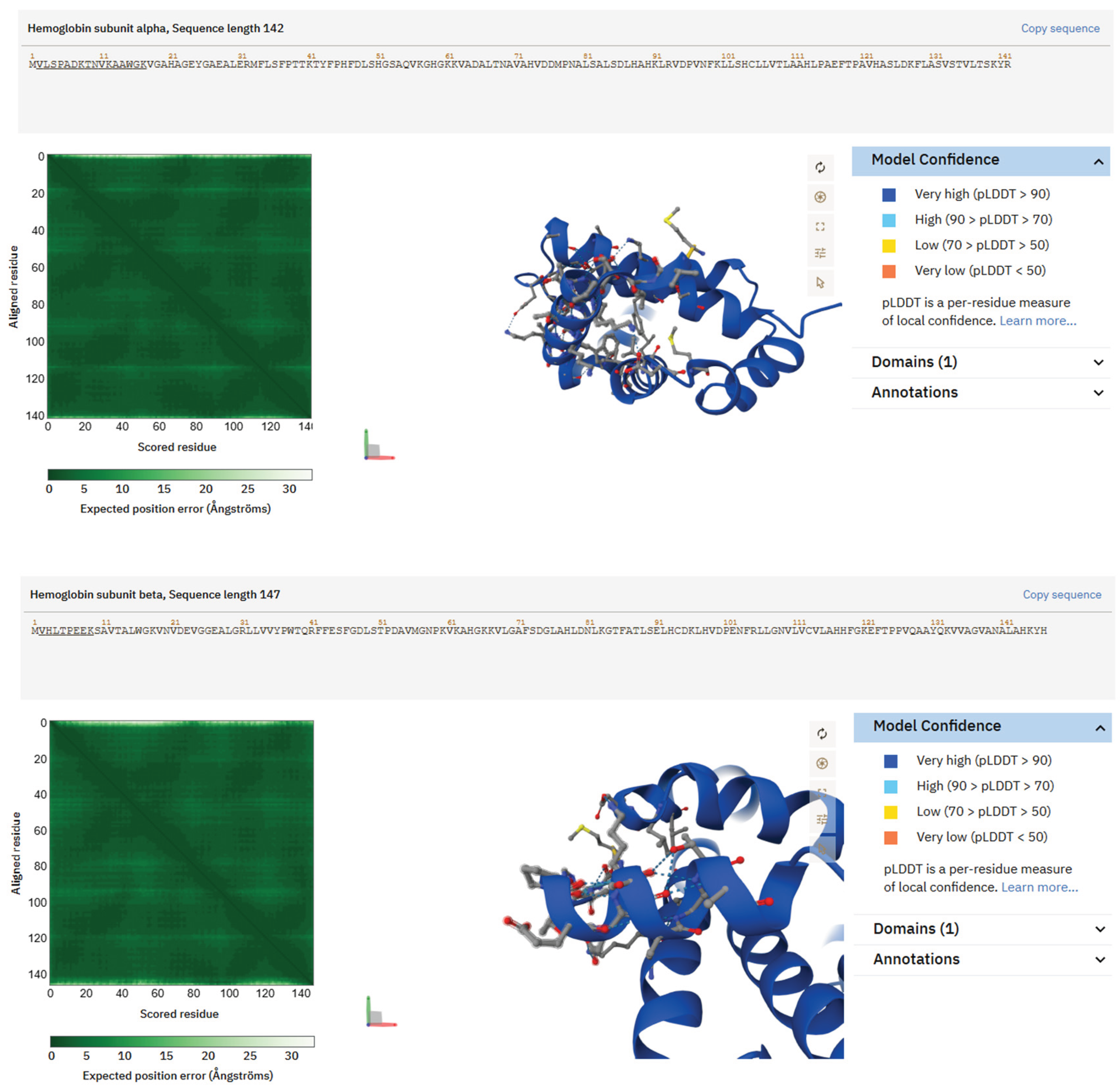
Figure 6.
Structures of hemoglobin-derived peptides predicted by AlphaFold, abundant in saliva from individuals with Type 2 Diabetes. Hb δ/β-like peptide VLGAFSDGLAHLDNLK exhibits a partly helical conformation typical of globin substructures, characterized by alternating hydrophobic and polar residues that create a structured contact surface. (B) The Hb β-chain peptide VHLTPEEK demonstrates a compact shape characterized by a short protruding helix and a heterogeneous distribution of hydrophobic and charged residues. These structural characteristics may facilitate redox regulation, oxygen management, or matrix interactions within bioprinting contexts.
Figure 6.
Structures of hemoglobin-derived peptides predicted by AlphaFold, abundant in saliva from individuals with Type 2 Diabetes. Hb δ/β-like peptide VLGAFSDGLAHLDNLK exhibits a partly helical conformation typical of globin substructures, characterized by alternating hydrophobic and polar residues that create a structured contact surface. (B) The Hb β-chain peptide VHLTPEEK demonstrates a compact shape characterized by a short protruding helix and a heterogeneous distribution of hydrophobic and charged residues. These structural characteristics may facilitate redox regulation, oxygen management, or matrix interactions within bioprinting contexts.