Submitted:
04 December 2025
Posted:
04 December 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Pharmaceutical Management and Biosafety
2.2. Cultivation of C. sowerbii and Morphological Observation
2.3. Sample Preparation for RNA-Seq Analysis
2.4. Bioinformatics Analysis Pipeline
2.5. Differential Expression Analysis
3. Results
3.1. The Symptoms and Features of C. sowerbii Under Water Pollution
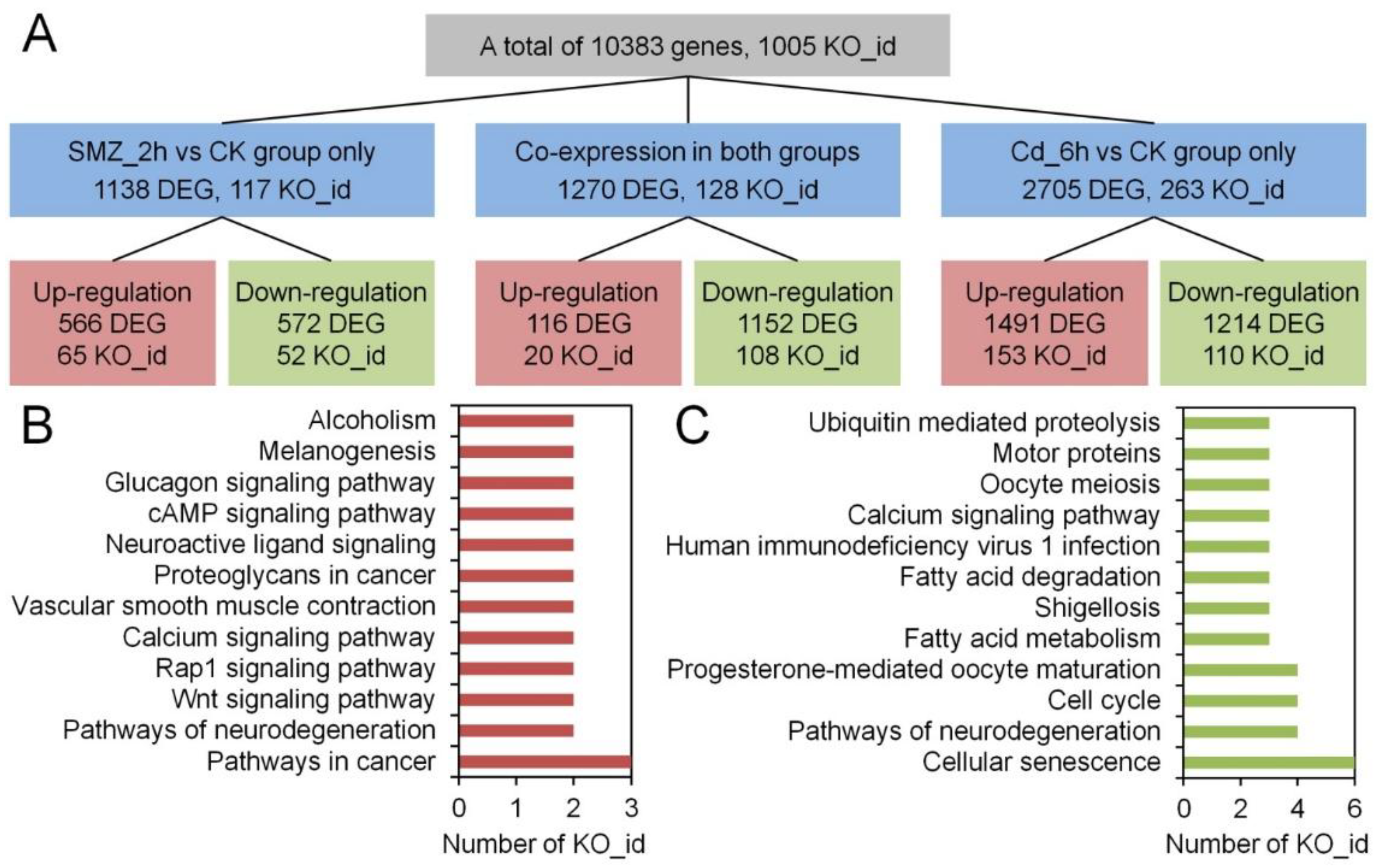
3.2. Transcriptomic Variations of C. sowerbii Under Water Pollution
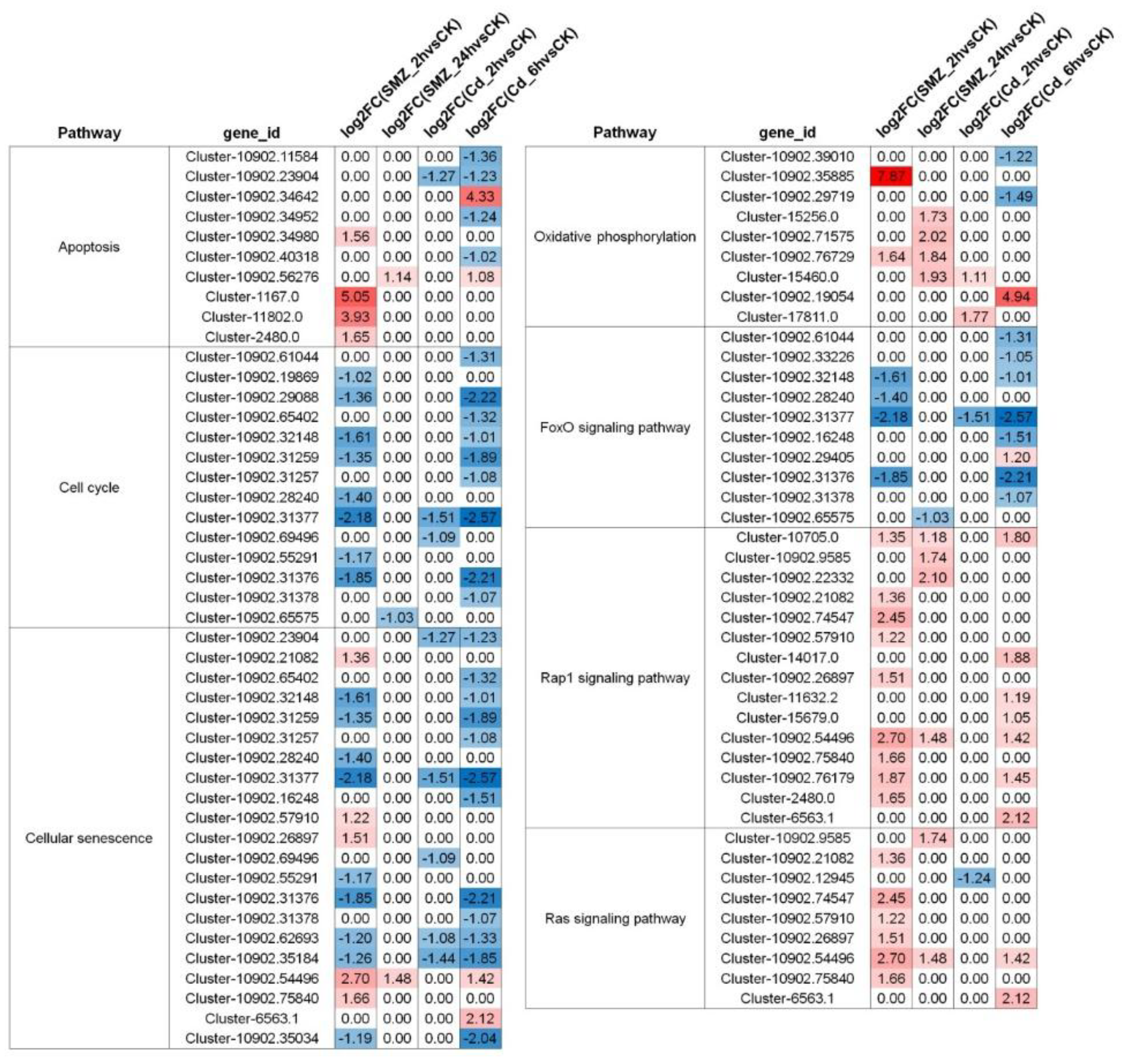
3.3. Metabolic Pathway Alterations in C. sowerbii in Response to Water Pollution
3.4. Alterations in Gene Expression Profiles of C. sowerbii in Response to Water Pollution
4. Discussion
4.1. Similarities and Differences of C. sowerbii Under Different Water Pollution Treatments
4.2. Adaptation and Evolution of Aquatic Lifes Under the Stress of Water Pollution
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- van Vliet, M.T.H.; Thorslund, J.; Strokal, M.; Hofstra, N.; Flörke, M.; Ehalt Macedo, H.; Nkwasa, A.; Tang, T.; Kaushal, S.S.; Kumar, R.; et al. Global river water quality under climate change and hydroclimatic extremes. Nature Reviews Earth & Environment 2023, 4, 687-702. [CrossRef]
- Manca, F.; Benedetti-Cecchi, L.; Bradshaw, C.J.A.; Cabeza, M.; Gustafsson, C.; Norkko, A.M.; Roslin, T.V.; Thomas, D.N.; White, L.; Strona, G. Projected loss of brown macroalgae and seagrasses with global environmental change. Nature Communications 2024, 15, 5344. [CrossRef]
- Kaijser, W.; Musiol, M.; Schneider, A.R.; Prati, S.; Brauer, V.S.; Bayer, R.; Birk, S.; Brauns, M.; Dunne, L.; Enss, J.; et al. Meta-analysis-derived estimates of stressor–response associations for riverine organism groups. Nature Ecology & Evolution 2025. [CrossRef]
- Xie, J.; Wang, T.; Zhang, P.; Zhang, H.; Wang, H.; Wang, K.; Zhang, M.; Xu, J. Effects of multiple stressors on freshwater food webs: Evidence from a mesocosm experiment. Environmental Pollution 2024, 348, 123819. [CrossRef]
- Jiang, X.; Kirsten, K.L.; Qadeer, A. Contaminants in the Water Environment: Significance from the Perspective of the Global Environment and Health. Water 2025, 17, 1257.
- Yu, K.; Mohapatra, S.; Chen, Y.; Jiang, P.; Tong, X. Interactive Effects of Climate Change and Contaminants in Aquatic Ecosystems on Environmental-Human Health. Current Pollution Reports 2025, 11, 46. [CrossRef]
- Ho, J.C.; Michalak, A.M.; Pahlevan, N. Widespread global increase in intense lake phytoplankton blooms since the 1980s. Nature 2019, 574, 667-670. [CrossRef]
- Rajak, P.; Ganguly, A.; Nanda, S.; Mandi, M.; Ghanty, S.; Das, K.; Biswas, G.; Sarkar, S. 14 - Toxic contaminants and their impacts on aquatic ecology and habitats. In Spatial Modeling of Environmental Pollution and Ecological Risk, Shit, P.K., Datta, D.K., Bera, B., Islam, A., Adhikary, P.P., Eds.; Woodhead Publishing: 2024; pp. 255-273.
- Madesh, S.; Gopi, S.; Sau, A.; Rajagopal, R.; Namasivayam, S.K.R.; Arockiaraj, J. Chemical contaminants and environmental stressors induced teratogenic effect in aquatic ecosystem – A comprehensive review. Toxicology Reports 2024, 13, 101819. [CrossRef]
- Jan, S.; Mishra, A.K.; Bhat, M.A.; Bhat, M.A.; Jan, A.T. Pollutants in aquatic system: a frontier perspective of emerging threat and strategies to solve the crisis for safe drinking water. Environmental Science and Pollution Research 2023, 30, 113242-113279. [CrossRef]
- Arambawatta-Lekamge, S.H.; Pathiratne, A.; Rathnayake, I.V.N. Sensitivity of freshwater organisms to cadmium and copper at tropical temperature exposures: Derivation of tropical freshwater ecotoxicity thresholds using species sensitivity distribution analysis. Ecotoxicology and Environmental Safety 2021, 211, 111891. [CrossRef]
- Wang, F.; Xiang, L.; Sze-Yin Leung, K.; Elsner, M.; Zhang, Y.; Guo, Y.; Pan, B.; Sun, H.; An, T.; Ying, G.; et al. Emerging contaminants: A One Health perspective. The Innovation 2024, 5, 100612. [CrossRef]
- Bashir, I.; Lone, F.A.; Bhat, R.A.; Mir, S.A.; Dar, Z.A.; Dar, S.A. Concerns and Threats of Contamination on Aquatic Ecosystems. In Bioremediation and Biotechnology: Sustainable Approaches to Pollution Degradation, Hakeem, K.R., Bhat, R.A., Qadri, H., Eds.; Springer International Publishing: Cham, 2020; pp. 1-26.
- Schar, D.; Klein, E.Y.; Laxminarayan, R.; Gilbert, M.; Van Boeckel, T.P. Global trends in antimicrobial use in aquaculture. Scientific Reports 2020, 10, 21878. [CrossRef]
- Singh, B.K.; Paul, S.; Das, I.; Singha, E.R.; Giri, A. Global Warming and Emerging Contaminants: Impacts on Aquatic Organisms and Their Responses. International Journal of Environmental Research 2025, 19, 186. [CrossRef]
- Grenni, P.; Ancona, V.; Barra Caracciolo, A. Ecological effects of antibiotics on natural ecosystems: A review. Microchemical Journal 2018, 136, 25-39. [CrossRef]
- Xiong, J.-Q.; Govindwar, S.; Kurade, M.B.; Paeng, K.-J.; Roh, H.-S.; Khan, M.A.; Jeon, B.-H. Toxicity of sulfamethazine and sulfamethoxazole and their removal by a green microalga, Scenedesmus obliquus. Chemosphere 2019, 218, 551-558. [CrossRef]
- Zhang, H.; Quan, H.; Song, S.; Sun, L.; Lu, H. Comprehensive assessment of toxicity and environmental risk associated with sulfamethoxazole biodegradation in sulfur-mediated biological wastewater treatment. Water Research 2023, 246, 120753. [CrossRef]
- Peng, P.; Yan, X.; Zhou, X.; Chen, L.; Li, X.; Miao, Y.; Zhao, F. Enhancing degradation of antibiotic-combined pollutants by a hybrid system containing advanced oxidation and microbial treatment, a review. Journal of Hazardous Materials 2024, 480, 136300. [CrossRef]
- Studziński, W.; Gackowska, A.; Kudlek, E.; Przybyłek, M. Environmental and toxicological aspects of sulfamethoxazole photodegradation in the presence of oxidizing agents. Environmental Science and Pollution Research 2025, 32, 4733-4753. [CrossRef]
- Bojarski, B.; Kot, B.; Witeska, M. Antibacterials in Aquatic Environment and Their Toxicity to Fish. Pharmaceuticals 2020, 13, 189.
- Galasso, F.; Frank, A.B.; Foster, W.J. Heavy metal toxicity and its role as a major driver of past biodiversity crises. Communications Earth & Environment 2025, 6, 780. [CrossRef]
- Haider, F.U.; Liqun, C.; Coulter, J.A.; Cheema, S.A.; Wu, J.; Zhang, R.; Wenjun, M.; Farooq, M. Cadmium toxicity in plants: Impacts and remediation strategies. Ecotoxicology and Environmental Safety 2021, 211, 111887. [CrossRef]
- Liu, Y.; Chen, Q.; Li, Y.; Bi, L.; Jin, L.; Peng, R. Toxic Effects of Cadmium on Fish. Toxics 2022, 10, 622. [CrossRef]
- Samarakoon, T.; Fujino, T.; Hagimori, M.; Saito, R. Cadmium uptake and oxidative-stress-induced DNA alterations in the freshwater cladoceran Moina macrocopa (Straus 1820) following consecutive short-term exposure assessments. Limnology 2023, 24, 9-23. [CrossRef]
- El-Sharkawy, M.; Alotaibi, M.O.; Li, J.; Du, D.; Mahmoud, E. Heavy Metal Pollution in Coastal Environments: Ecological Implications and Management Strategies: A Review. Sustainability 2025, 17, 701.
- Yan, H.; Wang, Y.; Wu, M.; Li, Y.; Wang, W.; Zhang, D.; Guo, J.; Fohrer, N.; Li, B.L. Feeding Behavior and Ecological Significance of Craspedacusta sowerbii in a Freshwater Reservoir: Insights from Prey Composition and Trophic Interactions. Biology 2025, 14, 665. [CrossRef]
- Jankowski, T. The freshwater medusae of the world – a taxonomic and systematic literature study with some remarks on other inland water jellyfish. Hydrobiologia 2001, 462, 91-113. [CrossRef]
- Acker, T.S.; Muscat, A.M. The Ecology of Craspedacusta sowerbii Lankester, a Freshwater Hydrozoan. The American Midland Naturalist 1976, 95, 323-336. [CrossRef]
- Lüskow, F.; Väinölä, R.; Lehtiniemi, M.; von Numers, M.; Pakhomov, E.A. Evidence for non-indigenous freshwater jellyfish Craspedacusta sowerbii spreading in Finland. Hydrobiologia 2025. [CrossRef]
- Gießler, S.; Strauss, T.; Schachtl, K.; Jankowski, T.; Klotz, R.; Stibor, H. Trophic Positions of Polyp and Medusa Stages of the Freshwater Jellyfish Craspedacusta sowerbii Based on Stable Isotope Analysis. Biology 2023, 12, 814.
- Marchessaux, G.; Lüskow, F.; Sarà, G.; Pakhomov, E.A. Predicting the current and future global distribution of the invasive freshwater hydrozoan Craspedacusta sowerbii. Scientific Reports 2021, 11, 23099. [CrossRef]
- Marchessaux, G.; Bejean, M. From frustules to medusae: A new culture system for the study of the invasive hydrozoan Craspedacusta sowerbii in the laboratory. Invertebrate Biology 2020, 139, e12308. [CrossRef]
- Folino-Rorem, N.C.; Reid, M.; Peard, T. Culturing the freshwater hydromedusa, Craspedacusta sowerbii under controlled laboratory conditions. Invertebrate Reproduction & Development 2016, 60, 17-27. [CrossRef]
- Winata, K.; Zhu, J.A.; Hanselman, K.M.; Zerbe, E.; Langguth, J.; Folino-Rorem, N.; Cartwright, P. Life Cycle Transitions in the Freshwater Jellyfish Craspedacusta sowerbii. Biology 2024, 13, 1069.
- Zhang, Y.W.; Pan, X.F.; Wang, X.A.; Jiang, W.S.; Liu, Q.; Yang, J.X. Effects of osmotic pressure, temperature and stocking density on survival and sexual reproduction of Craspedacusta sowerbii. Dongwuxue Yanjiu 2016, 37, 90-95. [CrossRef]
- Lüskow, F.; Polgári, B.; Stibor, H.; Schachtl, K.; Abonyi, A. Light increases surface occurrence of the freshwater jellyfish Craspedacusta sowerbii via positive phototaxis. Hydrobiologia 2025. [CrossRef]
- Luk, C.Y.L. The Chinese Freshwater Jellyfish Unbound: Evolution, Nomenclature, and Bioinvasion of Craspedacusta sowerbii, 1880–1941. Historical Studies in the Natural Sciences 2024, 54, 493-520. [CrossRef]
- Gasith, A.; Gafny, S.; Hershkovitz, Y.; Goldstein, H.; Galil, B. The invasive freshwater medusa Craspedacusta sowerbii Lankester, 1880 (Hydrozoa: Olindiidae) in Israel. Aquatic Invasions 2011, 6, S147-S152. [CrossRef]
- Caputo, L.; Fuentes, R.; Woelfl, S.; Castañeda, L.E.; Cárdenas, L. Phenotypic plasticity of clonal populations of the freshwater jellyfish Craspedacusta sowerbii (Lankester, 1880) in Southern Hemisphere lakes (Chile) and the potential role of the zooplankton diet. Austral Ecology 2021, 46, 1192-1197. [CrossRef]
- Lüskow, F.; Boersma, M.; López-González, P.J.; Pakhomov, E.A. Genetic variability, biomass parameters, elemental composition and energy content of the non-indigenous hydromedusa Craspedacusta sowerbii in North America. Journal of Plankton Research 2022, 45, 82-98. [CrossRef]
- Lüskow, F.; López-González, P.J.; Pakhomov, E.A. Freshwater jellyfish in northern temperate lakes: Craspedacusta sowerbii in British Columbia, Canada. Aquatic Biology 2021, 30, 69-84.
- Marchessaux, G.; Bejean, M. Growth and ingestion rates of the freshwater jellyfish Craspedacusta sowerbii. Journal of Plankton Research 2020, 42, 783-786. [CrossRef]
- Sreeram, M.P.; Prasad, R.; Sreekumar, K.M.; Raju, A.K.; Augustina, T.A.X.; Lüskow, F.; Saravanan, R. Post-flooding blooms of the non-indigenous freshwater jellyfish Craspedacusta sowerbii Lankester, 1880 in Kollam District of Kerala, India. Journal of Plankton Research 2024, 00, 1-4. [CrossRef]
- Lucas, K.; Colin, S.P.; Costello, J.H.; Katija, K.; Klos, E. Fluid Interactions That Enable Stealth Predation by the Upstream-Foraging Hydromedusa Craspedacusta sowerbyi. The Biological Bulletin 2013, 225, 60-70. [CrossRef]
- Xu, D.; Xie, Y.; Li, J. Toxic effects and molecular mechanisms of sulfamethoxazole on Scenedesmus obliquus. Ecotoxicology and Environmental Safety 2022, 232, 113258. [CrossRef]
- Diogo, B.S.; Rodrigues, S.; Golovko, O.; Antunes, S.C. From bacteria to fish: ecotoxicological insights into sulfamethoxazole and trimethoprim. Environmental Science and Pollution Research 2024, 31, 52233-52252. [CrossRef]
- Zhou, J.; Yun, X.; Wang, J.; Li, Q.; Wang, Y.; Zhang, W.; Fan, Z. Biological toxicity of sulfamethoxazole in aquatic ecosystem on adult zebrafish (Danio rerio). Scientific Reports 2024, 14, 9401. [CrossRef]
- Lin, T.; Yu, S.; Chen, Y.; Chen, W. Integrated biomarker responses in zebrafish exposed to sulfonamides. Environmental Toxicology and Pharmacology 2014, 38, 444-452. [CrossRef]
- Roy, D.; Mitra, A.; Sen, B.M.; Homechaudhuri, S. Biochemical Responses in Zebra Fish (Danio rerio) on Acute Cadmium Exposure Under Temperature Variations. Proceedings of the Zoological Society 2024, 77, 164-172. [CrossRef]
- Yuan, W.; Liang, Y.; Xia, X.; Xie, Y.; Lan, S.; Li, X. Protection of Danio rerio from cadmium (Cd2+) toxicity using biological iron sulfide composites. Ecotoxicology and Environmental Safety 2018, 161, 231-236. [CrossRef]
- Gao, C.-H.; Cao, H.; Ju, F.; Xiao, K.-Q.; Cai, P.; Wu, Y.; Huang, Q. Emergent transcriptional adaption facilitates convergent succession within a synthetic community. ISME Communications 2021, 1, 46. [CrossRef]
- Wang, H.; Xu, Y.; Zhang, Z.; Zhang, G.; Tan, C.; Ye, L. Development and application of transcriptomics technologies in plant science. Crop Design 2024, 3, 100057. [CrossRef]
- Jing, L.; Wang, H.; Xia, S.; Shao, Q. Wnt/Ca(2+) signaling: Dichotomous roles in regulating tumor progress (Review). Oncol Lett 2025, 30, 399. [CrossRef]
- Sanchez-Collado, J.; Lopez, J.J.; Jardin, I.; Salido, G.M.; Rosado, J.A. Cross-Talk Between the Adenylyl Cyclase/cAMP Pathway and Ca2+ Homeostasis. In Reviews of Physiology, Biochemistry and Pharmacology, Pedersen, S.H.F., Ed.; Springer International Publishing: Cham, 2021; pp. 73-116.
- Schmidt, M.F.; Gan, Z.Y.; Komander, D.; Dewson, G. Ubiquitin signalling in neurodegeneration: mechanisms and therapeutic opportunities. Cell Death & Differentiation 2021, 28, 570-590. [CrossRef]
- Dantuma, N.P.; Bott, L.C. The ubiquitin-proteasome system in neurodegenerative diseases: precipitating factor, yet part of the solution. Front Mol Neurosci 2014, 7, 70. [CrossRef]
- Ajoolabady, A.; Pratico, D.; Bahijri, S.; Tuomilehto, J.; Uversky, V.N.; Ren, J. Hallmarks of cellular senescence: biology, mechanisms, regulations. Experimental & Molecular Medicine 2025, 57, 1482-1491. [CrossRef]
- Minchin, D.; Caffrey, J.M.; Haberlin, D.; Germaine, D.; Walsh, C.; Boelens, R.; Doyle, T.K. First observations of the freshwater jellyfish Craspedacusta sowerbii Lankester, 1880 in Ireland coincides with unusually high water temperatures. Bioinvasions Records 2016, 5, 67-74. [CrossRef]
- Schifani, E.; Viviano, A.; Viviano, R.; Naselli-Flores, L.; Marrone, F. Different lineages of freshwater jellyfishes (Cnidaria, Olindiidae, Craspedacusta) invading Europe: another piece of the puzzle from Sicily, Italy. Limnology 2019, 20, 143-151. [CrossRef]
- Seçer, B. New locality records of invasive freshwater jellyfish Craspedacusta sowerbii (Lankester, 1880) in Türkiye. Limnology and Freshwater Biology 2025, 2025, 298-301.
- Moore, J.P.; Green, H.C.; Stewart, D.J.; Lüskow, F.; Wilder, M.L. Invasive freshwater jellyfish (Craspedacusta cf. sowerbii) in the Hudson River basin, NYS: comparisons of detection methods. Hydrobiologia 2025. [CrossRef]
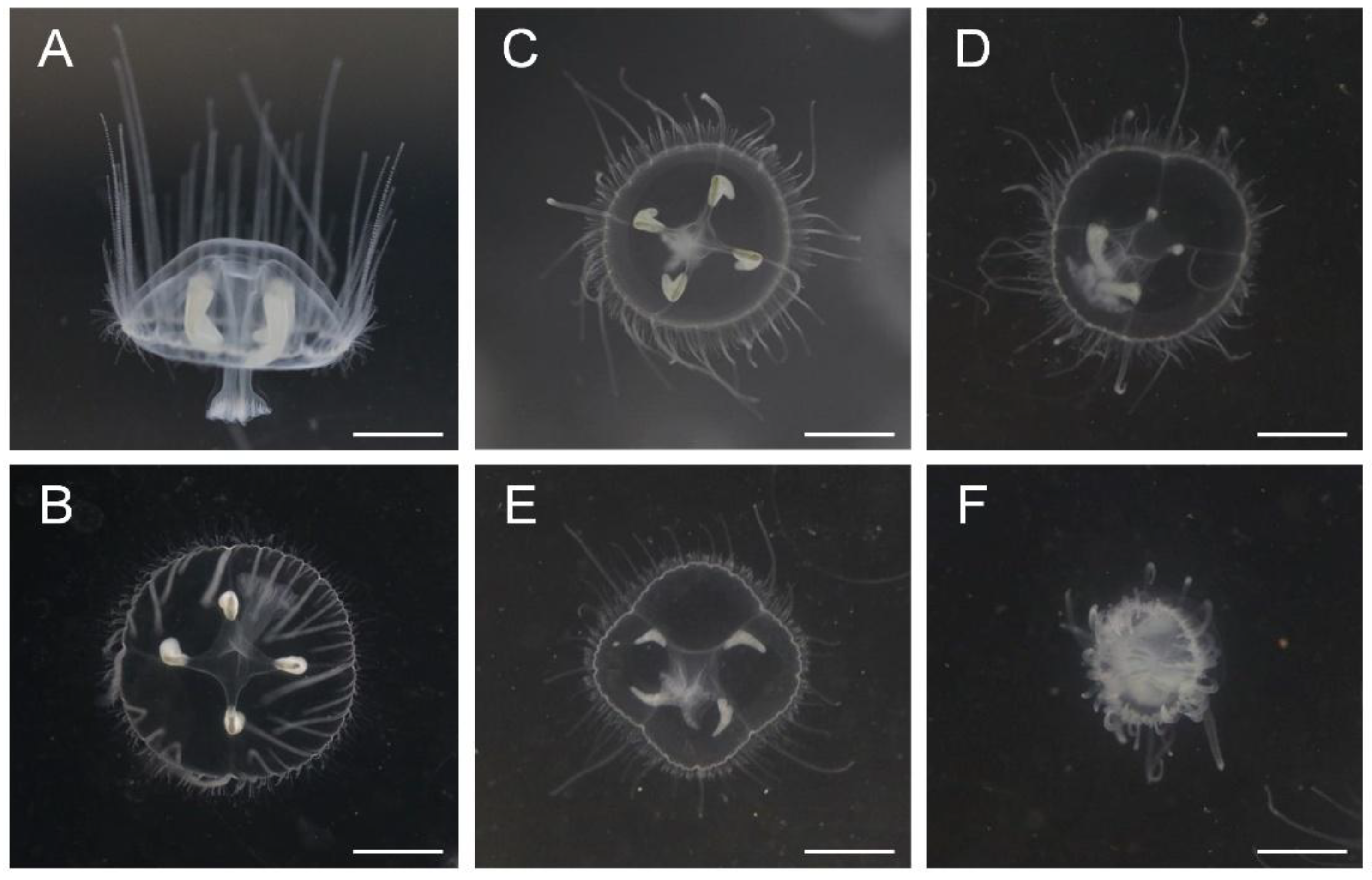
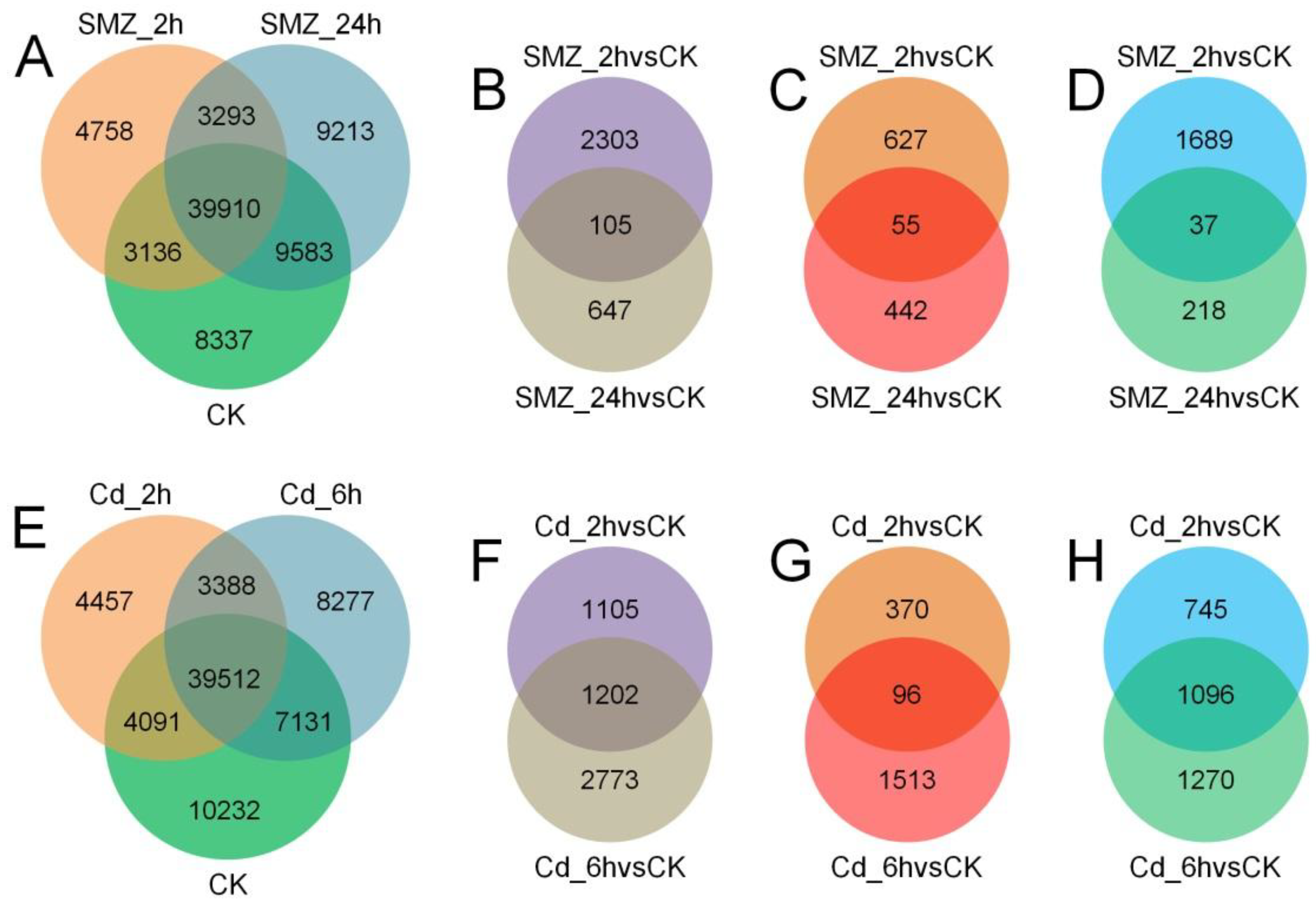
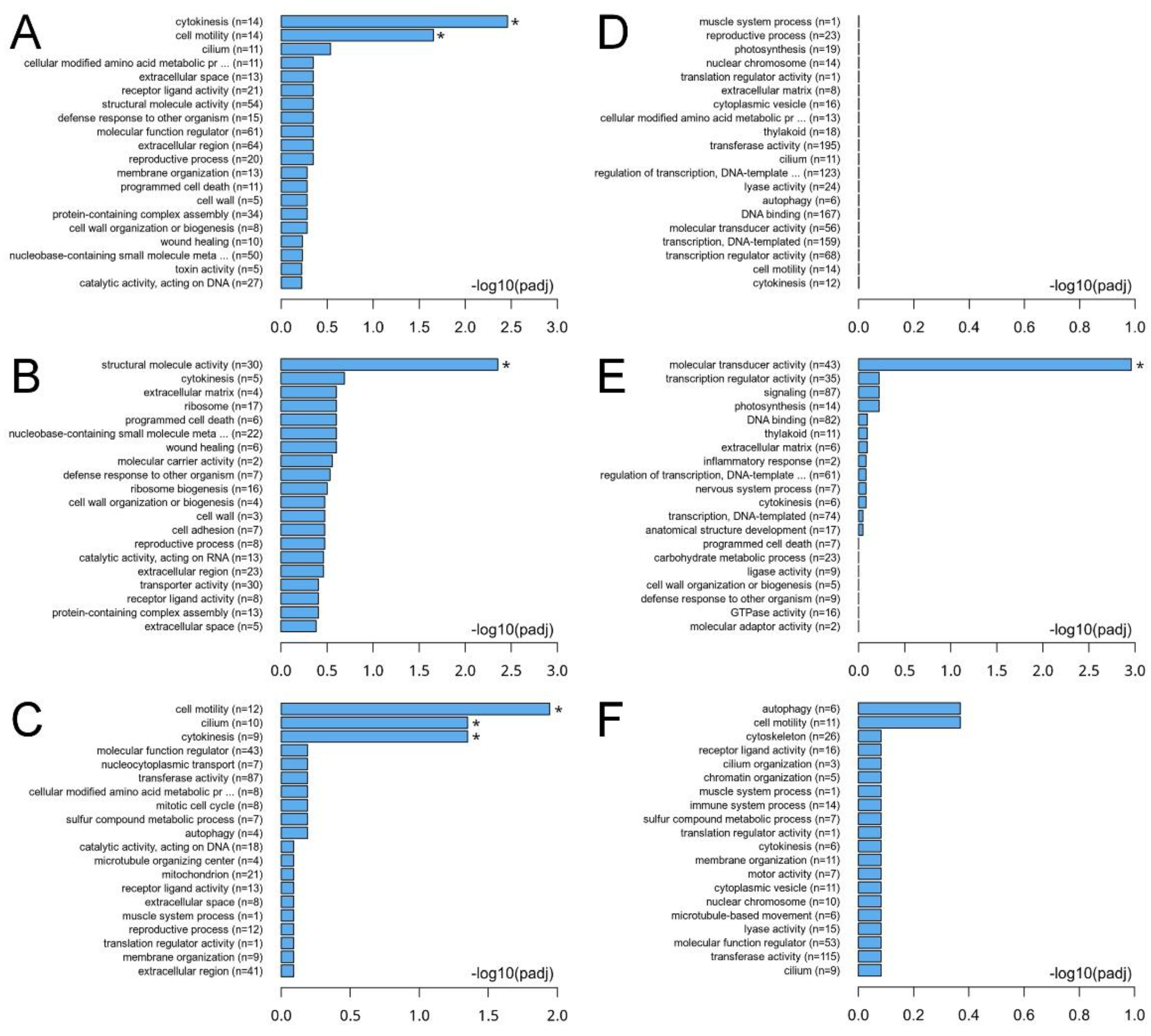
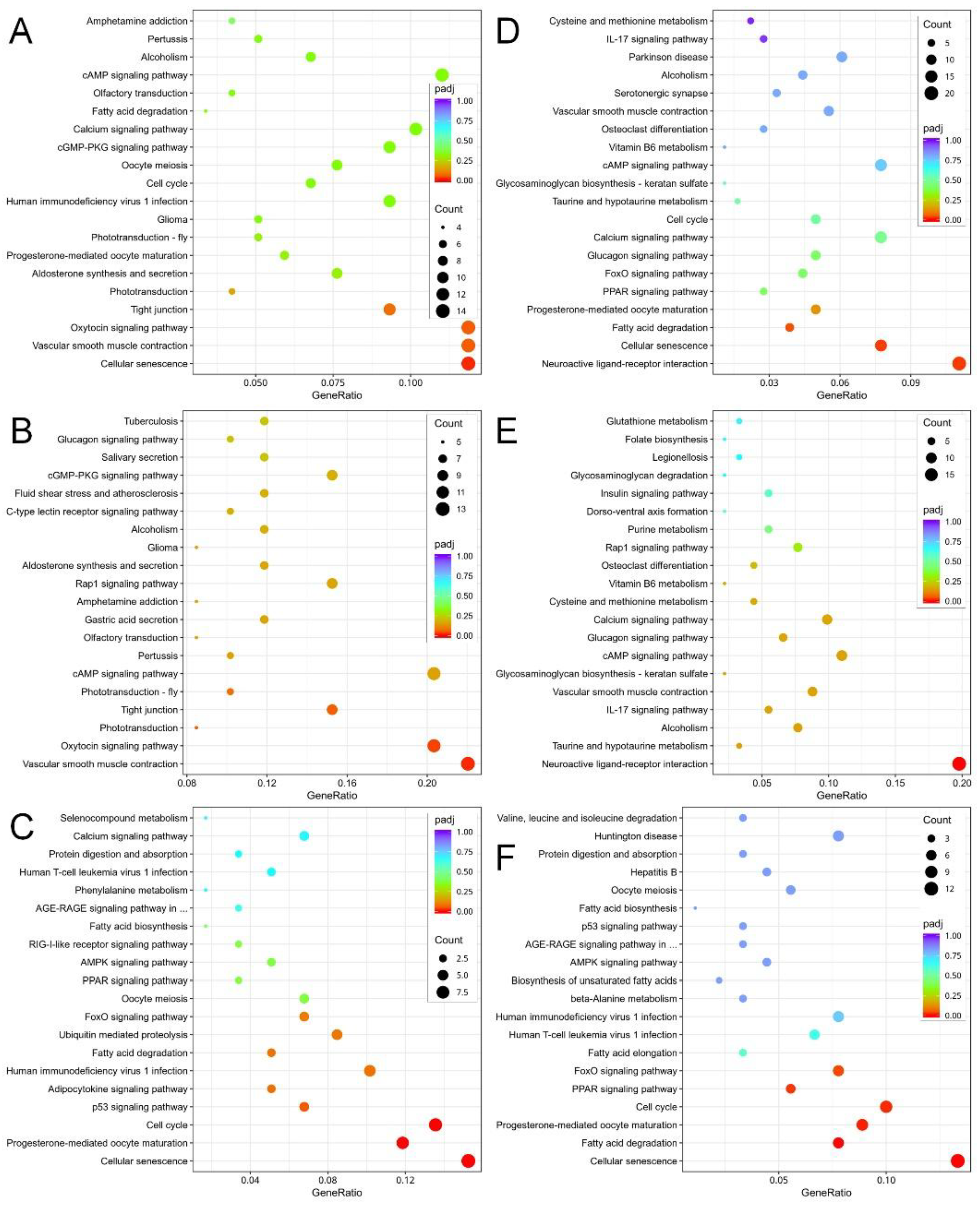

| Group | 1 h | 2 h | 6 h | 12 h | 24 h |
| CK | 42.3 ± 8.1 | 43.9 ± 5.6 | 45.5 ± 6.0 | 43.1 ± 7.2 | 43.5 ± 6.4 |
| SMZ | 43.9 ± 10.8 | 41.7 ± 13.8 | 40.4 ± 12.3 | 41.8 ± 7.5 | 23.4 ± 8.7 |
| Cd | 42.7 ± 6.9 | 20.1 ± 8.3 | 4.3 ± 5.0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





