Introduction
Mpox is a zoonotic disease caused by two distinct clades of mpox virus (MPXV); Clade I and Clade II, each divided into two clades a and b [
1,
2]. Although the virus can be transmitted through zoonotic spillover, in recent years, at least three different lineages have emerged and spread through sustained human-to-human transmission: Clade IIb/sh2017, Clade Ib/sh2023, and Clade Ia/sh2024 [
2,
3,
4].
The Democratic Republic of the Congo (DRC) has traditionally been the epicenter of Clade I MPXV. For several decades, the country has seen an increasing number of outbreaks caused by zoonotic spillover of Clade Ia [
5,
6]. In 2024, Clade Ib/sh2023 emerged, prompting the World Health Organization to declare mpox for the second time a public health emergency of international concern [
7]. Shortly thereafter, Clade Ia/sh2024 was detected in the capital Kinshasa, where both lineages continue to co-circulate [
3,
8]. Despite efforts to curb the epidemic, the DRC continues to report the highest number of overall suspected and confirmed mpox cases worldwide [
9]. Between 01 January and 20 July 2025, the Ministry of Public Health of DRC through the Institut National de Santé Publique reported 61,532 suspected cases of mpox (14,351 PCR confirmed) across all 26 provinces.
On the other hand, human cases of MPXV Clade II were first observed in West Africa [
10]. Following the emergence of Clade IIb/sh2017 in Nigeria in 2017 [
11], the virus spread internationally and caused the global 2022 mpox outbreak that affected more than 100 countries [
12,
13]. Although global case numbers have since subsided, Sierra Leone is currently experiencing a large Clade IIb/sh2017 outbreak [
14]. In contrast, throughout the global epidemic, the DRC had not reported any cases of Clade IIb/sh2017 to date.
Here, we report the first two Clade IIb/sh2017 mpox cases identified in the DRC, imported from West Africa by the index case and locally transmitted to his wife, with phylogenetic linkage to the ongoing outbreak in Sierra Leone. Samples were collected as part of a routine countrywide mpox surveillance program and were exempt from informed consent procedures. Permission to use anonymised data from the Institut National de Santé Publique for this report was granted by the Ethics Committee of the Kinshasa School of Public Health (ESP-UNIKIN, Ethics Approval Number ESP/CE/238/2024).
Case Investigations
Between 21 and 26 July 2025, an adult man presented himself to the General Referral Hospital of Kinkole (HGRK – Nsele Health Zone) in Kinshasa, DRC with vesicle lesions. The patient self-reported symptom onset two days before the medical examination, with fever and headache, followed by the appearance of vesicle lesions on the face, palms, and penis, as well as inguinal lymphadenopathy. He had self-medicated for the fever with acetaminophen. He reported no contact or exposure to animals during the preceding three weeks, nor any sexual activities or known contact with MPXV-infected individuals. He also reported a prior history of smallpox vaccination. He had recently traveled from Ivory Coast to DRC, via transit through Jomo Kenyatta International Airport, Kenya.
Suspecting mpox, the physician immediately transferred the patient into isolation at the mpox treatment unit. Blood and vesicle swab specimens were collected for laboratory analysis. The dual HIV/syphilis Combo rapid diagnostic test (SD Biosensor, South Korea) was negative for both infections. Initial testing with the Xpert Mpox assay on the GeneXpert system (Cepheid, US), which detects Clade II and non-variola Orthopoxviruses including Clade I MPXV, at HGRK detected MPXV Clade II in the vesicle swab sample. Subsequently, vesicle and blood specimens were shipped to the Institut National de Recherche Biomédicale (INRB), Kinshasa for diagnostic confirmation and whole-genome sequencing of MPXV.
As part of routine contact tracing, nine individuals were identified as high-risk contacts, including the patient’s wife and their three children, one uncle, one nephew, one hairdresser, one renter and one healthcare worker. Because of sexual contacts with the index case, a nasopharyngeal swab was collected from the patient’s wife on day 2 after her last exposure to the patient. PCR testing performed at HGRK with the Xpert Mpox assay (Cepheid, US) confirmed the presence of Clade II MPXV DNA in the sample. Consequently, she was immediately admitted to the mpox treatment unit and reported having a fever, swelling of the vulva and itching. She had vesicle lesions on the face, palms, vulva, and trunk. Vesicle and nasopharyngeal swabs were collected and shipped to INRB, Kinshasa. A follow-up among the other contacts for 21 days is still ongoing.
At INRB, vesicle swab samples of both patients were re-tested with the RADI RP022 mpox detection kit on the RADIONE system (KH Medical, Republic of Korea), a fully automated point-of-care molecular diagnostic device. In addition, real-time PCR was performed in triplicate on each specimen from both patients (vesicle and blood from the index case, and vesicle and nasopharyngeal swab from his wife) using the RADI FAST RV015R mpox detection kit (KH Medical, Republic of Korea), also following the manufacturer’s instructions.
Following PCR confirmation, multiplex tiling PCR was performed using the Clade IIb MPXV primer pools designed by Chen et al [
15]. The library was also prepared in triplicate using the rapid sequencing DNA V14 barcoding kit (SQK-RBK114.96; Oxford Nanopore Technologies (ONT), Oxford, UK), following the manufacturer’s instructions. Sequencing library was loaded on a R10.4.1 flow cell and run on the GridION sequencer.
MPXV consensus genomes were generated by processing concatenated FASTQ files using the artic-mpxv-nf workflow v2.1.0 (
https://github.com/artic-network/artic-mpxv-nf) and Clade II MPXV genome (GenBank ID: NC_063383.1) was used as reference. The Nextclade online tool (
https://clades.nextstrain.org/) was used to assign the clade of MPXV genomes. Multiple sequence alignment against the Clade II MPXV reference genome (GenBank ID: NC_063383.1) was performed using SQUIRREL (
https://github.com/aineniamh/squirrel). Phylogenetic tree was inferred using IQ-TREE v2.1.4 [
16] with the HKY substitution model [
17].
PCR results obtained from the RADIONE indicated amplification cycle threshold (Ct) values of 19.09 and 19.03 for Clade II MPXV in vesicle samples from the index case and his wife, respectively. The RADI FAST real-time PCR assay showed mean Ct values for Clade II of 21.47 and 28.62 in the vesicle and blood specimen triplicates from the first patient, and mean Ct of 21.23 and 34.68 in the vesicle and nasopharyngeal specimen triplicates from the patient’s wife (
Table 1).
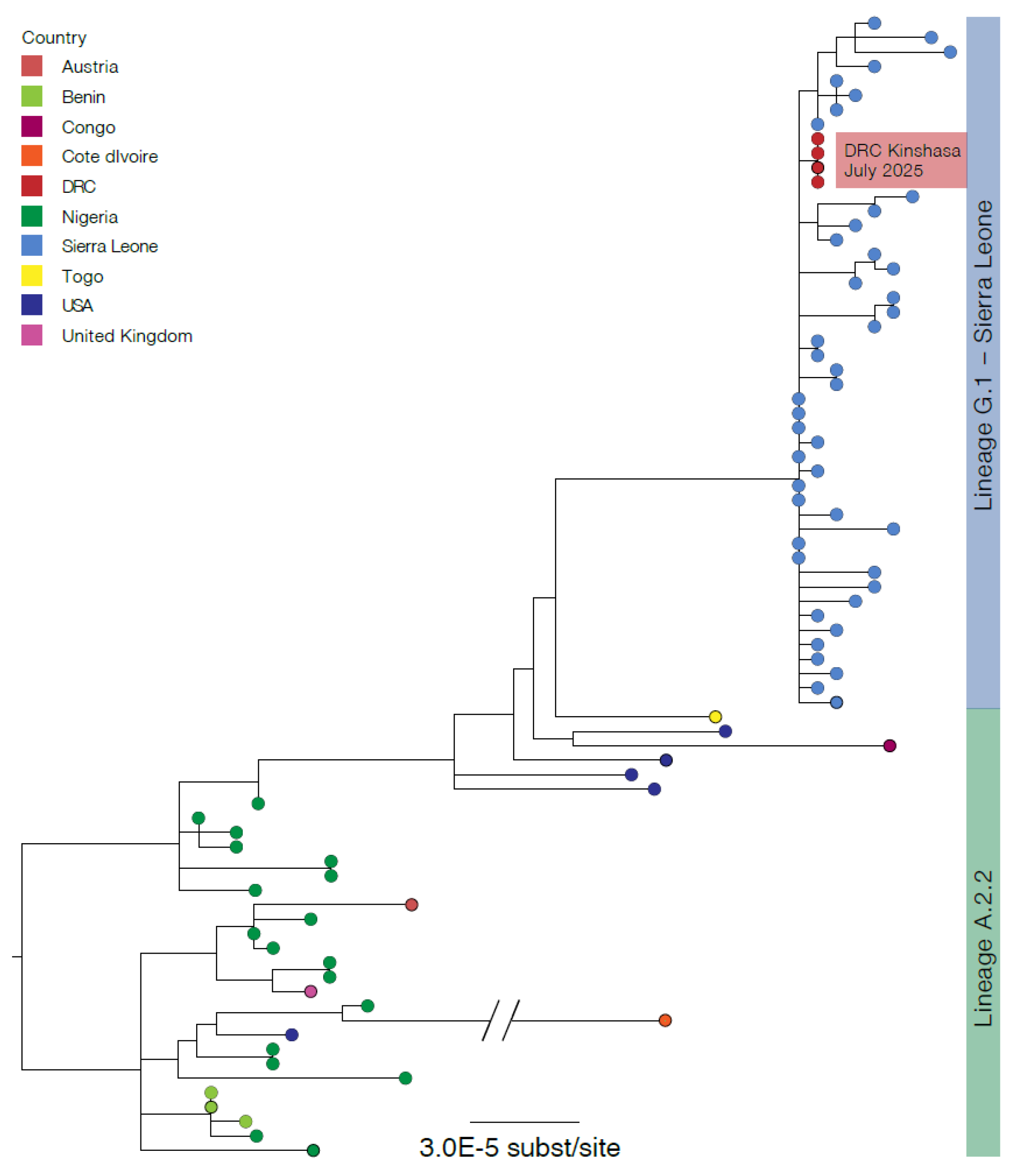
Of the index patient, MPXV genomes were generated from the vesicle swab and blood sample with horizontal genome coverages of 94.19% and 93.39%, respectively. Of the index patient’s wife, the vesicle and nasopharyngeal swabs generated MPXV genomes with horizontal genome coverages of 93.90% and 81.85%, respectively. The four MPXV genomes were similar, assigned to Clade IIb/sh2017 within the newly designated lineage G.1, and clustered with MPXV Clade IIb sequences recently identified in Sierra-Leone (
https://virological.org/t/genomic-epidemiology-of-mpox-virus-in-sierra-leone/995) (
Figure 1).
Conclusions
The co-circulation of Clade Ib/sh2023 and Clade Ia/sh2024, with the novel importation of Clade IIb/sh2017 and its potential spread in Kinshasa -the largest metropolitan area of the DRC, with international and national connections- warrants enhanced genomic and epidemiological surveillance in the region. This finding underscores the urgent need to improve rapid detection and isolation of mpox cases, enhance contact-tracing efforts as well as strengthening of genomic surveillance and implementation of vaccine strategies to mitigate the risk of widespread dissemination of Clade IIb/sh2017 and other MPXV variants in general across DRC provinces.
Dr. Tony Wawina-Bokalanga is a postdoct researcher at the Institute of Tropical Medicine in Antwerp, Belgium. His research interests include emerging and reemerging viral diseases, with a focus on whole and targeted genome sequencing, phylogenetics, immunogenetics, and public health interventions.
Author Contributions
TWB drafted the first manuscript; TWB, EKL, JJMT and PMK supervised the lab work; PAB, CMK, JSM, ICB, FCK and JTS performed the wet lab; RLN and EMB handled the shipment and preparation of specimens; EKL, PPT, GLN, and ELL conducted Bioinformatics analysis; CN, JKM, NN, STM, SJ, CK, AL, PA and DM contributed to the case investigations; TWB, EKL, JJMT and PMK contributed to data acquisition and interpretation; AR built the phylogenetic tree; TWB, EKL, CN, LL, LS, AAA, DJ, JCMC, ST, NL, AOT, AWR, NAH, JK, SAM, MP, KV, AR, JJMT, PMK edited and cross-reviewed the manuscript. All authors reviewed and approved the final version.
Funding
The project or effort depicted is sponsored in part by the Belgian Directorate-General for Development Cooperation and Humanitarian Aid; the Research Foundation Flanders (“Fonds voor Wetenschappelijk Onderzoek–Vlaanderen”, grant number G069725N and G096222N); Institute of Tropical Medicine Structurele Onderzoeksfinanciering (Flemish Government; Science, Technology, and Innovation); the Africa CDC (INV-018278), the Global Health European and Developing Countries Clinical Trials Partnership 3 (EDCTP3) Joint Undertaking (Grant 101195465; MBOTE-SK); the US Department of Defense Threat Reduction Agency; the US Department for Agriculture Research Service (HDTRA1-21-1-0040); the Canadian Institutes of Health Research; the International Mpox Research Consortium (IMReC) through funding from the Canadian Institutes of Health Research and International Development Research Centre (grant no. MRR-184813); the US NIAID/NIH grant number U01AI151799 through Center for Research in Emerging Infectious Disease-East and Central Africa (CREID-ECA) and the Agence Française de Développement through the AFROSCREEN project (grant agreement CZZ3209, coordinated by ANRS-MIE Maladies infectieuses émergentes in partnership with Institut de Recherche pour le Développement (IRD) and Pasteur Institute). We acknowledge the support of the Wellcome Trust (Collaborators Award 206298/Z/17/Z and 313694/Z/24/Z, ARTIC network). TWB acknowledges funding from the European Union (FORTIFIEDx project under the Horizon Europe research and innovation programme and grant agreement no. 101092049). Views and opinions expressed are those of the author(s) only and do not necessarily reflect those of the European Union or the granting authority European Union’s Horizon Europe research and innovation programme. Neither the European Union nor the granting authority can be held responsible for them.
Data Availability Statement
Genomic sequences recovered in this work have been submitted to Pathoplexus database and have the following accession numbers: PP_0033A47.1; PP_0033A55.1; PP_0033A63.1; and PP_0033A71.1.
Acknowledgments
We gratefully acknowledge all data contributors, including the authors and their originating laboratories, for sharing MPXV sequences and metadata via the GISAID Initiative (EPI_ISL_19561134, EPI_ISL_19842849, and EPI_ISL_19957792), the International Nucleotide Sequence Database Collaboration (OP331336.1, OP422631.1 – 633.1, OP450997.1, OR113690.1, PP852955.1, PP_000ZLHE.1, PP_000ZM62.1, PP_000ZM70.1, PP_000ZMGG.1, PP_000ZMWM.1, PP_000ZNEK.1, PP_000ZNM4.1, PP_000ZPPZ.1, PP_000ZPVM.1, PP_000ZPBQ.1, PP_000ZNZE.1, PP_000ZP52.1, PP_000ZPFG.1, PP_000ZPST.1, PP_000ZPZD.1, PP_000ZQBP.1, PP_000ZQRU.1, PQ159993.1, PQ834953.1, PV584374.1, and PV584375.1), and Pathoplexus (PP_002Y1RH.2, PP_002Y1SF.2, PP_002Y1TD.1, PP_002Y1UB.1, PP_002Y1V9.1, PP_002Y1W7.1, PP_002Y1X5.1, PP_002Y1Y3.1, PP_002Y1Z1.1, PP_002Y20Z.1, PP_002Y21X.1, PP_002Y22V.1, PP_002Y23T.1, PP_002Y24R.1, PP_002Y25P.1, PP_002Y26M.1, PP_002Y27K.1, PP_002Y28H.1, PP_002Y29F.1, PP_002Y2AD.1, PP_002Y2BB.1, PP_002Y2C9.1, PP_002Y2D7.1, PP_002Y2E5.1, PP_002Y2F3.1, PP_002Y2G1.1, PP_002Y2HY.1, PP_002Y2JW.1, PP_002Y2KU.1, PP_002Y2LS.1, PP_002Y2MQ.1, PP_002Y2NN.1, PP_002Y2PL.1, PP_002Y2QJ.1, PP_002Y2RG.1, PP_002Y2SE.1, PP_002Y2TC.1, PP_002Y2UA.1, PP_002Y2V8.1, PP_002Y2W6.1, PP_002Y2X4.1, PP_002Y2Y2.1, PP_002Y2Z0.1, and PP_002Y30Y.1), which were used in this study. We are also grateful for the support of the mpox investigation team at the Hôpital Général de Référence de Kinkole. Lastly, we thank Nelson Mapenzi-Kashali, Ange Ponga-Museme and the entire team from INRB’s genomic sequencing laboratory.
Conflicts of Interest
None declared.
References
- World Health Organization (WHO). Monkeypox: experts give virus variants new names. 2022 [cited September 20, 2024]; Available from: https://www.who.int/news/item/12-08-2022-monkeypox--experts-give-virus-variants-new-names.
- Vakaniaki EH, Kacita C, Kinganda-Lusamaki E, O'Toole A, Wawina-Bokalanga T, Mukadi-Bamuleka D, et al. Sustained human outbreak of a new MPXV clade I lineage in eastern Democratic Republic of the Congo. Nat Med. 2024 Jun 13.
- Wawina-Bokalanga T, Merritt S, Kinganda-Lusamaki E, Jansen D, Halbrook M, O'Toole A, et al. Epidemiology and phylogenomic characterisation of two distinct mpox outbreaks in Kinshasa, DR Congo, involving a new subclade Ia lineage: a retrospective, observational study. Lancet. 2025 Jul 5;406(10498):63-75.
- Ruis C, Lusamaki E, O'Toole A, Otieno JR, Colquhoun R, Roemer C, et al. A systematic nomenclature for mpox viruses causing outbreaks with sustained human-to-human transmission. Nat Med. 2025 Jul 29.
- Bangwen E, Diavita R, De Vos E, Vakaniaki EH, Nundu SS, Mutombo A, et al. Suspected and confirmed mpox cases in DR Congo: a retrospective analysis of national epidemiological and laboratory surveillance data, 2010-23. Lancet. 2025 Feb 1;405(10476):408-19.
- Kinganda-Lusamaki E, Amuri-Aziza A, Fernandez-Nunez N, Makangara-Cigolo JC, Pratt C, Vakaniaki EH, et al. Clade I mpox virus genomic diversity in the Democratic Republic of the Congo, 2018-2024: Predominance of zoonotic transmission. Cell. 2024 Oct 24.
- World Health Organization (WHO). WHO Director-General declares mpox outbreak a public health emergency of international concern. 2024 [cited August 21, 2024]; Available from: https://www.who.int/news/item/14-08-2024-who-director-general-declares-mpox-outbreak-a-public-health-emergency-of-international-concern.
- Wawina-Bokalanga T, Akil-Bandali P, Kinganda-Lusamaki E, Lokilo E, Jansen D, Amuri-Aziza A, et al. Co-circulation of monkeypox virus subclades Ia and Ib in Kinshasa Province, Democratic Republic of the Congo, July to 24. Euro Surveill. 2024 Sep;29(38). 20 August.
- World Health Organization (WHO). Global Mpox Trends. July 28, 2025 [cited July 28, 2025]; Available from: https://worldhealthorg.shinyapps.io/mpx_global/.
- Likos AM, Sammons SA, Olson VA, Frace AM, Li Y, Olsen-Rasmussen M, et al. A tale of two clades: monkeypox viruses. J Gen Virol. 2005 Oct;86(Pt 10):2661-72.
- Yinka-Ogunleye A, Aruna O, Ogoina D, Aworabhi N, Eteng W, Badaru S, et al. Reemergence of Human Monkeypox in Nigeria, 2017. Emerg Infect Dis. 2018 Jun;24(6):1149-51.
- World Health Organization. Multi-country monkeypox outbreak in non-endemic countries. [cited May 24, 2022]; Available from: https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON385.
- Wawina-Bokalanga T, Vanmechelen B, Logist AS, Bloemen M, Laenen L, Bontems S, et al. A retrospective genomic characterisation of the 2022 mpox outbreak in Belgium, and in vitro assessment of three antiviral compounds. EBioMedicine. 2024 Dec;110:105488.
- Mitja O, Watson-Jones D, Choi EM, Jalloh MB, Sahr F. Clade IIb mpox outbreak in Sierra Leone. Lancet. 2025 Jun 28;405(10497):2274-5.
- Chen NFG, Chaguza C, Gagne L, Doucette M, Smole S, Buzby E, et al. Development of an amplicon-based sequencing approach in response to the global emergence of mpox. PLoS Biol. 2023 Jun;21(6):e3002151.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol. 2020 ;37(5):1530-4. 1 May.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol Biol Evol. 2018 Feb 1;35(2):518-22.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).