Submitted:
14 November 2025
Posted:
17 November 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Monozygotic Twin TSD Cohort
2.2. RNA Sequencing
2.3. DNA Methylation
2.4. Identification of Differentially Expressed Genes and Differentially Methylated Probes
2.5. Expression Quantitative Trait Methylation (eQTM)
2.6. Gene Set Enrichment Analysis (GSEA)
3. Results
3.1. Data Exploration and Statistical Model Generation
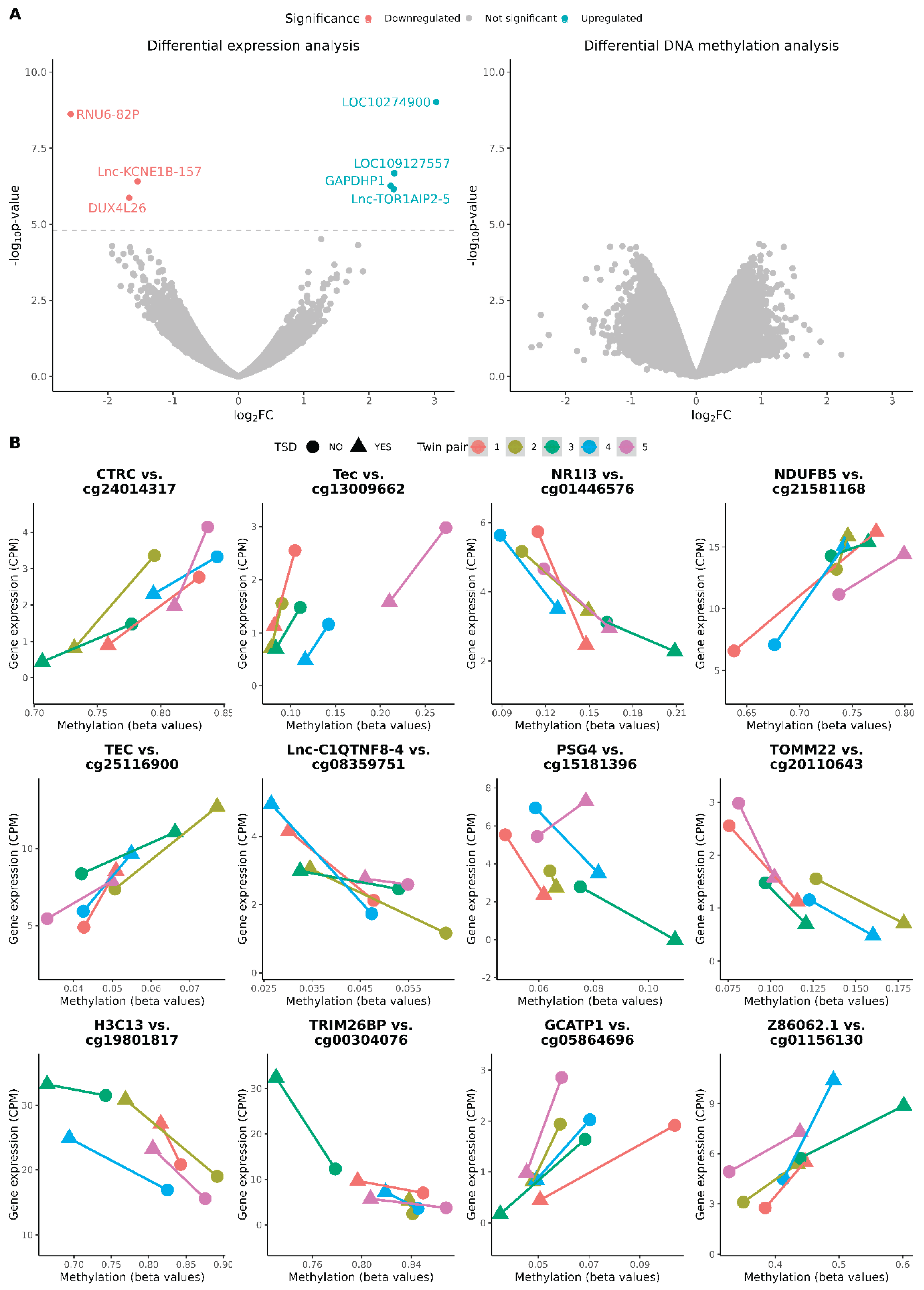
3.2. Differential Expression and Methylation Analyses
3.3. Expression Quantitative Trait Methylation (eQTM) Analysis
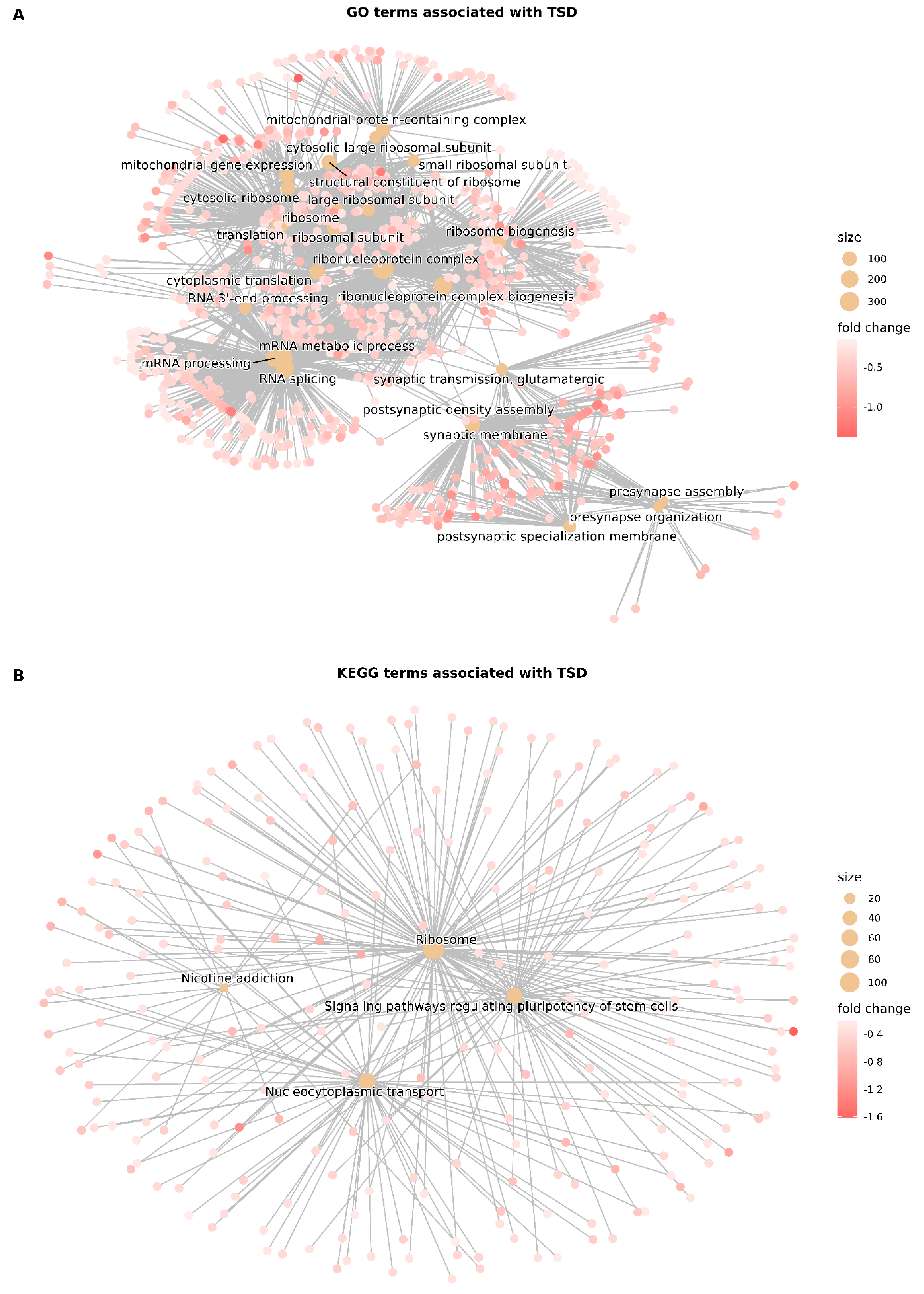
3.4. Gene Set Enrichment Analysis (GSEA)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Ethics Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | Meaning |
| ADHD | Attention-deficit hyperactivity disorder |
| ASD | Autism spectrum disorder |
| CPM | Counts per million |
| CTD | Chronic tic disorder |
| DEG | Differentially expressed gene |
| DMP | Differentially methylated probe |
| DNAm | DNA methylation |
| DSM-5 | Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition |
| eQTM | Expression quantitative trait methylation |
| FDR | False discovery rate |
| GO | Gene Ontology |
| GSEA | Gene set enrichment analysis |
| GTS | Gilles de la Tourette syndrome |
| KEGG | Kyoto Encyclopedia of Gens and Genomes |
| LCPM | Log-transformed counts per million |
| OCD | Obsessive-compulsive disorder |
| PCA | Principal component analysis |
| Q-Q | Quantile-quantile |
| SNP | Single-nucleotide polymorphism |
| TS | Tourette syndrome |
| TSD | Tic spectrum disorder |
| YGTSS | Yale Global Tic Severity Scale |
References
- Johnson, K.A.; Worbe, Y.; Foote, K.D.; Butson, C.R.; Gunduz, A.; Okun, M.S. Tourette Syndrome: Clinical Features, Pathophysiology, and Treatment. Lancet Neurol 2023, 22, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Martino, D.; Dale, R.C.; Gilbert, D.L.; Giovannoni, G.; Leckman, J.F. Immunopathogenic Mechanisms in Tourette Syndrome: A Critical Review. Movement Disorders 2009, 24, 1267–1279. [Google Scholar] [CrossRef]
- Hsu, C.-J.; Wong, L.-C.; Lee, W.-T. Immunological Dysfunction in Tourette Syndrome and Related Disorders. Int J Mol Sci 2021, 22, 853. [Google Scholar] [CrossRef]
- Müller-Vahl, K.R.; Sambrani, T.; Jakubovski, E. Tic Disorders Revisited: Introduction of the Term “Tic Spectrum Disorders. ” Eur Child Adolesc Psychiatry 2019, 28, 1129–1135. [Google Scholar] [CrossRef]
- Groth, C.; Mol Debes, N.; Rask, C.U.; Lange, T.; Skov, L. Course of Tourette Syndrome and Comorbidities in a Large Prospective Clinical Study. J Am Acad Child Adolesc Psychiatry 2017, 56, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Delgar, B.; Servera, M.; Coffey, B.J.; Lázaro, L.; Openneer, T.; Benaroya-Milshtein, N.; Steinberg, T.; Hoekstra, P.J.; Dietrich, A.; Morer, A.; et al. Tic Disorders in Children and Adolescents: Does the Clinical Presentation Differ in Males and Females? A Report by the EMTICS Group. Eur Child Adolesc Psychiatry 2022, 31, 1539–1548. [Google Scholar] [CrossRef]
- Mataix-Cols, D.; Isomura, K.; Pérez-Vigil, A.; Chang, Z.; Rück, C.; Johan Larsson, K.; Leckman, J.F.; Serlachius, E.; Larsson, H.; Lichtenstein, P. Familial Risks of Tourette Syndrome and Chronic Tic Disorders. A Population-Based Cohort Study. JAMA Psychiatry 2015, 72, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Sul, J.H.; Tsetsos, F.; Nawaz, M.S.; Huang, A.Y.; Zelaya, I.; Illmann, C.; Osiecki, L.; Darrow, S.M.; Hirschtritt, M.E.; et al. Interrogating the Genetic Determinants of Tourette’s Syndrome and Other Tic Disorders Through Genome-Wide Association Studies. Am J Psychiatry 2019, 176, 217–227. [Google Scholar] [CrossRef]
- Swain, J.E.; Scahill, L.; Lombroso, P.J.; King, R.A.; Leckman, J.F. Tourette Syndrome and Tic Disorders: A Decade of Progress. J Am Acad Child Adolesc Psychiatry 2007, 46, 947–968. [Google Scholar] [CrossRef] [PubMed]
- Hildonen, M.; Levy, A.M.; Hansen, C.S.; Bybjerg-Grauholm, J.; Skytthe, A.; Debes, N.M.; Tan, Q.; Tümer, Z. EWAS of Monozygotic Twins Implicate a Role of MTOR Pathway in Pathogenesis of Tic Spectrum Disorder. Genes (Basel) 2021, 12, 1510. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res 2015, 43, e47–e47. [Google Scholar] [CrossRef] [PubMed]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A Flexible and Comprehensive Bioconductor Package for the Analysis of Infinium DNA Methylation Microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The Sva Package for Removing Batch Effects and Other Unwanted Variation in High-Throughput Experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Series B Stat Methodol 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Shabalin, A.A. Matrix EQTL: Ultra Fast EQTL Analysis via Large Matrix Operations. Bioinformatics 2012, 28, 1353–1358. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Korotkevich, G.; Sukhov, V.; Budin, N.; Shpak, B.; Artyomov, M.N.; Sergushichev, A. Fast Gene Set Enrichment Analysis. 2016. [Google Scholar] [CrossRef]
- Filomena, E.; Picardi, E.; Tullo, A.; Pesole, G.; D’Erchia, A.M. Identification of Deregulated LncRNAs in Alzheimer’s Disease: An Integrated Gene Co-Expression Network Analysis of Hippocampus and Fusiform Gyrus RNA-Seq Datasets. Front Aging Neurosci 2024, 16, 1437278. [Google Scholar] [CrossRef]
- Claus, C.; Slavin, M.; Ansseau, E.; Lancelot, C.; Bah, K.; Lassche, S.; Fiévet, M.; Greco, A.; Tomaiuolo, S.; Tassin, A.; et al. The Double Homeodomain Protein DUX4c Is Associated with Regenerating Muscle Fibers and RNA-Binding Proteins. Skeletal Muscle 2023 13:1 2023, 13, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Ronchetti, D.; Agnelli, L.; Taiana, E.; Galletti, S.; Manzoni, M.; Todoerti, K.; Musto, P.; Strozzi, F.; Neri, A. Distinct LncRNA Transcriptional Fingerprints Characterize Progressive Stages of Multiple Myeloma. Oncotarget 2016, 7, 14814. [Google Scholar] [CrossRef]
- Boccitto, M.; Wolin, S.L. Ro60 and Y RNAs: Structure, Functions and Roles in Autoimmunity. Crit Rev Biochem Mol Biol 2019, 54, 133. [Google Scholar] [CrossRef]
- Chen, H.; Yang, K.; Pang, L.; Fei, J.; Zhu, Y.; Zhou, J. ANKRD22 Is a Potential Novel Target for Reversing the Immunosuppressive Effects of PMN-MDSCs in Ovarian Cancer. J Immunother Cancer 2023, 11, e005527. [Google Scholar] [CrossRef] [PubMed]
- FCGR1A - High Affinity Immunoglobulin Gamma Fc Receptor I - Homo Sapiens (Human) | UniProtKB | UniProt. Available online: https://www.uniprot.org/uniprotkb/P12314/entry#function (accessed on 31 October 2025).
- Le, D.T.; Florez, M.A.; Kus, P.; Tran, B.T.; Kain, B.; Zhu, Y.; Christensen, K.; Jain, A.; Malovannaya, A.; King, K.Y. BATF2 Promotes HSC Myeloid Differentiation by Amplifying IFN Response Mediators during Chronic Infection. iScience 2023, 26, 106059. [Google Scholar] [CrossRef]
- Roy, S.; Guler, R.; Parihar, S.P.; Schmeier, S.; Kaczkowski, B.; Nishimura, H.; Shin, J.W.; Negishi, Y.; Ozturk, M.; Hurdayal, R.; et al. Batf2/Irf1 Induces Inflammatory Responses in Classically Activated Macrophages, Lipopolysaccharides, and Mycobacterial Infection. J Immunol 2015, 194, 6035–6044. [Google Scholar] [CrossRef]
- Van Der Geest, R.; Penaloza, H.F.; Xiong, Z.; Gonzalez-Ferrer, S.; An, X.; Li, H.; Fan, H.; Tabary, M.; Nouraie, S.M.; Zhao, Y.; et al. BATF2 Enhances Proinflammatory Cytokine Responses in Macrophages and Improves Early Host Defense against Pulmonary Klebsiella Pneumoniae Infection. https://doi.org/10.1152/ajplung.00441.2022 2023, 325, L604–L616. [CrossRef]
- Tinkey, R.A.; Smith, B.C.; Habean, M.L.; Williams, J.L. BATF2 Is a Regulator of Interferon-γ Signaling in Astrocytes during Neuroinflammation. Cell Rep 2025, 44, 115393. [Google Scholar] [CrossRef]
- Li, C.; Liu, M.; Liu, K.; Li, M.; Liu, Y.; Li, T.; Wei, Y.; Long, Y.; He, W.; Shi, X.; et al. BATF2 Balances the T Cell-Mediated Immune Response of CADM with an Anti-MDA5 Autoantibody. Biochem Biophys Res Commun 2021, 551, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Chang, Y.; Huang, K.; Liu, J.; Zhao, Y. The Role of BATF2 Deficiency in Immune Microenvironment Rearrangement in Cervical Cancer - New Biomarker Benefiting from Combination of Radiotherapy and Immunotherapy. Int Immunopharmacol 2024, 126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Y.; Dai, L.; Shi, G.; Deng, J.; Luo, Q.; Xie, Q.; Cheng, L.; Li, C.; Lin, Y.; et al. BATF2 Prevents Glioblastoma Multiforme Progression by Inhibiting Recruitment of Myeloid-Derived Suppressor Cells. Oncogene 2021 40:8 2021, 40, 1516–1530. [Google Scholar] [CrossRef]
- Yu, X.; Tian, J.; Wang, Y.; Su, N.; Luo, J.; Duan, M.; Shi, N. The Pseudogene GBP1P1 Suppresses Influenza A Virus Replication by Acting as a Protein Decoy for DHX9. J Virol 2024, 98. [Google Scholar] [CrossRef]
- Kanemaru, H.; Yamane, F.; Fukushima, K.; Matsuki, T.; Kawasaki, T.; Ebina, I.; Kuniyoshi, K.; Tanaka, H.; Maruyama, K.; Maeda, K.; et al. Antitumor Effect of Batf2 through IL-12 P40 up-Regulation in Tumor-Associated Macrophages. Proc Natl Acad Sci U S A 2017, 114, E7331–E7340. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Fan, H.; Yang, D.; Zhang, J.; Shi, T.; Zhang, D.; Lu, G. Ribosomal RNA-Depleted RNA Sequencing Reveals the Pathogenesis of Refractory Mycoplasma Pneumoniae Pneumonia in Children. Mol Med Rep 2021, 24, 761. [Google Scholar] [CrossRef]
- Cheng, J.; Zhou, X.; Feng, W.; Jia, M.; Zhang, X.; An, T.; Luan, M.; Pan, Y.; Zhang, S.; Zhou, Z.; et al. Risk Stratification by Long Non-coding RNAs Profiling in COVID-19 Patients. J Cell Mol Med 2021, 25, 4753. [Google Scholar] [CrossRef]
- Xiao, X.Y.; Guo, Q.; Tong, S.; Wu, C.Y.; Chen, J.L.; Ding, Y.; Wan, J.H.; Chen, S.S.; Wang, S.H. TRAT1 Overexpression Delays Cancer Progression and Is Associated with Immune Infiltration in Lung Adenocarcinoma. Front Oncol 2022, 12. [Google Scholar] [CrossRef]
- Guo, Q.; Wang, S. hua; Ji, Y. mei; Tong, S.; Li, D.; Ding, X. chao; Wu, C. yan The Roles and Mechanisms of TRAT1 in the Progression of Non-Small Cell Lung Cancer. Curr Med Sci 2022, 42, 1186–1200. [Google Scholar] [CrossRef]
- Delesque-Touchard, N.; Pendaries, C.; Volle-Challier, C.; Millet, L.; Salel, V.; Hervé, C.; Pflieger, A.M.; Berthou-Soulie, L.; Prades, C.; Sorg, T.; et al. Regulator of G-Protein Signaling 18 Controls Both Platelet Generation and Function. PLoS One 2014, 9. [Google Scholar] [CrossRef]
- Greene, D.; Thys, C.; Berry, I.R.; Jarvis, J.; Ortibus, E.; Mumford, A.D.; Freson, K.; Turro, E. Mutations in the U4 SnRNA Gene RNU4-2 Cause One of the Most Prevalent Monogenic Neurodevelopmental Disorders. Nat Med 2024, 30, 2165–2169. [Google Scholar] [CrossRef] [PubMed]
- De Jonghe, J.; Kim, H.C.; Adedeji, A.; Leitão, E.; Dawes, R.; Chen, Y.; Blakes, A.J.; Simons, C.; Rius, R.; Alvi, J.R.; et al. Saturation Genome Editing of RNU4-2 Reveals Distinct Dominant and Recessive Neurodevelopmental Disorders. medRxiv 2025. [Google Scholar] [CrossRef]
- Zsurka, G.; Appel, M.L.T.; Nastaly, M.; Hallmann, K.; Hansen, N.; Nass, D.; Baumgartner, T.; Surges, R.; Hartmann, G.; Bartok, E.; et al. Loss of the Immunomodulatory Transcription Factor BATF2 in Humans Is Associated with a Neurological Phenotype. Cells 2023, 12. [Google Scholar] [CrossRef]
- Shmookler Reis, R.J.; Atluri, R.; Balasubramaniam, M.; Johnson, J.; Ganne, A.; Ayyadevara, S. “Protein Aggregates” Contain RNA and DNA, Entrapped by Misfolded Proteins but Largely Rescued by Slowing Translational Elongation. Aging Cell 2021, 20, e13326. [Google Scholar] [CrossRef]
- Ramos, E.I.; Bien-Willner, G.A.; Li, J.; Hughes, A.E.O.; Giacalone, J.; Chasnoff, S.; Kulkarni, S.; Parmacek, M.; Cole, F.S.; Druley, T.E. Genetic Variation in MKL2 and Decreased Downstream PCTAIRE1 Expression in Extreme, Fatal Primary Human Microcephaly. Clin Genet 2013, 85, 423. [Google Scholar] [CrossRef]
- Qiu, Y.; Yang, S.; Pan, T.; Yu, L.; Liu, J.; Zhu, Y.; Wang, H. ANKRD22 Is Involved in the Progression of Prostate Cancer. Oncol Lett 2019, 18, 4106–4113. [Google Scholar] [CrossRef]
- Yin, J.; Fu, W.; Dai, L.; Jiang, Z.; Liao, H.; Chen, W.; Pan, L.; Zhao, J. ANKRD22 Promotes Progression of Non-Small Cell Lung Cancer through Transcriptional up-Regulation of E2F1. Scientific Reports 2017 7:1 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Wang, R.; Wu, Y.; Zhu, Y.; Yao, S.; Zhu, Y. ANKRD22 Is a Novel Therapeutic Target for Gastric Mucosal Injury. Biomedicine & Pharmacotherapy 2022, 147, 112649. [Google Scholar] [CrossRef]
- Jafari, S.; Ravan, M.; Karimi-Sani, I.; Aria, H.; Hasan-Abad, A.M.; Banasaz, B.; Atapour, A.; Sarab, G.A. Screening and Identification of Potential Biomarkers for Pancreatic Cancer: An Integrated Bioinformatics Analysis. Pathol Res Pract 2023, 249. [Google Scholar] [CrossRef]
- Liu, J.; Wu, J.; Wang, R.; Zhong, D.; Qiu, Y.; Wang, H.; Song, Z.; Zhu, Y. ANKRD22 Drives Rapid Proliferation of Lgr5+ Cells and Acts as a Promising Therapeutic Target in Gastric Mucosal Injury. CMGH 2021, 12, 1433–1455. [Google Scholar] [CrossRef]
- Berhane, T.; Holm, A.; Karstensen, K.T.; Petri, A.; Ilieva, M.S.; Krarup, H.; Vyberg, M.; Løvendorf, M.B.; Kauppinen, S. Knockdown of the Long Noncoding RNA PURPL Induces Apoptosis and Sensitizes Liver Cancer Cells to Doxorubicin. Sci Rep 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Savinovskaya, Y.I.; Nushtaeva, A.A.; Savelyeva, A. V.; Morozov, V. V.; Ryabchikova, E.I.; Kuligina, E. V.; Richter, V.A.; Semenov, D. V. Human Blood Extracellular Vesicles Activate Transcription of NF-KB-Dependent Genes in A549 Lung Adenocarcinoma Cells. Curr Issues Mol Biol 2022, 44, 6028–6045. [Google Scholar] [CrossRef]
- Jaskiewicz, K.; Maleszka-Kurpiel, M.; Matuszewska, E.; Kabza, M.; Rydzanicz, M.; Malinowski, R.; Ploski, R.; Matysiak, J.; Gajecka, M. The Impaired Wound Healing Process Is a Major Factor in Remodeling of the Corneal Epithelium in Adult and Adolescent Patients With Keratoconus. Invest Ophthalmol Vis Sci 2023, 64, 22. [Google Scholar] [CrossRef]
- Mohebifar, H.; Sabbaghian, A.; Farazmandfar, T.; Golalipour, M. Construction and Analysis of Pseudogene-Related CeRNA Network in Breast Cancer. Sci Rep 2023, 13, 21874. [Google Scholar] [CrossRef]
- Shi, H.; Pan, M.; Sheng, Y.; Jia, E.; Wang, Y.; Dong, J.; Tu, J.; Bai, Y.; Cai, L.; Ge, Q. Extracellular Cell-Free RNA Profile in Human Large Follicles and Small Follicles. Front Cell Dev Biol 2022, 10, 940336. [Google Scholar] [CrossRef]
- Berhane, T.; Holm, A.; Karstensen, K.T.; Petri, A.; Ilieva, M.S.; Krarup, H.; Vyberg, M.; Løvendorf, M.B.; Kauppinen, S. Knockdown of the Long Noncoding RNA PURPL Induces Apoptosis and Sensitizes Liver Cancer Cells to Doxorubicin. Sci Rep 2022, 12. [Google Scholar] [CrossRef]
- Hara, T.; Meng, S.; Tsuji, Y.; Arao, Y.; Saito, Y.; Sato, H.; Motooka, D.; Uchida, S.; Ishii, H. RN7SL1 May Be Translated under Oncogenic Conditions. Proc Natl Acad Sci U S A 2024, 121. [Google Scholar] [CrossRef]
- Gómez-Matas, J.; Duran-Sanchon, S.; Lozano, J.J.; Ferrero, G.; Tarallo, S.; Pardini, B.; Naccarati, A.; Castells, A.; Gironella, M. SnoRNA Profiling in Colorectal Cancer and Assessment of Non-Invasive Biomarker Capacity by DdPCR in Fecal Samples. iScience 2024, 27, 109283. [Google Scholar] [CrossRef]
- Ramos, E.I.; Veerapandian, R.; Das, K.; Chacon, J.A.; Gadad, S.S.; Dhandayuthapani, S. Pathogenic Mycoplasmas of Humans Regulate the Long Noncoding RNAs in Epithelial Cells. Noncoding RNA Res 2023, 8, 282. [Google Scholar] [CrossRef]
- Zhang, T.; Nie, Y.; Xia, H.; Zhang, Y.; Cai, K.; Chen, X.; Li, H.; Wang, J. Identification of Immune-Related Prognostic Genes and LncRNAs Biomarkers Associated With Osteosarcoma Microenvironment. Front Oncol 2020, 10, 1109. [Google Scholar] [CrossRef]
- Itami-Matsumoto, S.; Hayakawa, M.; Uchida-Kobayashi, S.; Enomoto, M.; Tamori, A.; Mizuno, K.; Toyoda, H.; Tamura, T.; Akutsu, T.; Ochiya, T.; et al. Circulating Exosomal MiRNA Profiles Predict the Occurrence and Recurrence of Hepatocellular Carcinoma in Patients with Direct-Acting Antiviral-Induced Sustained Viral Response. Biomedicines 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Quintanilha, J.C.F.; Cursino, M.A.; Borges, J.B.; Torso, N.G.; Bastos, L.B.; Oliveira, J.M.; Cobaxo, T.S.; Pincinato, E.C.; Hirata, M.H.; Geraldo, M. V.; et al. MiR-3168, MiR-6125, and MiR-4718 as Potential Predictors of Cisplatin-Induced Nephrotoxicity in Patients with Head and Neck Cancer. BMC Cancer 2021, 21, 575. [Google Scholar] [CrossRef]
- She, J.; Tan, K.; Liu, J.; Cao, S.; Li, Z.; Peng, Y.; Xiao, Z.; Diao, R.; Wang, L. The Alteration of M6A Modification at the Transcriptome-Wide Level in Human Villi During Spontaneous Abortion in the First Trimester. Front Genet 2022, 13, 861853. [Google Scholar] [CrossRef]
- Claus, C.; Slavin, M.; Ansseau, E.; Lancelot, C.; Bah, K.; Lassche, S.; Fiévet, M.; Greco, A.; Tomaiuolo, S.; Tassin, A.; et al. The Double Homeodomain Protein DUX4c Is Associated with Regenerating Muscle Fibers and RNA-Binding Proteins. Skeletal Muscle 2023 13:1 2023, 13, 1–30. [Google Scholar] [CrossRef]
- Kumar, S.; Shih, C.M.; Tsai, L.W.; Dubey, R.; Gupta, D.; Chakraborty, T.; Sharma, N.; Singh, A.V.; Swarup, V.; Singh, H.N. Transcriptomic Profiling Unravels Novel Deregulated Gene Signatures Associated with Acute Myocardial Infarction: A Bioinformatics Approach. Genes (Basel) 2022, 13, 2321. [Google Scholar] [CrossRef]
- Alsugair, Z.; Perrot, J.; Descotes, F.; Lopez, J.; Champagnac, A.; Pissaloux, D.; Castain, C.; Onea, M.; Céruse, P.; Philouze, P.; et al. Characterization of a Molecularly Distinct Subset of Oncocytic Pleomorphic Adenomas/Myoepitheliomas Harboring Recurrent ZBTB47-AS1::PLAG1 Gene Fusion. Am J Surg Pathol 2024, 48, 551–561. [Google Scholar] [CrossRef]
- Meng, Y.; Qiu, S.Q.; Wang, Q.; Zuo, J.L. Regulator of G Protein Signalling 18 Promotes Osteocyte Proliferation by Activating the Extracellular Signal-regulated Kinase Signalling Pathway. Int J Mol Med 2024, 53. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Li, H.; Peng, Z.; Ke, D.; Fu, H.; Zheng, X. Identification of Plasma RGS18 and PPBP MRNAs as Potential Biomarkers for Gastric Cancer Using Transcriptome Arrays. Oncol Lett 2019, 17, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Dale, R.C.; Merheb, V.; Pillai, S.; Wang, D.; Cantrill, L.; Murphy, T.K.; Ben-Pazi, H.; Varadkar, S.; Aumann, T.D.; Horne, M.K.; et al. Antibodies to Surface Dopamine-2 Receptor in Autoimmune Movement and Psychiatric Disorders. Brain 2012, 135, 3453–3468. [Google Scholar] [CrossRef]
- Addabbo, F.; Baglioni, V.; Schrag, A.; Schwarz, M.J.; Dietrich, A.; Hoekstra, P.J.; Martino, D.; Buttiglione, M.; Anastasiou, Z.; Apter, A.; et al. Anti-Dopamine D2 Receptor Antibodies in Chronic Tic Disorders. Dev Med Child Neurol 2020, 62, 1205–1212. [Google Scholar] [CrossRef]
- Fernández de la Cruz, L.; Mataix-Cols, D. General Health and Mortality in Tourette Syndrome and Chronic Tic Disorder: A Mini-Review. Neurosci Biobehav Rev 2020, 119, 514–520. [Google Scholar] [CrossRef]
- Murphy, T.K.; Storch, E.A.; Turner, A.; Reid, J.M.; Tan, J.; Lewin, A.B. Maternal History of Autoimmune Disease in Children Presenting with Tics and/or Obsessive-Compulsive Disorder. J Neuroimmunol 2010, 229, 243–247. [Google Scholar] [CrossRef]
- Xue, D.; Shi, H.; Smith, J.D.; Chen, X.; Noe, D.A.; Cedervall, T.; Yangt, D.D.; Eynon, E.; Brash, D.E.; Kashgarian, M.; et al. A Lupus-like Syndrome Develops in Mice Lacking the Ro 60-KDa Protein, a Major Lupus Autoantigen. Proc Natl Acad Sci U S A 2003, 100, 7503–7508. [Google Scholar] [CrossRef] [PubMed]
- Shmookler Reis, R.J.; Atluri, R.; Balasubramaniam, M.; Johnson, J.; Ganne, A.; Ayyadevara, S. “Protein Aggregates” Contain RNA and DNA, Entrapped by Misfolded Proteins but Largely Rescued by Slowing Translational Elongation. Aging Cell 2021, 20, e13326. [Google Scholar] [CrossRef]
- Zsurka, G.; Appel, M.L.T.; Nastaly, M.; Hallmann, K.; Hansen, N.; Nass, D.; Baumgartner, T.; Surges, R.; Hartmann, G.; Bartok, E.; et al. Loss of the Immunomodulatory Transcription Factor BATF2 in Humans Is Associated with a Neurological Phenotype. Cells 2023, 12, 227. [Google Scholar] [CrossRef]
- Chen, Y.; Dawes, R.; Kim, H.C.; Ljungdahl, A.; Stenton, S.L.; Walker, S.; Lord, J.; Lemire, G.; Martin-Geary, A.C.; Ganesh, V.S.; et al. De Novo Variants in the RNU4-2 SnRNA Cause a Frequent Neurodevelopmental Syndrome. Nature 2024, 632, 832. [Google Scholar] [CrossRef]
- Ramos, E.I.; Bien-Willner, G.A.; Li, J.; Hughes, A.E.O.; Giacalone, J.; Chasnoff, S.; Kulkarni, S.; Parmacek, M.; Cole, F.S.; Druley, T.E. Genetic Variation in MKL2 and Decreased Downstream PCTAIRE1 Expression in Extreme, Fatal Primary Human Microcephaly. Clin Genet 2013, 85, 423. [Google Scholar] [CrossRef]
- Karlsson Linnér, R.; Mallard, T.T.; Barr, P.B.; Sanchez-Roige, S.; Madole, J.W.; Driver, M.N.; Poore, H.E.; de Vlaming, R.; Grotzinger, A.D.; Tielbeek, J.J.; et al. Multivariate Analysis of 1.5 Million People Identifies Genetic Associations with Traits Related to Self-Regulation and Addiction. Nat Neurosci 2021, 24, 1367. [Google Scholar] [CrossRef]
- Nemoda, Z.; Sahin-Tóth, M. Chymotrypsin C (Caldecrin) Stimulates Autoactivation of Human Cationic Trypsinogen. J Biol Chem 2006, 281, 11879–11886. [Google Scholar] [CrossRef] [PubMed]
- Szmola, R.; Bence, M.; Carpentieri, A.; Szabó, A.; Costello, C.E.; Samuelson, J.; Sahin-Tóth, M. Chymotrypsin C Is a Co-Activator of Human Pancreatic Procarboxypeptidases A1 and A2. J Biol Chem 2010, 286, 1819. [Google Scholar] [CrossRef] [PubMed]
- Arnold, G.L.; Hyman, S.L.; Mooney, R.A.; Kirby, R.S. Plasma Amino Acids Profiles in Children with Autism: Potential Risk of Nutritional Deficiencies. J Autism Dev Disord 2003, 33, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Pearson, D.A.; Arnold, L.E.; Bostrom, S.; Aman, M.G. 2017 International Meeting for Autism Research: Chymotrypsin: Evidence of a Novel Pancreatic Insufficiency in Children with ASD? Available online: https://insar.confex.com/imfar/2017/webprogram/Paper23908.html (accessed on 24 April 2025).
- Heil, M.; Pearson, D.A.; Fallon, J. 2014 International Meeting for Autism Research: Low Endogenous Fecal Chymotrypsin: A Possible Biomarker for Autism? Available online: https://www.researchgate.net/publication/268131323_Low_Endogenous_Fecal_Chymotrypsin_A_Possible_Biomarker_for_Autism (accessed on 24 April 2025).
- Pearson, D.A.; Hendren, R.L.; Heil, M.F.; McIntyre, W.R.; Raines, S.R. Pancreatic Replacement Therapy for Maladaptive Behaviors in Preschool Children With Autism Spectrum Disorder. JAMA Netw Open 2023, 6, e2344136. [Google Scholar] [CrossRef]
- Sullivan, M.A.; Rudnik-Leven, F. Attention Deficit/Hyperactivity Disorder and Substance Abuse. Ann N Y Acad Sci 2001, 931, 251–270. [Google Scholar] [CrossRef]
- Srichawla, B.S.; Telles, C.C.; Schweitzer, M.; Darwish, B. Attention Deficit Hyperactivity Disorder and Substance Use Disorder: A Narrative Review. Cureus 2022. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




